The Role of MSI Testing Methodology and Its Heterogeneity in Predicting Colorectal Cancer Immunotherapy Response
Abstract
1. Introduction
2. Results
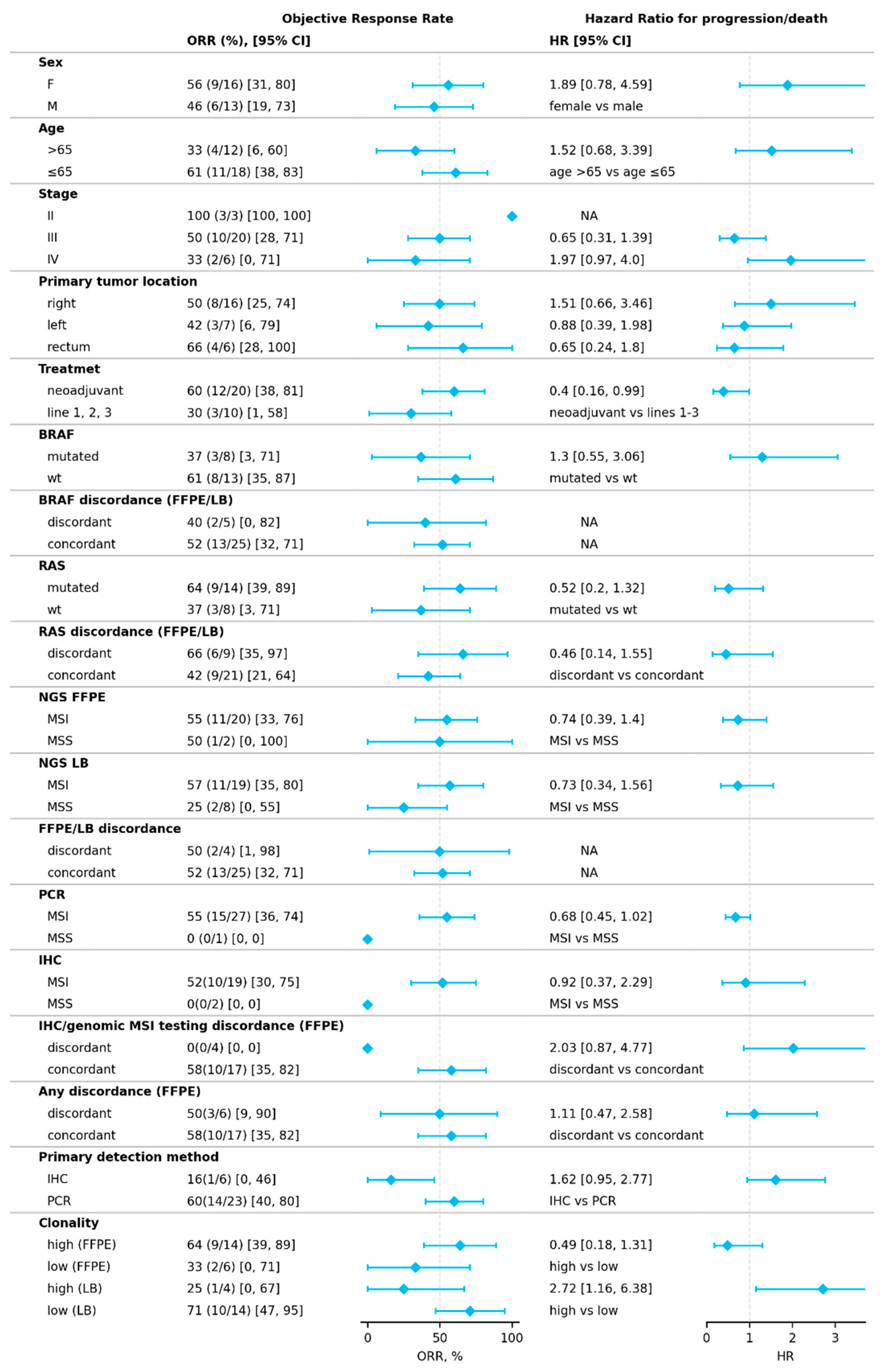
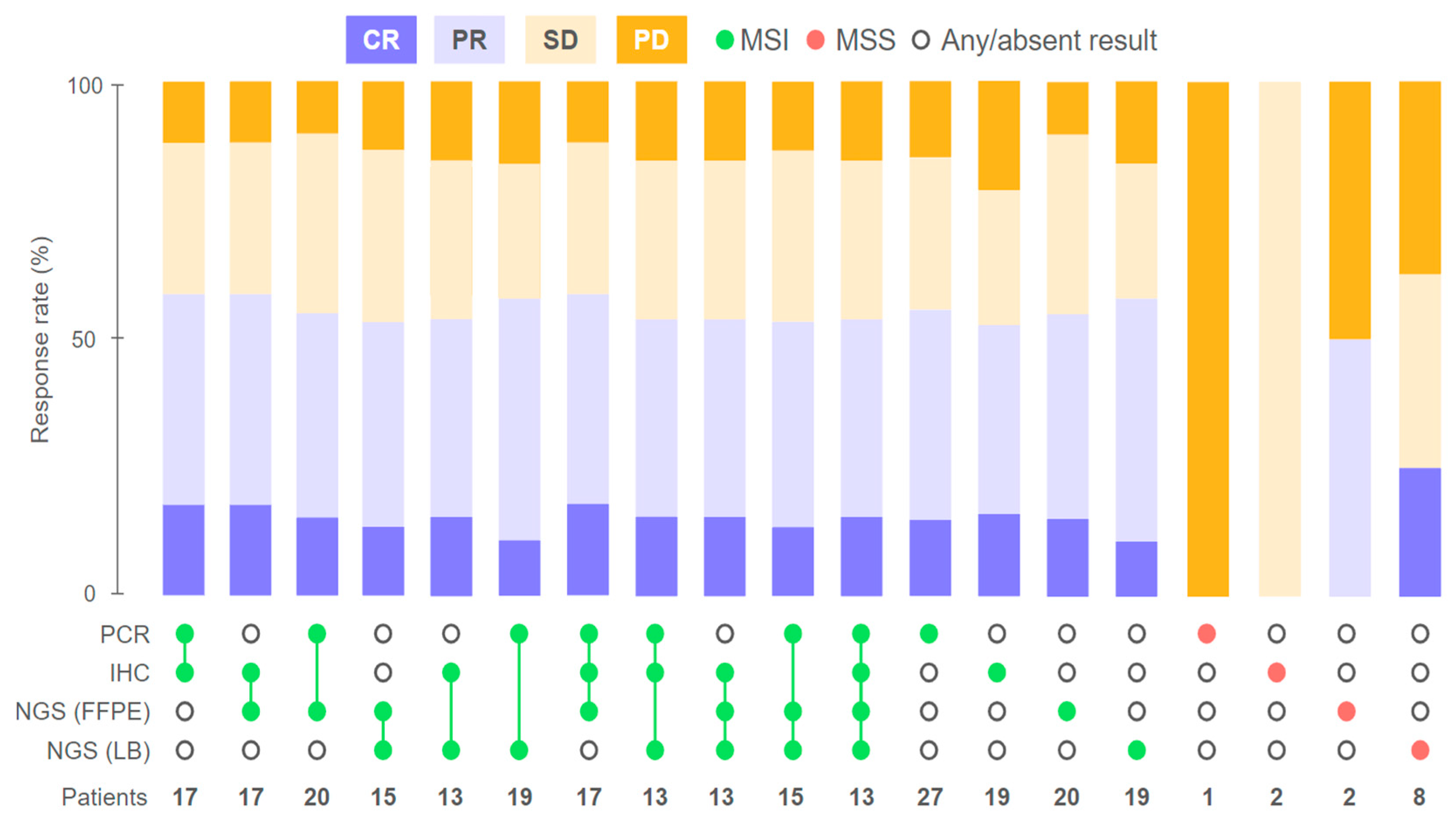
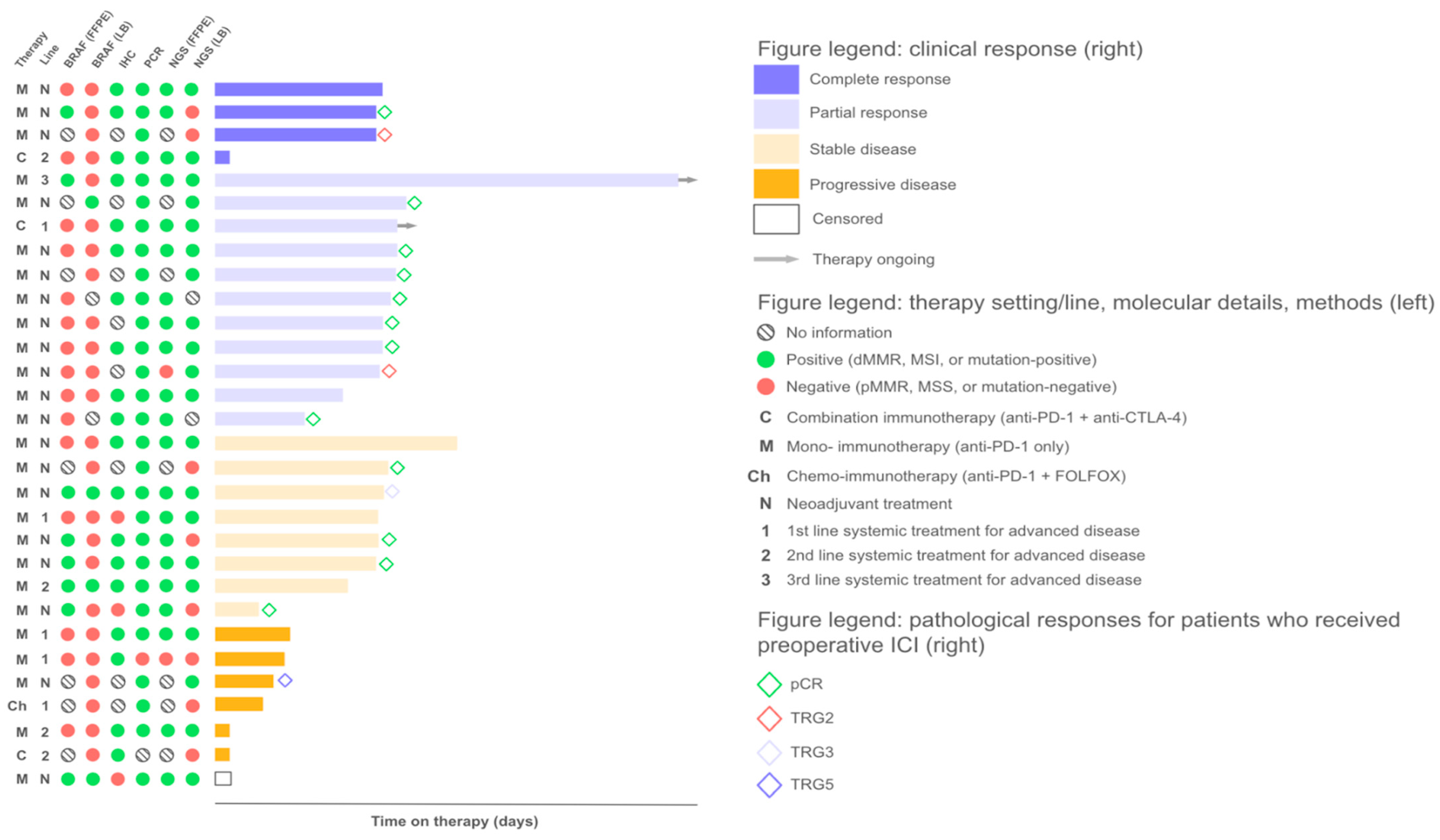
2.1. Response to Treatment
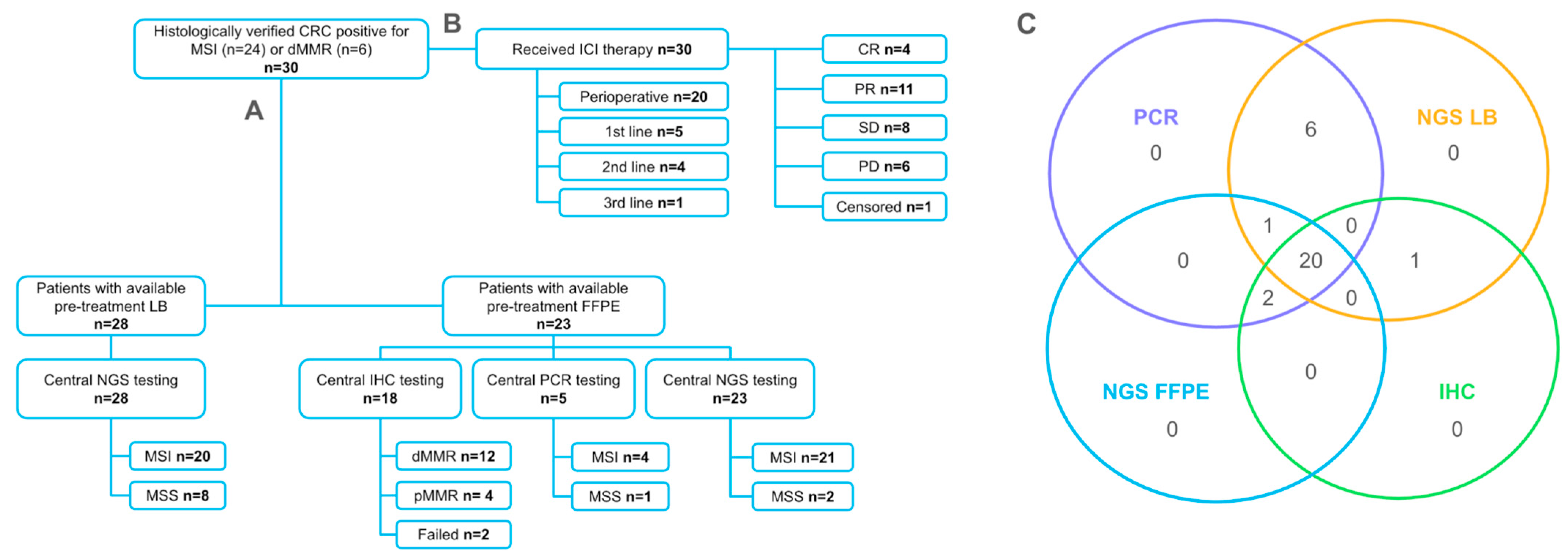
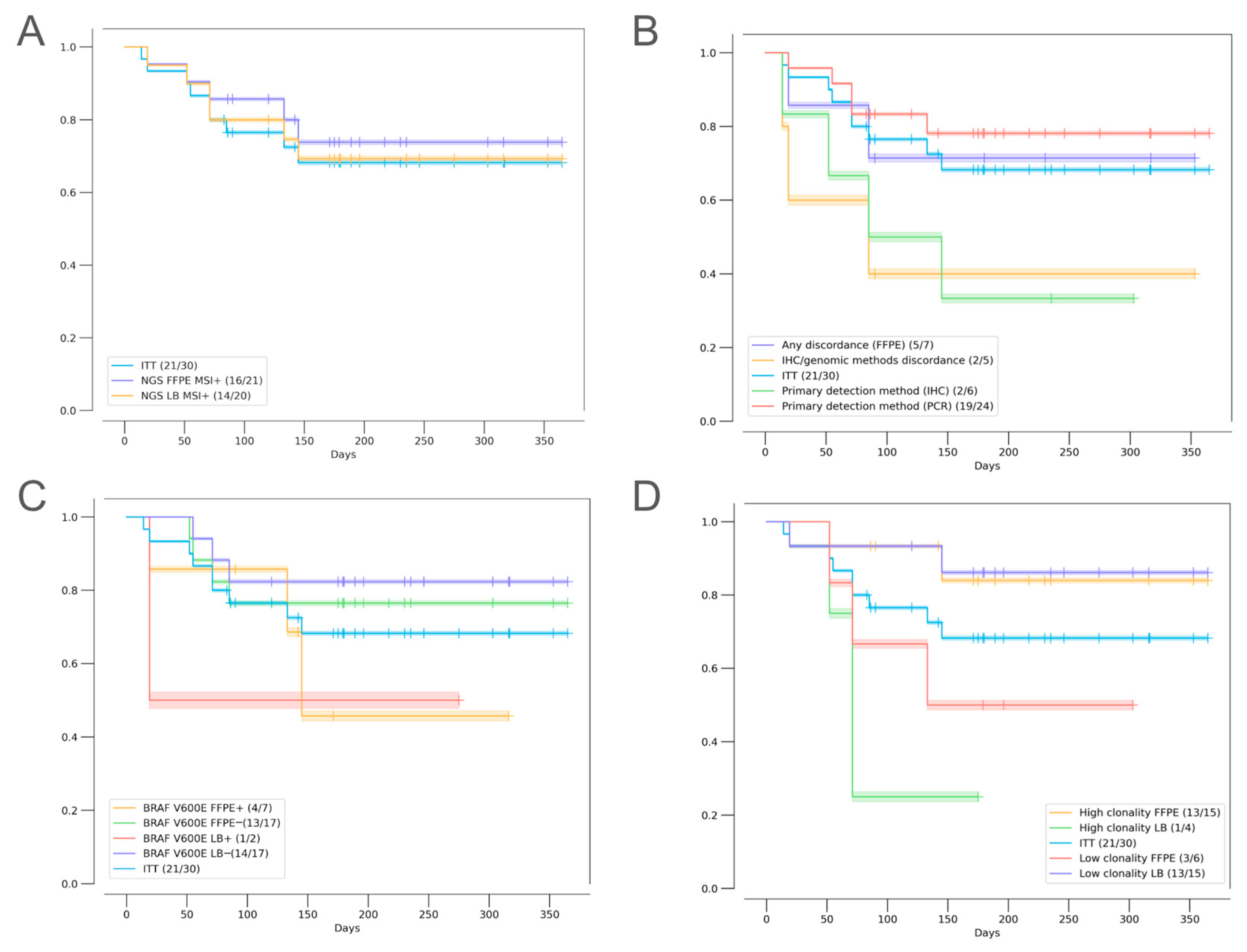
2.2. Agreement of Methods for MSI/dMMR Detection
2.3. Factors Influencing Treatment Outcomes
2.3.1. Discordant MSI/dMMR Results
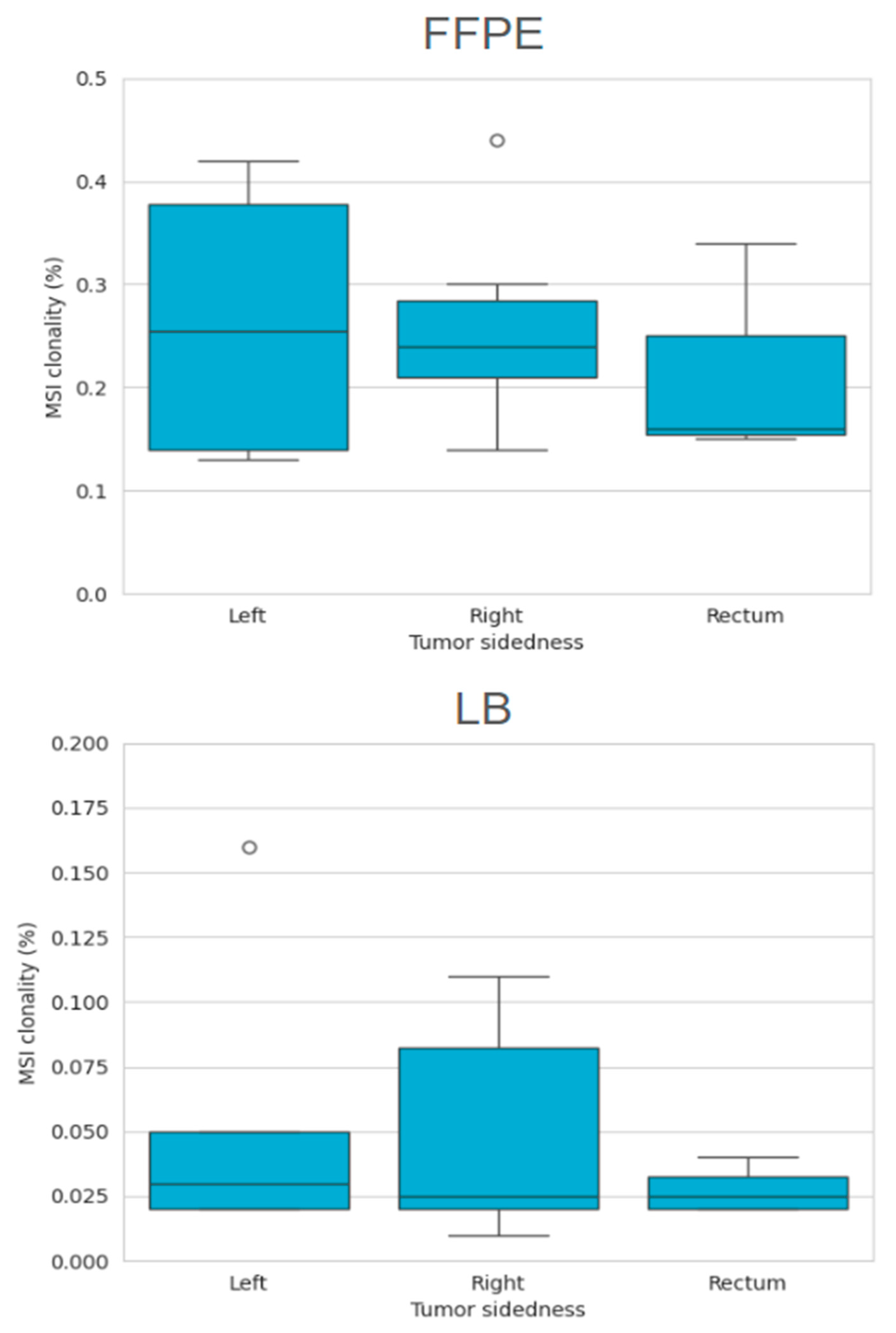
2.3.2. MSI Clonality
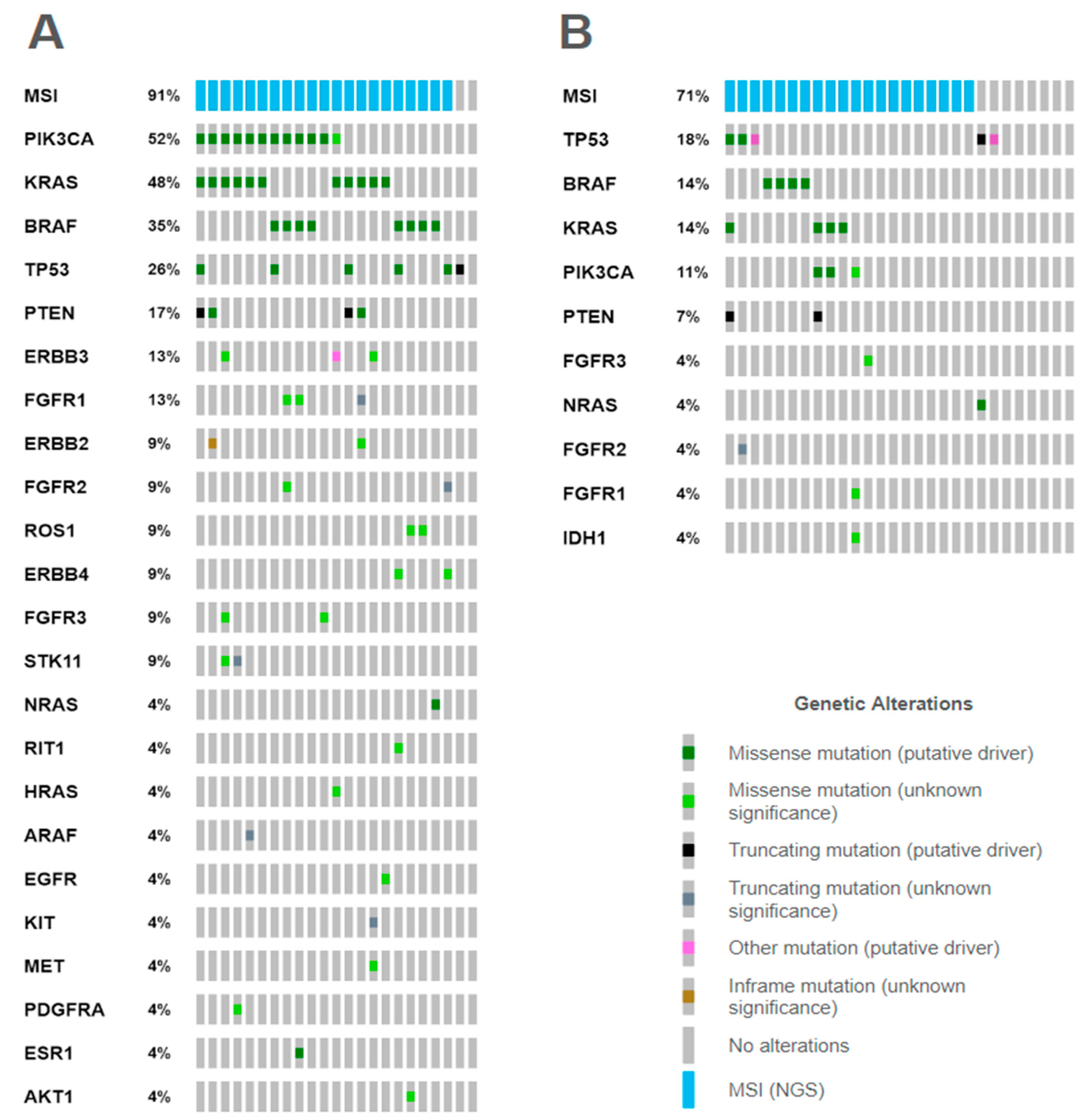
2.3.3. Mutational Correlates
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients
4.2. Central MSI/dMMR Testing
4.3. Next-Generation Sequencing
4.3.1. DNA Extraction
4.3.2. Library Preparation and Sequencing
4.3.3. NGS Data Analysis
4.3.4. MSI Analysis and Clonality Estimation
4.4. Treatment Outcomes
4.5. Statistical Analysis and Data Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CI | confidence interval |
| CNV | copy number variation |
| CR | complete response |
| CRC | colorectal cancer |
| ctDNA | circulating tumor DNA |
| DCR | disease control rate |
| dMMR | mismatch repair deficiency |
| ECOG | Eastern Cooperative Oncology Group |
| ESMO | European Society for Medical Oncology |
| FFPE | formalin-fixed paraffin-embedded |
| FOLFOX | folinic acid (leucovorin, FOL), fluorouracil (5-FU, F), and oxaliplatin (Eloxatin, OX) |
| HR | hazard ratio |
| ICI | immune checkpoint inhibitor |
| IHC | immunohistochemistry |
| ITT | intention-to-treat (population) |
| LB | liquid biopsy |
| MSI | microsatellite instability |
| NGS | next-generation sequencing |
| ORR | objective response rate |
| PCR | polymerase chain reaction |
| PD | progressive disease |
| PFS | progression-free survival |
| PR | partial response |
| SD | stable disease |
| STR | short tandem repeats |
Appendix A
References
- Baretti, M.; Le, D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018, 189, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Setton, J.; Lee, N.Y.; Riaz, N.; Powell, S.N. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018, 9, 3292. [Google Scholar] [CrossRef] [PubMed]
- Lepore Signorile, M.; Disciglio, V.; Di Carlo, G.; Pisani, A.; Simone, C.; Ingravallo, G. From Genetics to Histomolecular Characterization: An Insight into Colorectal Carcinogenesis in Lynch Syndrome. Int. J. Mol. Sci. 2021, 22, 6767. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Diao, Z.; Han, Y.; Chen, Y.; Zhang, R.; Li, J. The clinical utility of microsatellite instability in colorectal cancer. Crit. Rev. Oncol. Hematol. 2021, 157, 103171. [Google Scholar] [CrossRef]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.-Y.; Andre, F.; et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef]
- Müller, M.F.; Ibrahim, A.E.K.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef]
- Cornista, A.M.; Giolito, M.V.; Baker, K.; Hazime, H.; Dufait, I.; Datta, J.; Khumukcham, S.S.; De Ridder, M.; Roper, J.; Abreu, M.T.; et al. Colorectal Cancer Immunotherapy: State of the Art and Future Directions. Gastro. Hep. Adv. 2023, 2, 1103–1119. [Google Scholar] [CrossRef]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B., III; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. JCO 2023, 41, 678–700. [Google Scholar] [CrossRef]
- Yakushina, V.; Kavun, A.; Veselovsky, E.; Grigoreva, T.; Belova, E.; Lebedeva, A.; Mileyko, V.; Ivanov, M. Microsatellite Instability Detection: The Current Standards, Limitations, and Misinterpretations. JCO Precis Oncol. 2023, 7, e2300010. [Google Scholar] [CrossRef]
- Amato, M.; Franco, R.; Facchini, G.; Addeo, R.; Ciardiello, F.; Berretta, M.; Vita, G.; Sgambato, A.; Pignata, S.; Caraglia, M.; et al. Microsatellite Instability: From the Implementation of the Detection to a Prognostic and Predictive Role in Cancers. Int. J. Mol. Sci. 2022, 23, 8726. [Google Scholar] [CrossRef]
- Dedeurwaerdere, F.; Claes, K.B.; Van Dorpe, J.; Rottiers, I.; Van der Meulen, J.; Breyne, J.; Swaerts, K.; Martens, G. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; Chapelle, A.d.l.; Ruschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Nagasaka, T.; Hamelin, R.; Boland, C.R. An Optimized Pentaplex PCR for Detecting DNA Mismatch Repair-Deficient Colorectal Cancers. PLoS ONE 2010, 5, e9393. [Google Scholar] [CrossRef]
- Gilson, P.; Merlin, J.-L.; Harlé, A. Detection of Microsatellite Instability: State of the Art and Future Applications in Circulating Tumour DNA (ctDNA). Cancers 2021, 13, 1491. [Google Scholar] [CrossRef]
- Cisneros-Villanueva, M.; Hidalgo-Pérez, L.; Rios-Romero, M.; Cedro-Tanda, A.; Ruiz-Villavicencio, C.A.; Page, K.; Hastings, R.; Fernandez-Garcia, D.; Allsopp, R.; Fonseca-Montaño, M.A.; et al. Cell-free DNA analysis in current cancer clinical trials: A review. Br. J. Cancer 2022, 126, 391–400. [Google Scholar] [CrossRef]
- Yu, F.; Makrigiorgos, A.; Leong, K.W.; Makrigiorgos, G.M. Sensitive detection of microsatellite instability in tissues and liquid biopsies: Recent developments and updates. CSBJ 2021, 19, 4931–4940. [Google Scholar] [CrossRef]
- Malapelle, U.; Parente, P.; Pepe, F.; De Luca, C.; Cerino, P.; Covelli, C.; Balestrieri, M.; Russo, G.; Bonfitto, A.; Pisapia, P.; et al. Impact of Pre-Analytical Factors on MSI Test Accuracy in Mucinous Colorectal Adenocarcinoma: A Multi-Assay Concordance Study. Cells 2020, 9, 2019. [Google Scholar] [CrossRef]
- Shia, J. Immunohistochemistry versus Microsatellite Instability Testing For Screening Colorectal Cancer Patients at Risk For Hereditary Nonpolyposis Colorectal Cancer Syndrome. JMD 2008, 10, 293–300. [Google Scholar] [CrossRef]
- Guyot D’Asnières De Salins, A.; Tachon, G.; Cohen, R.; Karayan-Tapon, L.; Junca, A.; Frouin, E.; Godet, J.; Evrard, C.; Randrian, V.; Duval, A.; et al. Discordance between Immunochemistry of Mismatch Repair Proteins and Molecular Testing of Microsatellite Instability in Colorectal Cancer. ESMO Open 2021, 6, 100120. [Google Scholar] [CrossRef]
- de Abreu, A.R.; Op de Beeck, K.; Laurent-Puig, P.; Taly, V.; Benhaim, L. The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer. Cancers 2023, 15, 1284. [Google Scholar] [CrossRef] [PubMed]
- Geurts, B.S.; Zeverijn, L.J.; van Berge Henegouwen, J.M.; van der Wijngaart, H.; Hoes, L.R.; de Wit, G.F.; Spiekman, I.A.; Battaglia, T.W.; van Beek, D.M.; Roepman, P.; et al. Characterization of discordance between mismatch repair deficiency and microsatellite instability testing may prevent inappropriate treatment with immunotherapy. J. Pathol. 2024, 263, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Hain, E.; Buhard, O.; Guilloux, A.; Bardier, A.; Kaci, R.; Bertheau, P.; Renaud, F.; Bibeau, F.; Fléjou, J.-F.; et al. Association of Primary Resistance to Immune Checkpoint Inhibitors in Metastatic Colorectal Cancer With Misdiagnosis of Microsatellite Instability or Mismatch Repair Deficiency Status. JAMA Oncol. 2019, 5, 551. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; Swanton, C. Tumour Heterogeneity and the Evolution of Polyclonal Drug Resistance. Mol. Oncol. 2014, 8, 1095–1111. [Google Scholar] [CrossRef]
- Bonneville, R.; Paruchuri, A.; Wing, M.R.; Krook, M.A.; Reeser, J.W.; Chen, H.-Z.; Dao, T.; Samorodnitsky, E.; Smith, A.M.; Yu, L.; et al. Characterization of Clonal Evolution in Microsatellite Unstable Metastatic Cancers through Multiregional Tumor Sequencing. Mol. Cancer Res. 2021, 19, 465–474. [Google Scholar] [CrossRef]
- Chapusot, C.; Martin, L.; Bouvier, A.M.; Bonithon-Kopp, C.; Ecarnot-Laubriet, A.; Rageot, D.; Ponnelle, T.; Laurent Puig, P.; Faivre, J.; Piard, F. Microsatellite instability and intratumoural heterogeneity in 100 right-sided sporadic colon carcinomas. Br. J. Cancer 2002, 87, 400–404. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, M.S.; An, C.H.; Yoo, N.J.; Lee, S.H. Regional Bias of Intratumoral Genetic Heterogeneity of Nucleotide Repeats in Colon Cancers with Microsatellite Instability. Pathol. Oncol. Res. 2014, 20, 965–971. [Google Scholar] [CrossRef]
- Ai, X.; Cui, J.; Zhang, J.; Chen, R.; Lin, W.; Xie, C.; Liu, A.; Zhang, J.; Yang, W.; Hu, X.; et al. Clonal Architecture of EGFR Mutation Predicts the Efficacy of EGFR-Tyrosine Kinase Inhibitors in Advanced NSCLC: A Prospective Multicenter Study (NCT03059641). Clin. Cancer Res. 2021, 27, 704–712. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef]
- O’Connor, C.A.; Harrold, E.; Lin, D.; Walch, H.; Gazzo, A.; Ranganathan, M.; Kane, S.; Keane, F.; Schoenfeld, J.; Drew Moss, M.; et al. Lynch Syndrome and Somatic Mismatch Repair Variants in Pancreas Cancer. JAMA Oncol. 2024, 10, 1511–1518. [Google Scholar] [CrossRef]
- Tjulandin, S.; Demidov, L.; Moiseyenko, V.; Protsenko, S.; Semiglazova, T.; Odintsova, S.; Zukov, R.; Lazarev, S.; Makarova, Y.; Nechaeva, M.; et al. Novel PD-1 inhibitor prolgolimab: Expanding non-resectable/metastatic melanoma therapy choice. Eur. J. 2021, 149, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Bartley, A.N.; Hamilton, S.R.; Alsabeh, R.; Ambinder, E.P.; Berman, M.; Collins, E.; Fitzgibbons, P.L.; Gress, D.M.; Nowak, J.A.; Samowitz, W.S.; et al. Template for Reporting Results of Biomarker Testing of Specimens From Patients With Carcinoma of the Colon and Rectum. Arch. Path. Lab. 2014, 138, 166–170. [Google Scholar] [CrossRef]
- Lebedeva, A.; Veselovsky, E.; Kavun, A.; Belova, E.; Grigoreva, T.; Orlov, P.; Subbotovskaya, A.; Shipunov, M.; Mashkov, O.; Bilalov, F.; et al. Untapped Potential of Poly(ADP-Ribose) Polymerase Inhibitors: Lessons Learned From the Real-World Clinical Homologous Recombination Repair Mutation Testing. World J. Oncol. 2024, 15, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kockan, C.; Hach, F.; Sarrafi, I.; Bell, R.H.; McConeghy, B.; Beja, K.; Haegert, A.; Wyatt, A.W.; Volik, S.V.; Chi, K.N.; et al. SiNVICT: Ultra-sensitive detection of single nucleotide variants and indels in circulating tumour DNA. Bioinformatics 2016, 33, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Simpson, J.T.; Durbin, R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2011, 22, 549–556. [Google Scholar] [CrossRef]
- Horak, P.; Griffith, M.; Danos, A.M.; Pitel, B.A.; Madhavan, S.; Liu, X.; Chow, C.; Williams, H.; Carmody, L.; Barrow-Laing, L.; et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC). GIM 2022, 24, 986–998. [Google Scholar] [CrossRef]
- Lebedeva, A.; Belova, E.; Grigoreva, T.; Kuznetsova, O.A.; Kavun, A.; Aliyarova, S.; Veselovsky, E.; Iudina, V.; Nikulin, V.; Kravchuk, D.A.; et al. 103P MSI detection by NGS using tumor samples and liquid biopsy for patients with solid tumors: A single institution experience. ESMO Open 2023, 8, 101913. [Google Scholar] [CrossRef]

| Sex | Female | 14 (46.7%) |
| Male | 16 (53.3%) | |
| Median age (range), years | 60.5 (28–85) | |
| Primary tumor location | Right | 17 (56.7%) |
| Left | 7 (23.3%) | |
| Rectum | 6 (20%) | |
| Histology | Intestinal adenocarcinoma | 27 (90%) |
| Signet cell carcinoma | 2 (6.7%) | |
| Mucinous adenocarcinoma | 1 (3.3%) | |
| Stage (AJCC) | II | 3 (10%) |
| III | 21 (70%) | |
| IV | 6 (20%) | |
| T | T1 | 0 (0%) |
| T2 | 0 (0%) | |
| T3 | 22 (73.3%) | |
| T4 | 8 (26.7%) | |
| N | N0 | 6 (20%) |
| N1 | 11 (36.7%) | |
| N2 | 13 (43.3%) | |
| M | M0 | 23 (76.7%) |
| M1 | 6 (20%) | |
| Mx | 1 (3.3%) | |
| Therapy | Prolgolimab | 18 (60%) |
| Pembrolizumab | 5 (16.7%) | |
| Nivolumab | 3 (10%) | |
| Nivolumab + Ipilimumab | 3 (10%) | |
| Nivolumab + FOLFOX | 1 (3.3%) | |
| BRAF p.V600 (FFPE, NGS) * | Positive | 8 (26.7%) |
| Negative | 15 (50%) | |
| Unknown | 7 (23.3%) | |
| RAS mutations (FFPE, NGS) * | Positive | 14 (46.7%) |
| Negative | 9 (30%) | |
| Unknown | 7 (23.3%) | |
| Variable | HR (95% CI) | p-Value |
|---|---|---|
| MSI clonality (FFPE) | 0.63 (0.39–0.99) | 0.0487 * |
| MSI clonality (LB) | 3.05 (2.01–4.65) | <0.00001 * |
| Discordance between IHC and genomic methods (present vs. absent) | 2.16 (1.39–3.33) | 0.0005 * |
| Age | 3.69 (1.84–7.43) | 0.0002 * |
| Sex (female vs. male) | 1.12 (0.84–1.51) | 0.4353 |
| Metastatic disease (metastatic vs. localized) | 0.76 (0.54–1.09) | 0.1326 |
| Primary method used for MSI detection (IHC vs. PCR) | 1.08 (0.77–1.49) | 0.666 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedeva, A.; Taraskina, A.; Grigoreva, T.; Belova, E.; Kuznetsova, O.; Ivanilova, D.; Sergeeva, A.; Kavun, A.; Veselovsky, E.; Nikulin, V.; et al. The Role of MSI Testing Methodology and Its Heterogeneity in Predicting Colorectal Cancer Immunotherapy Response. Int. J. Mol. Sci. 2025, 26, 3420. https://doi.org/10.3390/ijms26073420
Lebedeva A, Taraskina A, Grigoreva T, Belova E, Kuznetsova O, Ivanilova D, Sergeeva A, Kavun A, Veselovsky E, Nikulin V, et al. The Role of MSI Testing Methodology and Its Heterogeneity in Predicting Colorectal Cancer Immunotherapy Response. International Journal of Molecular Sciences. 2025; 26(7):3420. https://doi.org/10.3390/ijms26073420
Chicago/Turabian StyleLebedeva, Alexandra, Anastasiia Taraskina, Tatiana Grigoreva, Ekaterina Belova, Olesya Kuznetsova, Daria Ivanilova, Anastasia Sergeeva, Alexandra Kavun, Egor Veselovsky, Vladislav Nikulin, and et al. 2025. "The Role of MSI Testing Methodology and Its Heterogeneity in Predicting Colorectal Cancer Immunotherapy Response" International Journal of Molecular Sciences 26, no. 7: 3420. https://doi.org/10.3390/ijms26073420
APA StyleLebedeva, A., Taraskina, A., Grigoreva, T., Belova, E., Kuznetsova, O., Ivanilova, D., Sergeeva, A., Kavun, A., Veselovsky, E., Nikulin, V., Aliyarova, S., Belyaeva, L., Tryakin, A., Fedyanin, M., Mileyko, V., & Ivanov, M. (2025). The Role of MSI Testing Methodology and Its Heterogeneity in Predicting Colorectal Cancer Immunotherapy Response. International Journal of Molecular Sciences, 26(7), 3420. https://doi.org/10.3390/ijms26073420






