Harnessing the Potential of CRISPR/Cas in Targeted Alfalfa Improvement for Stress Resilience
Abstract
1. Introduction
2. Leveraging Genomic Resources for Alfalfa CRISPR Editing
3. Examples of CRISPR Target Traits and Editing Modalities
3.1. Knockout of Biotic Stress Tolerance Genes
3.2. Multiplexing Towards Abiotic Stress Tolerance
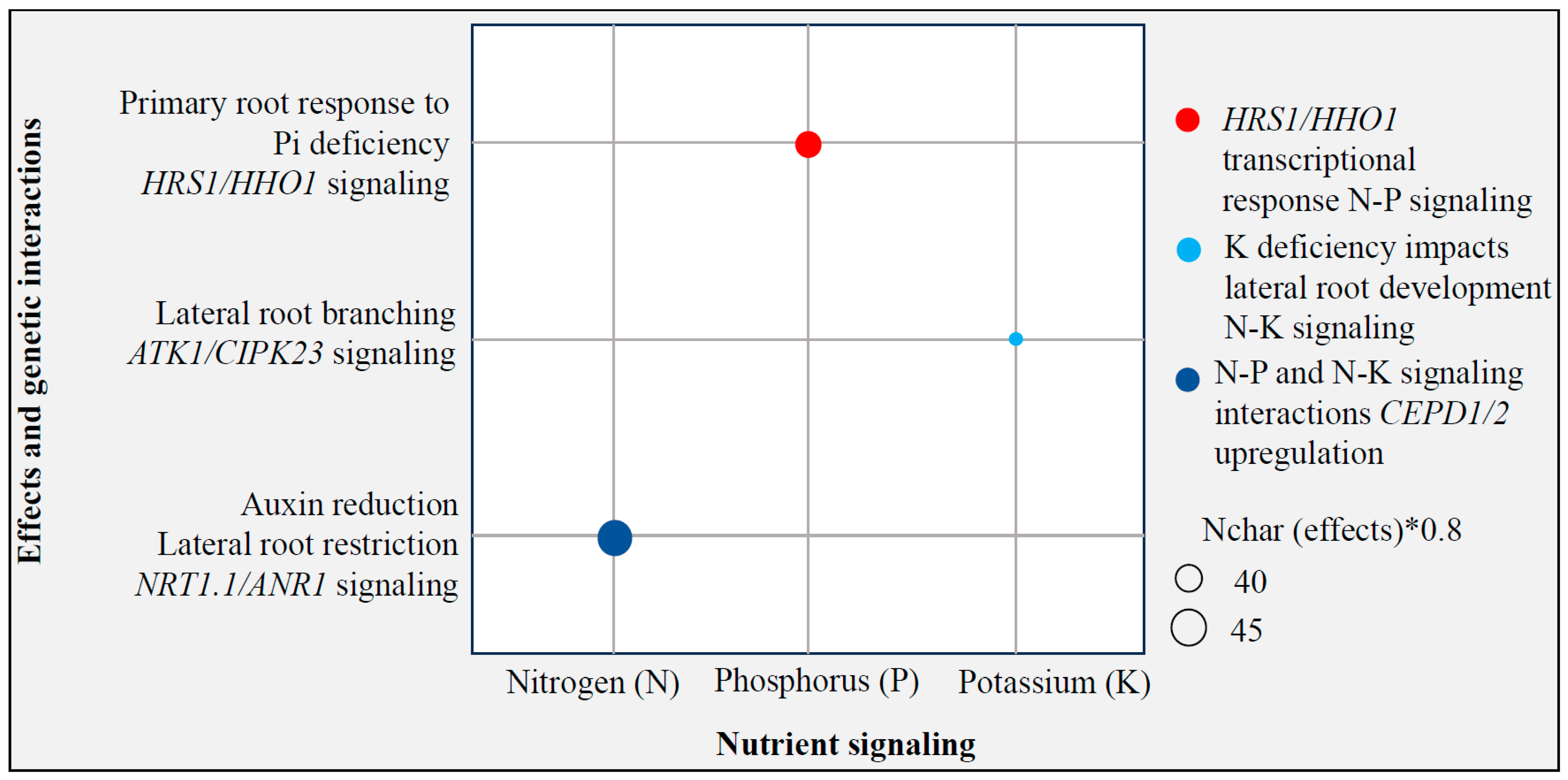
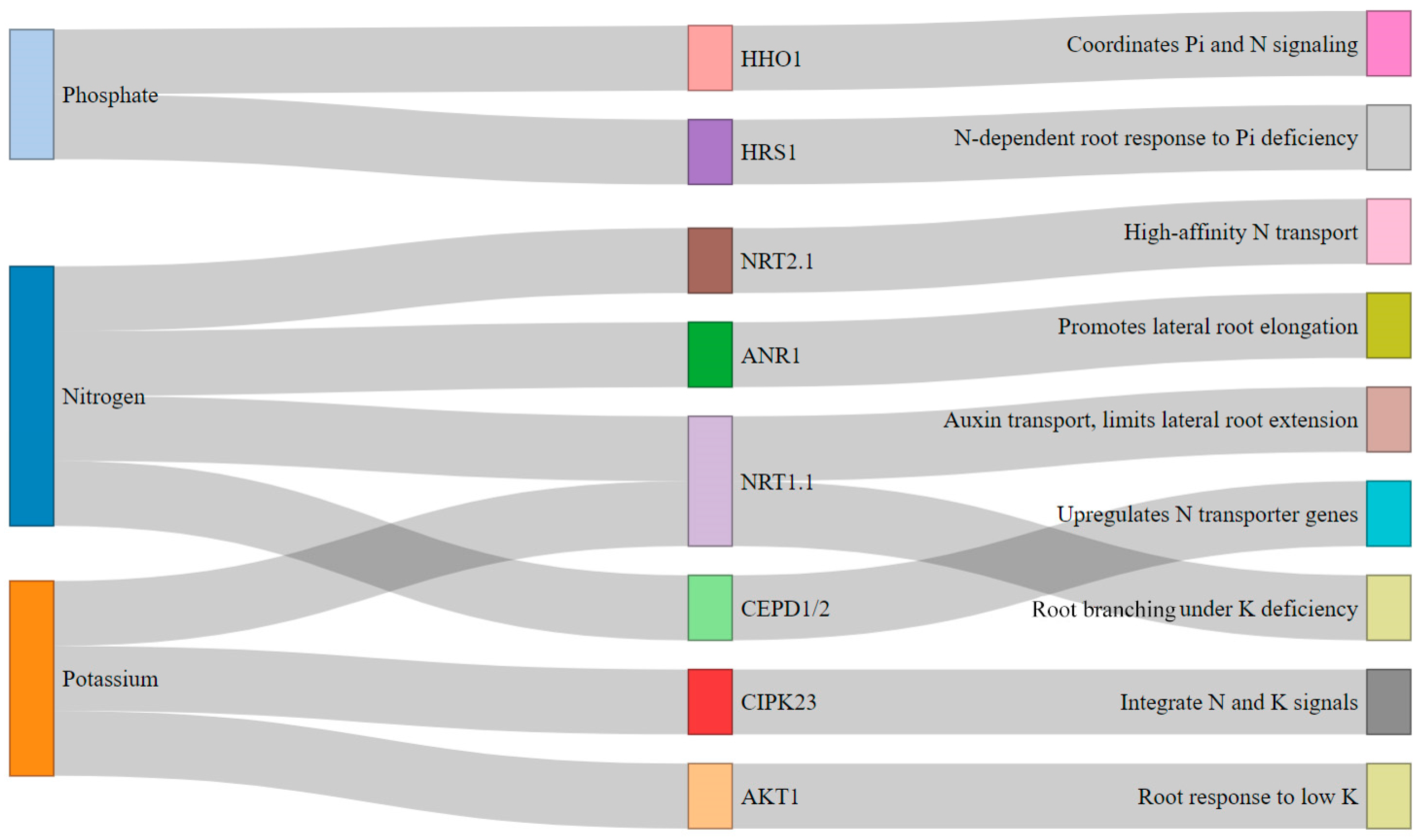
3.3. Editing for Nutrient Uptake Efficiency
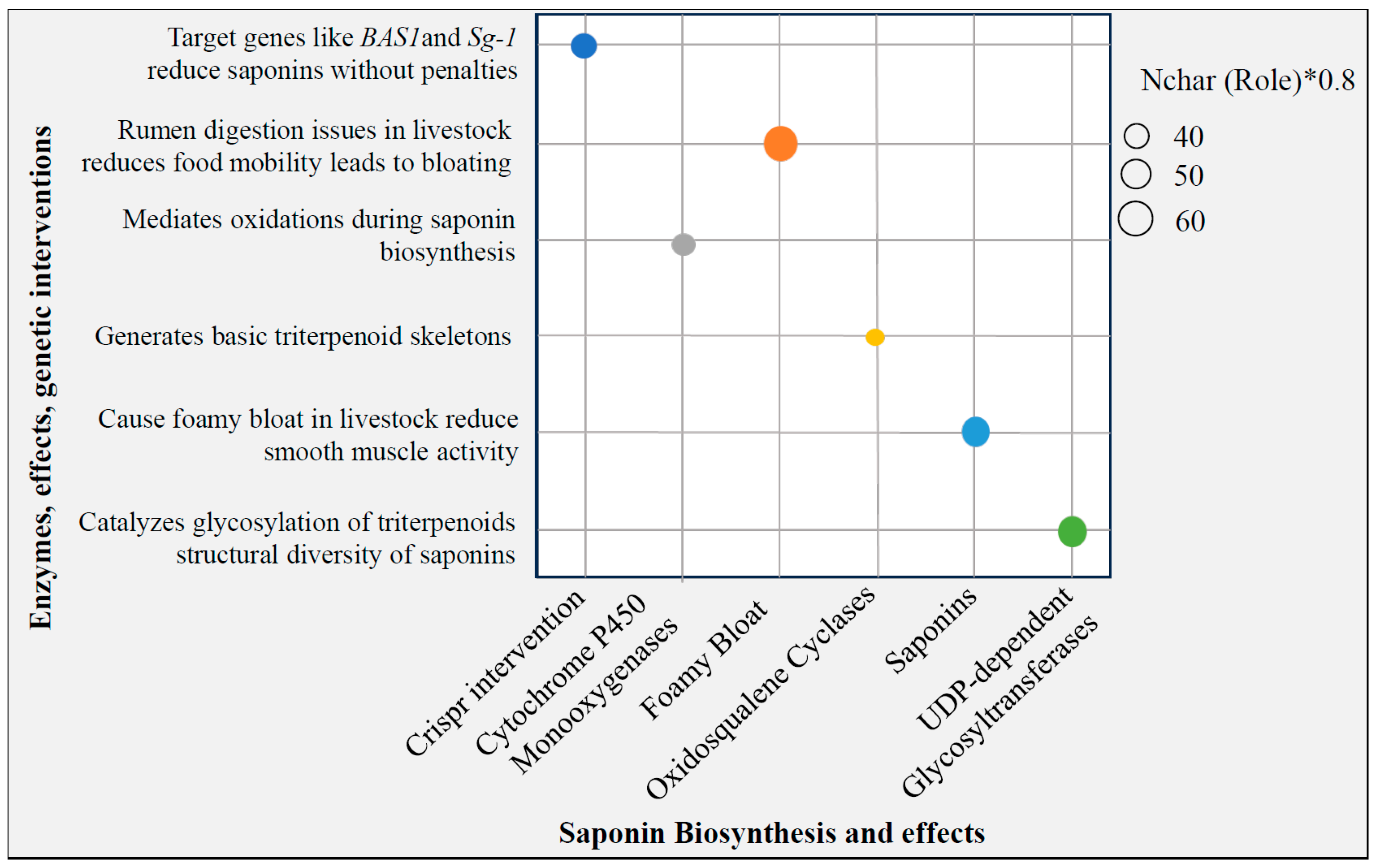
3.4. Antinutrient-Free Knockouts
4. Technical Bottlenecks and Advances in CRISPR/CAS Application in Alfalfa
5. Conclusion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soto-Zarazúa, M.G.; Bah, M.; Costa, A.S.G.; Rodrigues, F.; Pimentel, F.B.; Rojas-Molina, I.; Rojas, A.; Oliveira, M.B.P.P. Nutraceutical potential of new alfalfa (Medicago sativa) ingredients for beverage preparations. J. Med. Food 2017, 20, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, M.; Orellana Palacios, J.C.; McClements, D.J.; Mahfouzi, M.; Moreno, A. Alfalfa as a sustainable source of plant-based food proteins. Trends Food Sci. Technol. 2023, 135, 202–214. [Google Scholar] [CrossRef]
- Bouton, J. The economic benefits of forage improvement in the United States. Euphytica 2007, 154, 263–270. [Google Scholar] [CrossRef]
- Fan, S.; Chen, J.; Mu, J.; Zhang, M. Genetic diversity and salt tolerance assessment of 51 alfalfa (Medicago sativa) varieties under saline soil conditions. Front. Sustain. Food Syst. 2023, 7, 1278913. [Google Scholar] [CrossRef]
- Saini, H.; Thakur, R.; Gill, R.; Tyagi, K.; Goswami, M. CRISPR/Cas9-gene editing approaches in plant breeding. GM Crops Food 2023, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, G.; Padaria, J.C. Application of CRISPR/Cas9 in the improvement of industrially important crops. In Industrial Crops Improvement: Biotechnological Approaches for Sustainable Agricultural Development; Springer Nature: Cham, Switzerland, 2025; pp. 239–257. [Google Scholar]
- Palumbo, F.; Pasquali, E.; Albertini, E.; Barcaccia, G. A review of unreduced gametes and neopolyploids in alfalfa: How to fill the gap between well-established meiotic mutants and next-generation genomic resources. Plants 2021, 10, 999. [Google Scholar] [CrossRef]
- Wang, H.; Coulman, B.; Bai, Y.; Tar’an, B.; Biligetu, B. Genetic diversity and local adaption of alfalfa populations (Medicago sativa L.) under long-term grazing. Sci. Rep. 2023, 13, 1632. [Google Scholar] [CrossRef]
- Hrbáčková, M.; Dvořák, P.; Takáč, T.; Tichá, M.; Luptovčiak, I.; Šamajová, O.; Ovečka, M.; Šamaj, J. Biotechnological perspectives of omics and genetic engineering methods in alfalfa. Front. Plant Sci. 2020, 11, 592. [Google Scholar] [CrossRef]
- He, X.; Zhang, F.; He, F.; Shen, Y.; Yu, L.X.; Zhang, T.; Kang, J. Accuracy of genomic selection for alfalfa biomass yield in two full-sib populations. Front. Plant Sci. 2022, 13, 1037272. [Google Scholar] [CrossRef]
- Long, R.; Zhang, F.; Zhang, Z.; Li, M.; Chen, L.; Wang, X.; Liu, W.; Zhang, T.; Yu, L.X.; He, F.; et al. Genome assembly of alfalfa cultivar Zhongmu-4 and identification of SNPs associated with agronomic traits. Genom. Proteom. Bioinform. 2022, 20, 14–28. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Maureira, I.; Osborn, T. Molecular markers in genetics and breeding: Improvement of alfalfa (Medicago sativa L.). In Molecular Marker Systems in Plant Breeding and Crop Improvement; Lörz, H., Wenzel, G., Eds.; Biotechnology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 2004; Volume 55, pp. 239–257. [Google Scholar] [CrossRef]
- Mejia-Guerra, M.; Zhao, D.; Sheehan, M. Genomic resources for breeding in alfalfa: Availability, utility, and adoption. In The Alfalfa Genome; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Yang, B.; Zhao, Y.; Guo, Z. Research progress and prospect of alfalfa resistance to pathogens and pests. Plants 2022, 11, 2008. [Google Scholar] [CrossRef]
- McNeill, M.; Tu, X.; Altermann, E.; Wu, B.; Shi, S. Sustainable management of Medicago sativa for future climates: Insect pests, endophytes, and multitrophic interactions in a complex environment. Front. Agron. 2022, 4, 825087. [Google Scholar] [CrossRef]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Näpflin, K.; O’Connor, E.A.; Becks, L.; Bensch, S.; Ellis, V.A.; Hafer-Hahmann, N.; Harding, K.C.; Lindén, S.K.; Olsen, M.T.; Roved, J.; et al. Genomics of host-pathogen interactions: Challenges and opportunities across ecological and spatiotemporal scales. PeerJ 2019, 7, e8013. [Google Scholar] [CrossRef]
- Pfeilmeier, S.; George, J.; Morel, A.; Roy, S.; Smoker, M.; Stransfeld, L.; Downie, J.A.; Peeters, N.; Malone, J.G.; Zipfel, C. Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol. J. 2019, 17, 569–579. [Google Scholar] [CrossRef]
- Stefanova, G.; Slavov, S.; Gecheff, K.; Vlahova, M.; Atanassov, A. Expression of recombinant human lactoferrin in transgenic alfalfa plants. Biol. Plant 2013, 57, 457–464. [Google Scholar] [CrossRef]
- He, F.; Xu, M.; Liu, H.; Xu, Y.; Long, R.; Kang, J.; Yang, Q.; Chen, L. Unveiling alfalfa root rot resistance genes through an integrative GWAS and transcriptome study. BMC Plant Biol. 2025, 25, 58. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Marzougui, A.; Coyne, C.J.; Sankaran, S.; Main, D.; Porter, L.D.; Mugabe, D.; Smitchger, J.A.; Zhang, C.; Amin, M.N.; et al. Dissecting the genetic architecture of aphanomyces root rot resistance in lentil by QTL mapping and genome-wide association study. Int. J. Mol. Sci. 2020, 21, 2129. [Google Scholar] [CrossRef]
- Dai, H.; Zhang, X.; Zhao, B.; Shi, J.; Zhang, C.; Wang, G.; Yu, N.; Wang, E. Colonization of mutualistic mycorrhizal and parasitic blast fungi requires OsRAM2-regulated fatty acid biosynthesis in rice. Mol. Plant-Microbe Interact. 2022, 35, 178–186. [Google Scholar] [CrossRef]
- Zamboni, A.; Zanin, L.; Tomasi, N.; Pezzotti, M.; Pinton, R.; Varanini, Z.; Cesco, S. Genome-wide microarray analysis of tomato roots showed defined responses to iron deficiency. BMC Genom. 2012, 13, 1. [Google Scholar] [CrossRef]
- Fan, S.; Xu, X.; Chen, J.; Yin, Y.; Zhao, Y. Genome-wide identification, characterization, and expression analysis of m6A readers-YTH domain-containing genes in alfalfa. BMC Genom. 2024, 25, 18. [Google Scholar]
- Greveniotis, V.; Bouloumpasi, E.; Skendi, A.; Korkovelos, A.; Kantas, D.; Zotis, S.; Ipsilandis, C.G. Modeling stability of alfalfa yield and main quality traits. Agriculture 2024, 14, 542. [Google Scholar] [CrossRef]
- Buxton, D.R. Quality-related characteristics of forages as influenced by plant environment and agronomic factors. Anim. Feed Sci. Technol. 1996, 59, 37–49. [Google Scholar] [CrossRef]
- McCarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Ramegowda, V.; Kumar, A.; Pereira, A. Plant adaptation to drought stress. F1000Research 2016, 5, 1554. [Google Scholar] [CrossRef]
- Gao, R.; Wan, Z.-Y.; Jiang, C.; Zhang, J.-F.; Li, Y.-Y.; Wei, Z.-H.; Zhang, X.-Q. Quantitative trait locus analysis of freezing tolerance in alfalfa. PLoS ONE 2016, 11, e0161670. [Google Scholar] [CrossRef]
- Yang, P.; Li, X.; Wang, X.; Chen, H.; Chen, F.; Shen, S.; Li, Y. Proteomic analysis of alfalfa response to heat stress. PLoS ONE 2015, 10, e0121853. [Google Scholar] [CrossRef]
- Najera, V.A.; Twyman, R.M.; Christou, P.; Zhu, C. Applications of multiplex genome editing in higher plants. Curr. Opin. Biotechnol. 2019, 59, 93–102. [Google Scholar] [CrossRef]
- Rahman, M.A.; Song, Y.; Hasan, M.M.; Jahan, M.S.; Siddiqui, M.H.; Park, H.S.; Lee, S.-H.; Singh, D.; Corpas, F.L.; Kabir, A.H.; et al. Mechanistic basis of silicon-mediated cold stress tolerance in alfalfa (Medicago sativa L.). Silicon 2024, 16, 1057–1069. [Google Scholar] [CrossRef]
- Fernandes, C.; Barrios, S.; Santos, M.; Nunes, J.A.; Borges do Valle, C.; Jank, L.; Rios, E. Assessing genotype adaptability and stability in perennial forage breeding trials using random regression models for longitudinal dry matter yield data. G3: Genes Genome Genet. 2023, 15, jkae306. [Google Scholar] [CrossRef]
- Sinha, D.; Maurya, A.K.; Abdi, G.; Majeed, M.; Agarwal, R.; Mukherjee, R.; Ganguly, S.; Aziz, R.; Bhatia, M.; Majgaonkar, A.; et al. Integrated genomic selection for accelerating breeding programs of climate-smart cereals. Genes 2023, 14, 1484. [Google Scholar] [CrossRef]
- Govindasamy, P.; Muthusamy, S.K.; Bagavathiannan, M.; Mowrer, J.; Jagannadham, P.T.K.; Maity, A.; Halli, H.M.; Sujayanand, G.K.; Vadivel, R.; Das, T.K.; et al. Nitrogen use efficiency—A key to enhance crop productivity under a changing climate. Front. Plant Sci. 2023, 14, 1121073. [Google Scholar] [CrossRef]
- Wan, W.; Liu, Q.; Li, K.; Zhao, K.; Qi, F.; Li, Y.; Sun, Z.; Li, H. Nitrogen fertilizer application for improving the biomass, quality, and nitrogen fixation of alfalfa (Medicago sativa L.) at different growth stages in a saline-alkali soil. PeerJ 2025, 13, e18796. [Google Scholar] [CrossRef] [PubMed]
- Chrysargyris, A.; Tzortzakis, N. Optimizing nitrogen, phosphorus, and potassium requirements to improve Origanum dubium Boiss. growth, nutrient and water use efficiency, essential oil yield, and composition. Ind. Crops Prod. 2025, 224, 120291. [Google Scholar]
- Wang, W.; Hu, B.; Li, A.; Chu, C. NRT1.1s in plants: Functions beyond nitrate transport. J. Exp. Bot. 2020, 71, 4373–4379. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubeš, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zažímalová, E.; et al. Multiple mechanisms of nitrate sensing by arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef]
- Datta, T.; Kumar, R.S.; Sinha, H.; Trivedi, P.K. Small but mighty: Peptides regulating abiotic stress responses in plants. Plant Cell Environ. 2024, 47, 1207–1223. [Google Scholar]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef]
- Wang, X.; Kang, W.; Wu, F.; Miao, J.; Shi, S. Comparative transcriptome analysis reveals new insight of alfalfa (Medicago sativa L.) cultivars in response to abrupt freezing stress. Front. Plant Sci. 2022, 13, 798118. [Google Scholar] [CrossRef]
- Kellermeier, F.; Armengaud, P.; Seditas, T.J.; Danku, J.; Salt, D.E.; Amtmann, A. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.Y.; Luo, J.T.; Zheng, J.M.; Tan, W.F.; Pu, Z.J.; Wang, F. Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency. BMC Plant Biol. 2023, 23, 353. [Google Scholar] [CrossRef]
- Hirsch, R.E.; Lewis, B.D.; Spalding, E.P.; Sussman, M.R. A role for the AKT1 potassium channel in plant nutrition. Science 1998, 280, 918–921. [Google Scholar] [CrossRef]
- Singer, S.D.; Hannoufa, A.; Acharya, S. Molecular improvement of alfalfa for enhanced productivity and adaptability in a changing environment. Plant Cell Environ. 2018, 41, 1955–1971. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.; Ludewig, U.; Neuhäuser, B. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 2017, 29, 409–422. [Google Scholar] [CrossRef]
- Patra, A.K.; Saxena, J. A new perspective on the use of plant secondary metabolites to inhibit methanogenesis in the rumen. Phytochemistry 2010, 71, 1198–1222. [Google Scholar] [CrossRef]
- Sawai, S.; Saito, K. Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2011, 2, 25. [Google Scholar] [CrossRef]
- Tamura, K.; Teranishi, Y.; Ueda, S.; Suzuki, H.; Kawano, N.; Yoshimatsu, K.; Saito, K.; Kawahara, N.; Muranaka, T.; Seki, H. Cytochrome P450 monooxygenase CYP716A141 is a unique β-amyrin C-16β oxidase involved in triterpenoid saponin biosynthesis in Platycodon grandiflorus. Plant Cell Physiol. 2017, 58, 874–884. [Google Scholar] [CrossRef]
- Sayama, T.; Ono, E.; Takagi, K.; Takada, Y.; Horikawa, M.; Nakamoto, Y.; Hirose, A.; Sasama, H.; Ohashi, M.; Hasegawa, H.; et al. The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 2012, 24, 2123–2138. [Google Scholar] [CrossRef] [PubMed]
- Sundaramoorthy, J.; Park, G.T.; Komagamine, K.; Tsukamoto, C.; Chang, J.H.; Lee, J.D.; Kim, J.H.; Seo, H.S.; Song, J.T. Biosynthesis of DDMP saponins in soybean is regulated by a distinct UDP-glycosyltransferase. New Phytol. 2019, 222, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Mengstie, M.A.; Azezew, M.T.; Dejenie, T.A.; Teshome, A.A.; Admasu, F.T.; Teklemariam, A.B.; Mulu, A.T.; Agidew, M.M.; Adugna, D.G.; Geremew, H.; et al. Recent advancements in reducing the off-target effect of CRISPR-Cas9 genome editing. Biol. Targets Ther. 2024, 18, 21–28. [Google Scholar] [CrossRef]
- Schaart, J.G.; van de Wiel, C.C.M.; Smulders, M.J.M. Genome editing of polyploid crops: Prospects, achievements, and bottlenecks. Transgenic Res. 2021, 30, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, S.; Cao, Y.; Jiang, X.; Zhao, L.; Li, Z.; Wang, M.; Yang, R.; Zhou, C.; Wang, Z.; et al. Development of a single transcript CRISPR/Cas9 toolkit for efficient genome editing in autotetraploid alfalfa. Crop J. 2024, 12, 788–795. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Mahmood, K.; Jerez, I.T.; Cong, L.; Yun, J.; Udvardi, M.; Tadege, M.; Wang, Z.; Wen, J. Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 2023, 21, 1383–1392. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, F.; Wang, X.; Han, J.; Zhao, H.; Liu, G.; Wang, Z. Genomic prediction for 25 agronomic and quality traits in alfalfa (Medicago sativa). Front. Plant Sci. 2018, 9, 1220. [Google Scholar] [CrossRef]
- Bottero, E.; Massa, G.; González, M.; Stritzler, M.; Tajima, H.; Gómez, C.; Frare, R.; Feingold, S.; Blumwald, E.; Ayub, N.; et al. Efficient CRISPR/Cas9 genome editing in alfalfa using a public germplasm. Front. Agron. 2021, 3, 661526. [Google Scholar] [CrossRef]
- Shi, K.; Dong, H.; Du, H.; Li, Y.; Zhou, L.; Liang, C.; Şakiroğlu, M.; Wang, Z. The chromosome-level assembly of the wild diploid alfalfa genome provides insights into the full landscape of genomic variations between cultivated and wild alfalfa. Plant Biotechnol. J. 2024, 22, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, C.; Li, J.; Gao, C. Targeted genome-modification tools and their advanced applications in crop breeding. Nat. Rev. Genet. 2024, 25, 603–622. [Google Scholar] [PubMed]
- Che, P.; Wu, E.; Simon, M.K.; Anand, A.; Lowe, K.; Gao, H.; Sigmund, A.L.; Yang, M.; Albertsen, M.C.; Gordon-Kamm, W.; et al. Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun. Biol. 2022, 5, 344. [Google Scholar] [CrossRef]
- Subedi, U.; Burton Hughes, K.; Chen, G.; Hannoufa, A.; Singer, S.D. Eliciting targeted mutations in Medicago sativa using CRISPR/Cas9-mediated genome editing: A potential tool for the improvement of disease resistance. Methods Mol. Biol. 2023, 2659, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Feyissa, B.A.; Croft, M.; Hannoufa, A. Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta 2018, 247, 1043–1050. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Cong, L.; Park, J.J.; Bao, Q.; Chen, M.; Sun, J.; Xu, B.; Ge, Y.; Chai, M.; Liu, Z.; et al. Development of a highly efficient multiplex genome editing system in outcrossing tetraploid alfalfa (Medicago sativa). Front. Plant Sci. 2020, 11, 1063. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Mahmood, K.; Chen, F.; Torres-Jerez, I.; Udvardi, M.; Tadege, M.; Cong, L.; Wang, Z.; Wen, J. Mutating alfalfa COUMARATE 3-HYDROXYLASE using multiplex CRISPR/Cas9 leads to reduced lignin deposition and improved forage quality. Front. Plant Sci. 2024, 15, 1363182. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Jia, L.; Wu, J.; Zhao, Y. Harnessing the Potential of CRISPR/Cas in Targeted Alfalfa Improvement for Stress Resilience. Int. J. Mol. Sci. 2025, 26, 3311. https://doi.org/10.3390/ijms26073311
Fan S, Jia L, Wu J, Zhao Y. Harnessing the Potential of CRISPR/Cas in Targeted Alfalfa Improvement for Stress Resilience. International Journal of Molecular Sciences. 2025; 26(7):3311. https://doi.org/10.3390/ijms26073311
Chicago/Turabian StyleFan, Shugao, Linyan Jia, Jiawei Wu, and Ying Zhao. 2025. "Harnessing the Potential of CRISPR/Cas in Targeted Alfalfa Improvement for Stress Resilience" International Journal of Molecular Sciences 26, no. 7: 3311. https://doi.org/10.3390/ijms26073311
APA StyleFan, S., Jia, L., Wu, J., & Zhao, Y. (2025). Harnessing the Potential of CRISPR/Cas in Targeted Alfalfa Improvement for Stress Resilience. International Journal of Molecular Sciences, 26(7), 3311. https://doi.org/10.3390/ijms26073311






