microRNAs in Type 1 Diabetes: Roles, Pathological Mechanisms, and Therapeutic Potential
Abstract
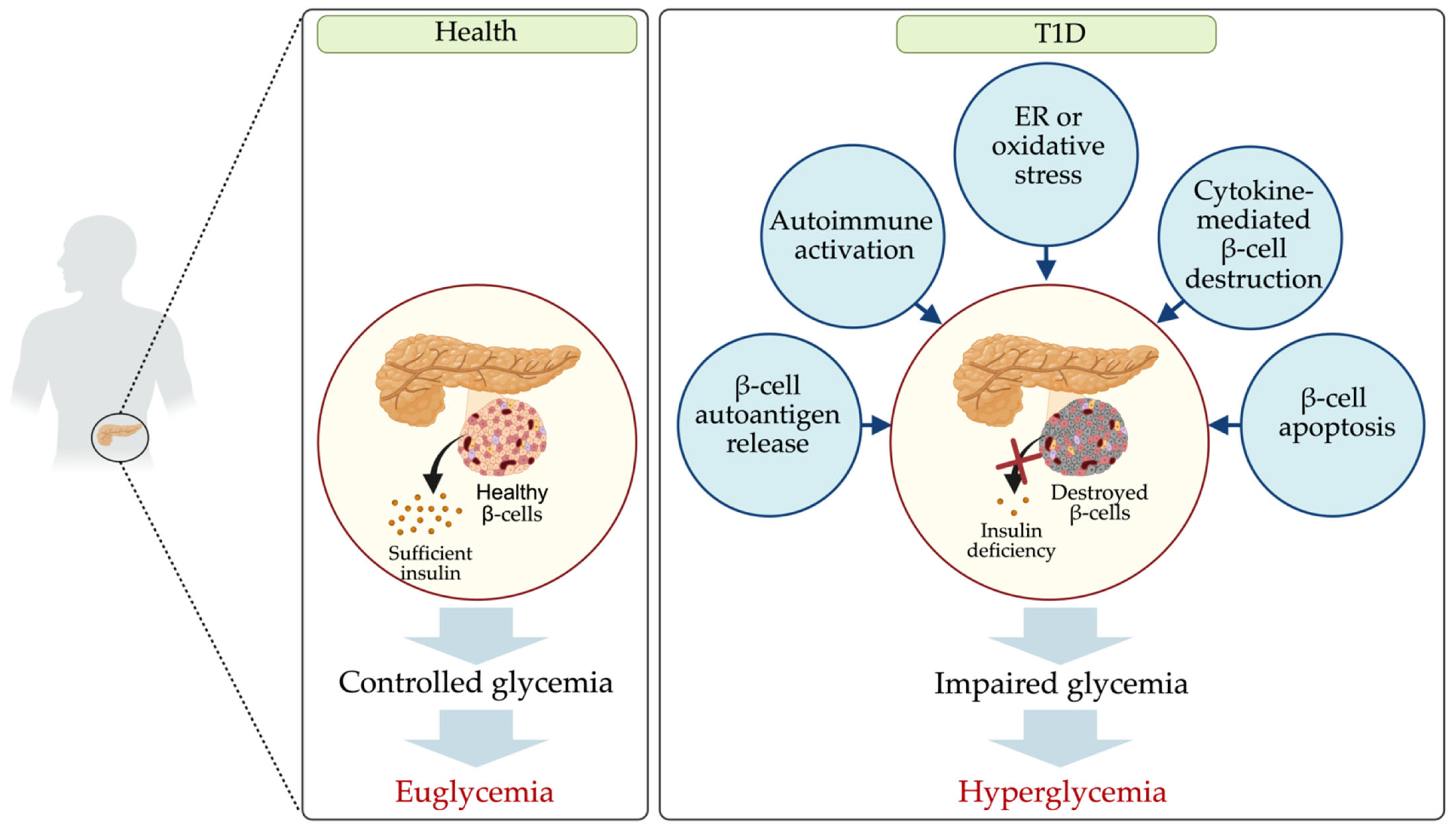
1. Introduction
2. Dysregulated miRNAs in T1D Patients
2.1. Dysregulated miRNAs in Various Samples from T1D Patients
| Sample | Expression | miRNA | Fold Change (vs HC *) | Ref. |
|---|---|---|---|---|
| Serum | Up | miR-21-5p miR-24-3p miR-25 miR-27a miR-29a miR-140-5p miR-144-5p miR-148a miR-152 miR-181a miR-199a miR-200a miR-208a-3p miR-210 miR-222-3p miR-323a-3p miR-345-5p miR-454-3p | NA ** 1.6–2.25 1.53 2.79 2.39 1.7 5.1 2.25 2.09 2.30 NA 1.23–2.73 1.97 1.65 2.7 1.8–5.6 2.3 2.6 | [29] [18,30] [30] [30] [30] [18] [18] [29,30] [30] [30] [30] [30,31] [31] [30] [18] [31] [18] [18] |
| Down | miR-16-5p miR-19a-3p miR-25-3p miR-155-5p 2 miR-195-5p miR-590-5p | 0.38 0.42 0.40 0.26-0.54 0.42 0.46 | [31] [31] [31] [31,32] [31] [31] | |
| Plasma | Up | miR-10b-5p 1 miR-21 miR-24 miR-103a-3p miR-125b-5p miR-146a-5p 2 miR-155-5p 1 miR-200a-3p miR-210-3p miR-365a-3p miR-770-5p | NA NA NA 121.0 NA NA 1.88 59.90 4.79 NA NA | [33] [34] [3] [32] [17] [3,32] [32] [32] [32,34] [17] [17] |
| Down | miR-10b-5p 2 miR-146a-5p 1 miR-5190 miR-409-3p | NA 0.29 NA NA | [33] [32] [17] [35] | |
| Peripheral blood cells | Up | miR-17-5p miR-21-5p miR-26a-5p miR-181a-5p miR-221-3p | NA NA NA NA NA | [36] [36] [36] [36] [36] |
| Down | miR-16-5p miR-17-5p miR-126-3p/5p miR-143-3p | NA NA NA NA | [36] [36] [36] [36] | |
| PBMC 3 | Up | miR-21 miR-22 miR-26b miR-32 miR-92-3p miR-126 miR-142-5p miR-143-3p miR-148a/b miR-186 miR-195 | 3.46 NA 2.21 2.11 NA 3.36 NA NA 1.60–2.01 1.65 2.16 | [19] [37] [19] [19] [38] [19] [38] [38] [19] [19] [19] |
| Down | miR-140-3p miR-146a/b miR-150 miR-423-5p miR-424 miR-720 | 0.63 NA NA 0.55 NA 0.56 | [19] [39,40] [37,38,40] [19] [40] [19] | |
| Plasma-derived exosome | Up | miR-25-3p | NA | [41] |
| Down | miR-16 miR-302d-3p miR-378e | NA NA NA | [41] [41] [41] |
2.2. Dysregulated miRNAs in Serum, Plasma, and Exosomes Derived from Plasma
2.3. Dysregulated miRNAs in Blood Cells
2.4. Dysregulated miRNAs in T-Cells
3. Dysregulated miRNAs in T1D Rodents
| Diabetic Animal Model | Sample | miRNA Expression Alteration | Ref. | |
|---|---|---|---|---|
| Expression | miRNA | |||
| Prediabetic NOD * mice | Pancreatic islet, cultured islet, infiltrating lymphocytes | Up | miR-29a miR-142-5p | [96,97] [96,97] |
| Down | miR-142-3p miR-150 | [96,97] [96,97] | ||
| Pancreatic β-cells | Up | miR-142-5p | [97] | |
| Down | miR-150 miR-155 | [97] [97] | ||
| Diabetic NOD mice | Pancreatic islet/plasma | Up | miR-21 | [98] |
| Down | miR-126a-3p/5p miR-155 miR-409-3p | [35] [35] [35] | ||
| C57BL/6J mice induced with STZ * | Pancreatic tissue | Up | miR-323-3p | [99] |
| Down | miR-10b-5p miR-16-5p miR-17-5p miR-126-3p/5p miR-143-3p | [99] [99] [99] [99] [99] | ||
| Pancreatic islet | Up | miR-21 | [98] | |
4. Dysregulated miRNAs in T1D and Their Potential Targets
| Group | Pathway | Upregulated miRNA | Downregulated miRNA |
|---|---|---|---|
| Apoptosis | β-cell apoptosis | miR-21-5p [102,103] miR-24 [104] miR-34a [105] miR-155 [106] miR-181a-5p [8] miR-200a-3p [32,107] miR-375 [108] miR-424 [19] | miR-100-5p [89] miR-126 [109] miR-146a-5p [32,40] miR-150-5p [40] miR-320a-3p [110] miR-424 [40] |
| TP53 signaling | miR-145 [111] | miR-324-5p [19,99] miR-342-3p [19,89] miR-423-5p [19] | |
| Wnt signaling | miR-143-3p [112] miR-144-5p [18] miR-148a-3p [29,32] miR-365a-3p [17] | miR-140-3p [19] miR-766 [19] miR-940 [19] | |
| TGF-β or mTOR signaling | miR-26b [32,113] miR-323-3p [99] miR-382-5p [38] | miR-10b-5p [114] | |
| ER or oxidative stress | FOXO or Notch signaling | miR-21-5p [29,115] miR-148a-3p [32] miR-323-3p [99] miR-486-5p [31,116] | miR-140-3p [19] miR-146a-5p [32] miR-324-5p [19] miR-423-5p [19] |
| NF-κβ signaling | miR-24-3p [117,118] miR-155-5p [32,119,120,121] | miR-146a-5p [32] miR-342 [122] | |
| Immune system activation | Immune system-related | miR-103a-3p [32] miR-200a-3p [32] | |
| T-cell regulation | miR-31 [89] miR-342 [89] | ||
| Chemokine signaling [19] | miR-18b miR-20b miR-101 miR-186 | miR-940 | |
| β-cell autoantigen release | Jak-STAT signaling | miR-21-5p [8] miR-24-3p [8,95] miR-125b-5p [17,123] miR-181-5p [8] miR-323-3p [99] miR-210-5p [8] | |
| MAPK signaling | miR-199a [19] miR-342 [32] miR-450a [19] miR-548c-3p [19] | miR-100-5p [8] miR-150-5p [8] | |
| β-cell insulin release | Insulin signaling | miR-21 [19] miR-26b [36] miR-32 [19] miR-103a-3p [32] miR-143-3p [36] miR-148a [19] miR-200a-3p [32] miR-210-3p [32] miR-320c [124] miR-424 [19] miR-1225-5p [124] | miR-29a [125] miR-146a-5p [32] miR-324-5p [19] miR-342-3p [19] miR-423-5p [19] |
4.1. β-Cell Autoantigen Release
4.2. Autoimmune Activation
4.3. Endoplasmic Reticulum and Oxidative Stress
4.4. Apoptosis
4.5. Insulin Signaling
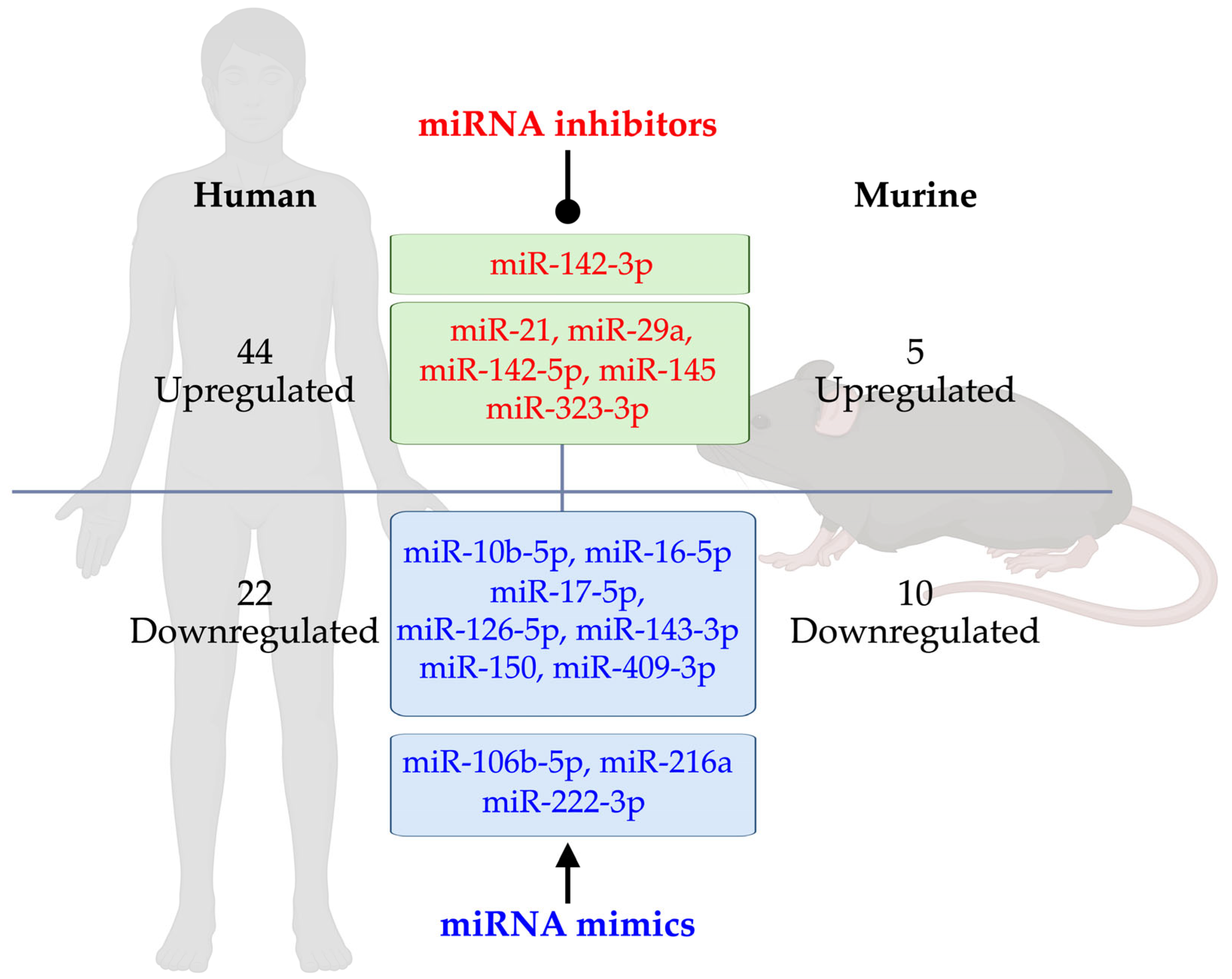
5. miRNA-Based Therapeutic Strategies for T1D
6. Conclusions and Future Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Banday, M.Z.; Sameer, A.S.; Nissar, S. Pathophysiology of diabetes: An overview. Avicenna J. Med. 2020, 10, 174–188. [Google Scholar] [PubMed]
- Tuomi, T.; Santoro, N.; Caprio, S.; Cai, M.; Weng, J.; Groop, L. The many faces of diabetes: A disease with increasing heterogeneity. Lancet 2014, 383, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479. [Google Scholar]
- Gregory, G.A.; Robinson, T.I.G.; Linklater, S.E.; Wang, F.; Colagiuri, S.; de Beaufort, C.; Donaghue, K.C.; Magliano, D.J.; Maniam, J.; Orchard, T.J.; et al. Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: A modelling study. Lancet Diabetes Endocrinol. 2022, 10, 741–760. [Google Scholar]
- Katsarou, A.; Gudbjornsdottir, S.; Rawshani, A.; Dabelea, D.; Bonifacio, E.; Anderson, B.J.; Jacobsen, L.M.; Schatz, D.A.; Lernmark, A. Type 1 diabetes mellitus. Nat. Rev. Dis. Primers 2017, 3, 17016. [Google Scholar]
- Daneman, D. Type 1 diabetes. Lancet 2006, 367, 847–858. [Google Scholar]
- Margaritis, K.; Margioula-Siarkou, G.; Giza, S.; Kotanidou, E.P.; Tsinopoulou, V.R.; Christoforidis, A.; Galli-Tsinopoulou, A. Micro-RNA Implications in Type-1 Diabetes Mellitus: A Review of Literature. Int. J. Mol. Sci. 2021, 22, 12165. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790. [Google Scholar]
- Pirot, P.; Cardozo, A.K.; Eizirik, D.L. Mediators and mechanisms of pancreatic beta-cell death in type 1 diabetes. Arq. Bras. Endocrinol. Metabol. 2008, 52, 156–165. [Google Scholar]
- Ounissi-Benkalha, H.; Polychronakos, C. The molecular genetics of type 1 diabetes: New genes and emerging mechanisms. Trends Mol. Med. 2008, 14, 268–275. [Google Scholar]
- Ziegler, A.G.; Rewers, M.; Simell, O.; Simell, T.; Lempainen, J.; Steck, A.; Winkler, C.; Ilonen, J.; Veijola, R.; Knip, M.; et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013, 309, 2473–2479. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed]
- von Scholten, B.J.; Kreiner, F.F.; Gough, S.C.L.; von Herrath, M. Current and future therapies for type 1 diabetes. Diabetologia 2021, 64, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Zogg, H.; Singh, R.; Ro, S. Current Advances in RNA Therapeutics for Human Diseases. Int. J. Mol. Sci. 2022, 23, 2736. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Satake, E.; Pezzolesi, M.G.; Md Dom, Z.I.; Smiles, A.M.; Niewczas, M.A.; Krolewski, A.S. Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes. Diabetes 2018, 67, 1013–1023. [Google Scholar] [CrossRef]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef]
- Takahashi, P.; Xavier, D.J.; Evangelista, A.F.; Manoel-Caetano, F.S.; Macedo, C.; Collares, C.V.; Foss-Freitas, M.C.; Foss, M.C.; Rassi, D.M.; Donadi, E.A.; et al. MicroRNA expression profiling and functional annotation analysis of their targets in patients with type 1 diabetes mellitus. Gene 2014, 539, 213–223. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Lu, Q.; Wu, R.; Zhao, M.; Garcia-Gomez, A.; Ballestar, E. miRNAs as Therapeutic Targets in Inflammatory Disease. Trends Pharmacol. Sci. 2019, 40, 853–865. [Google Scholar] [PubMed]
- Singh, R.; Ha, S.E.; Park, H.S.; Debnath, S.; Cho, H.; Baek, G.; Yu, T.Y.; Ro, S. Sustained Effectiveness and Safety of Therapeutic miR-10a/b in Alleviating Diabetes and Gastrointestinal Dysmotility without Inducing Cancer or Inflammation in Murine Liver and Colon. Int. J. Mol. Sci. 2024, 25, 2266. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Pollet, N. Synexpression groups in eukaryotes. Nature 1999, 402, 483–487. [Google Scholar] [PubMed]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Sayed, A.S.; Xia, K.; Salma, U.; Yang, T.; Peng, J. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ. 2014, 23, 503–510. [Google Scholar]
- Zampetaki, A.; Willeit, P.; Drozdov, I.; Kiechl, S.; Mayr, M. Profiling of circulating microRNAs: From single biomarkers to re-wired networks. Cardiovasc. Res. 2012, 93, 555–562. [Google Scholar]
- Grieco, G.E.; Cataldo, D.; Ceccarelli, E.; Nigi, L.; Catalano, G.; Brusco, N.; Mancarella, F.; Ventriglia, G.; Fondelli, C.; Guarino, E.; et al. Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA 2018, 4, 37. [Google Scholar]
- Nielsen, L.B.; Wang, C.; Sorensen, K.; Bang-Berthelsen, C.H.; Hansen, L.; Andersen, M.L.; Hougaard, P.; Juul, A.; Zhang, C.Y.; Pociot, F.; et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: Evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp. Diabetes Res. 2012, 2012, 896362. [Google Scholar]
- Santos, A.S.; Ferreira, L.R.P.; da Silva, A.C.; Alves, L.I.; Damasceno, J.G.; Kulikowski, L.; Cunha-Neto, E.; da Silva, M.E.R. Progression of Type 1 Diabetes: Circulating MicroRNA Expression Profiles Changes from Preclinical to Overt Disease. J. Immunol. Res. 2022, 2022, 2734490. [Google Scholar] [PubMed]
- Assmann, T.S.; Recamonde-Mendoza, M.; Punales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res. Clin. Pract. 2018, 141, 35–46. [Google Scholar] [PubMed]
- Samandari, N.; Mirza, A.H.; Kaur, S.; Hougaard, P.; Nielsen, L.B.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Influence of Disease Duration on Circulating Levels of miRNAs in Children and Adolescents with New Onset Type 1 Diabetes. Noncoding RNA 2018, 4, 35. [Google Scholar] [PubMed]
- Osipova, J.; Fischer, D.C.; Dangwal, S.; Volkmann, I.; Widera, C.; Schwarz, K.; Lorenzen, J.M.; Schreiver, C.; Jacoby, U.; Heimhalt, M.; et al. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: A cross-sectional cohort study. J. Clin. Endocrinol. Metab. 2014, 99, E1661–E1665. [Google Scholar]
- Ventriglia, G.; Mancarella, F.; Sebastiani, G.; Cook, D.P.; Mallone, R.; Mathieu, C.; Gysemans, C.; Dotta, F. miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2020, 63, 124–136. [Google Scholar]
- Ferraz, R.S.; Santos, L.C.B.; da-Silva-Cruz, R.L.; Braga-da-Silva, C.H.; Magalhaes, L.; Ribeiro-Dos-Santos, A.; Vidal, A.; Vinasco-Sandoval, T.; Reis-das-Merces, L.; Sena-Dos-Santos, C.; et al. Global miRNA expression reveals novel nuclear and mitochondrial interactions in Type 1 diabetes mellitus. Front. Endocrinol. 2022, 13, 1033809. [Google Scholar]
- Estrella, S.; Garcia-Diaz, D.F.; Codner, E.; Camacho-Guillen, P.; Perez-Bravo, F. Expression of miR-22 and miR-150 in type 1 diabetes mellitus: Possible relationship with autoimmunity and clinical characteristics. Med. Clin. 2016, 147, 245–247. [Google Scholar]
- Massaro, J.D.; Polli, C.D.; Costa, E.S.M.; Alves, C.C.; Passos, G.A.; Sakamoto-Hojo, E.T.; Rodrigues de Holanda Miranda, W.; Bispo Cezar, N.J.; Rassi, D.M.; Crispim, F.; et al. Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol. Cell Endocrinol. 2019, 490, 1–14. [Google Scholar]
- Yang, M.; Ye, L.; Wang, B.; Gao, J.; Liu, R.; Hong, J.; Wang, W.; Gu, W.; Ning, G. Decreased miR-146 expression in peripheral blood mononuclear cells is correlated with ongoing islet autoimmunity in type 1 diabetes patients 1miR-146. J. Diabetes 2015, 7, 158–165. [Google Scholar]
- Wang, G.; Gu, Y.; Xu, N.; Zhang, M.; Yang, T. Decreased expression of miR-150, miR146a and miR424 in type 1 diabetic patients: Association with ongoing islet autoimmunity. Biochem. Biophys. Res. Commun. 2018, 498, 382–387. [Google Scholar]
- Garcia-Contreras, M.; Shah, S.H.; Tamayo, A.; Robbins, P.D.; Golberg, R.B.; Mendez, A.J.; Ricordi, C. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci. Rep. 2017, 7, 5998. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Z.; Wang, J.; Ji, X.; Yao, Z.; Wang, X. miR-21 promotes osteoclastogenesis through activation of PI3K/Akt signaling by targeting Pten in RAW264.7 cells. Mol. Med. Rep. 2020, 21, 1125–1132. [Google Scholar] [PubMed]
- Aghaei-Zarch, S.M. Crosstalk between MiRNAs/lncRNAs and PI3K/AKT signaling pathway in diabetes mellitus: Mechanistic and therapeutic perspectives. Noncoding RNA Res. 2024, 9, 486–507. [Google Scholar] [PubMed]
- Melkman-Zehavi, T.; Oren, R.; Kredo-Russo, S.; Shapira, T.; Mandelbaum, A.D.; Rivkin, N.; Nir, T.; Lennox, K.A.; Behlke, M.A.; Dor, Y.; et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J. 2011, 30, 835–845. [Google Scholar]
- Xu, G.; Thielen, L.A.; Chen, J.; Grayson, T.B.; Grimes, T.; Bridges, S.L., Jr.; Tse, H.M.; Smith, B.; Patel, R.; Li, P.; et al. Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E723–E730. [Google Scholar]
- Angelescu, M.A.; Andronic, O.; Dima, S.O.; Popescu, I.; Meivar-Levy, I.; Ferber, S.; Lixandru, D. miRNAs as Biomarkers in Diabetes: Moving towards Precision Medicine. Int. J. Mol. Sci. 2022, 23, 12843. [Google Scholar] [CrossRef]
- Zi, Y.; Zhang, Y.; Wu, Y.; Zhang, L.; Yang, R.; Huang, Y. Downregulation of microRNA-25-3p inhibits the proliferation and promotes the apoptosis of multiple myeloma cells via targeting the PTEN/PI3K/AKT signaling pathway. Int. J. Mol. Med. 2021, 47, 1. [Google Scholar]
- Kong, R.; Gao, J.; Ji, L.; Zhao, D. MicroRNA-126 promotes proliferation, migration, invasion and endothelial differentiation while inhibits apoptosis and osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Cell Cycle 2020, 19, 2119–2138. [Google Scholar]
- Fang, S.; Ma, X.; Guo, S.; Lu, J. MicroRNA-126 inhibits cell viability and invasion in a diabetic retinopathy model via targeting IRS-1. Oncol. Lett. 2017, 14, 4311–4318. [Google Scholar]
- Rosell, R.; Wei, J.; Taron, M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2009, 10, 8–9. [Google Scholar]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [PubMed]
- Cortez, M.A.; Calin, G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert. Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [PubMed]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar]
- Wang, C.; Hu, J.; Lu, M.; Gu, H.; Zhou, X.; Chen, X.; Zen, K.; Zhang, C.Y.; Zhang, T.; Ge, J.; et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci. Rep. 2015, 5, 7610. [Google Scholar] [CrossRef]
- Gui, J.; Tian, Y.; Wen, X.; Zhang, W.; Zhang, P.; Gao, J.; Run, W.; Tian, L.; Jia, X.; Gao, Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011, 120, 183–193. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar]
- Olivieri, F.; Rippo, M.R.; Procopio, A.D.; Fazioli, F. Circulating inflamma-miRs in aging and age-related diseases. Front. Genet. 2013, 4, 121. [Google Scholar] [CrossRef]
- Du, M.; Liu, S.; Gu, D.; Wang, Q.; Zhu, L.; Kang, M.; Shi, D.; Chu, H.; Tong, N.; Chen, J.; et al. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis 2014, 35, 2723–2730. [Google Scholar] [CrossRef]
- Wang, W.; Sun, G.; Zhang, L.; Shi, L.; Zeng, Y. Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J. Stroke Cerebrovasc. Dis. 2014, 23, 2607–2613. [Google Scholar]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [PubMed]
- Sohel, M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [PubMed]
- Zhang, L.; Zhang, Y.; Zhao, Y.; Wang, Y.; Ding, H.; Xue, S.; Li, P. Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert. Opin. Ther. Pat. 2018, 28, 591–601. [Google Scholar]
- de Gonzalo-Calvo, D.; Vea, A.; Bar, C.; Fiedler, J.; Couch, L.S.; Brotons, C.; Llorente-Cortes, V.; Thum, T. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: A novel tool for personalized medicine? Eur. Heart J. 2019, 40, 1643–1650. [Google Scholar]
- Gao, W.; Liu, L.; Lu, X.; Shu, Y. Circulating microRNAs: Possible prediction biomarkers for personalized therapy of non-small-cell lung carcinoma. Clin. Lung Cancer 2011, 12, 14–17. [Google Scholar]
- Lu, J.; Xie, F.; Geng, L.; Shen, W.; Sui, C.; Yang, J. Potential Role of MicroRNA-210 as Biomarker in Human Cancers Detection: A Meta-Analysis. Biomed. Res. Int. 2015, 2015, 303987. [Google Scholar]
- Tang, Y.; Luo, X.; Cui, H.; Ni, X.; Yuan, M.; Guo, Y.; Huang, X.; Zhou, H.; de Vries, N.; Tak, P.P.; et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009, 60, 1065–1075. [Google Scholar]
- Zhu, H.; Leung, S.W. Identification of microRNA biomarkers in type 2 diabetes: A meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–911. [Google Scholar]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. CCS 2021, 19, 47. [Google Scholar]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar]
- Yang, J.; Wei, F.; Schafer, C.; Wong, D.T. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS ONE 2014, 9, e110641. [Google Scholar]
- Guay, C.; Menoud, V.; Rome, S.; Regazzi, R. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun. Signal 2015, 13, 17. [Google Scholar]
- Setyowati Karolina, D.; Sepramaniam, S.; Tan, H.Z.; Armugam, A.; Jeyaseelan, K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013, 10, 1365–1378. [Google Scholar]
- Knezevic, I.; Patel, A.; Sundaresan, N.R.; Gupta, M.P.; Solaro, R.J.; Nagalingam, R.S.; Gupta, M. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: Implications in postnatal cardiac remodeling and cell survival. J. Biol. Chem. 2012, 287, 12913–12926. [Google Scholar]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar]
- Luty, W.H.; Rodeberg, D.; Parness, J.; Vyas, Y.M. Antiparallel segregation of notch components in the immunological synapse directs reciprocal signaling in allogeneic Th:DC conjugates. J. Immunol. 2007, 179, 819–829. [Google Scholar]
- Pattu, V.; Qu, B.; Schwarz, E.C.; Strauss, B.; Weins, L.; Bhat, S.S.; Halimani, M.; Marshall, M.; Rettig, J.; Hoth, M. SNARE protein expression and localization in human cytotoxic T lymphocytes. Eur. J. Immunol. 2012, 42, 470–475. [Google Scholar]
- Pelayo, R.; Hirose, J.; Huang, J.; Garrett, K.P.; Delogu, A.; Busslinger, M.; Kincade, P.W. Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood 2005, 105, 4407–4415. [Google Scholar]
- Barutta, F.; Tricarico, M.; Corbelli, A.; Annaratone, L.; Pinach, S.; Grimaldi, S.; Bruno, G.; Cimino, D.; Taverna, D.; Deregibus, M.C.; et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS ONE 2013, 8, e73798. [Google Scholar]
- Murray, A.R.; Chen, Q.; Takahashi, Y.; Zhou, K.K.; Park, K.; Ma, J.X. MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Investig. Ophthalmol. Vis. Sci. 2013, 54, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Baseler, W.A.; Thapa, D.; Jagannathan, R.; Dabkowski, E.R.; Croston, T.L.; Hollander, J.M. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am. J. Physiol. Cell Physiol. 2012, 303, C1244–C1251. [Google Scholar] [CrossRef]
- Silva, V.A.; Polesskaya, A.; Sousa, T.A.; Correa, V.M.; Andre, N.D.; Reis, R.I.; Kettelhut, I.C.; Harel-Bellan, A.; De Lucca, F.L. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol. Vis. 2011, 17, 2228–2240. [Google Scholar] [PubMed]
- Xiong, F.; Du, X.; Hu, J.; Li, T.; Du, S.; Wu, Q. Altered retinal microRNA expression profiles in early diabetic retinopathy: An in silico analysis. Curr. Eye Res. 2014, 39, 720–729. [Google Scholar] [CrossRef]
- Zheng, D.; Ma, J.; Yu, Y.; Li, M.; Ni, R.; Wang, G.; Chen, R.; Li, J.; Fan, G.C.; Lacefield, J.C.; et al. Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia 2015, 58, 1949–1958. [Google Scholar] [CrossRef]
- Dieter, C.; Assmann, T.S.; Costa, A.R.; Canani, L.H.; de Souza, B.M.; Bauer, A.C.; Crispim, D. MiR-30e-5p and MiR-15a-5p Expressions in Plasma and Urine of Type 1 Diabetic Patients With Diabetic Kidney Disease. Front. Genet. 2019, 10, 563. [Google Scholar] [CrossRef]
- Tsai, S.; Shameli, A.; Santamaria, P. CD8+ T cells in type 1 diabetes. Adv. Immunol. 2008, 100, 79–124. [Google Scholar]
- ElEssawy, B.; Li, X.C. Type 1 diabetes and T regulatory cells. Pharmacol. Res. 2015, 98, 22–30. [Google Scholar] [CrossRef]
- Hezova, R.; Slaby, O.; Faltejskova, P.; Mikulkova, Z.; Buresova, I.; Raja, K.R.; Hodek, J.; Ovesna, J.; Michalek, J. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010, 260, 70–74. [Google Scholar] [CrossRef]
- Garzon, R.; Volinia, S.; Liu, C.G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.; Alder, H.; Nakamura, T.; et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008, 111, 3183–3189. [Google Scholar] [CrossRef]
- Hui, A.B.; Shi, W.; Boutros, P.C.; Miller, N.; Pintilie, M.; Fyles, T.; McCready, D.; Wong, D.; Gerster, K.; Waldron, L.; et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab. Investig. 2009, 89, 597–606. [Google Scholar] [PubMed]
- Gao, S.; Zhao, Z.Y.; Wu, R.; Zhang, Y.; Zhang, Z.Y. Prognostic value of microRNAs in colorectal cancer: A meta-analysis. Cancer Manag. Res. 2018, 10, 907–929. [Google Scholar] [PubMed]
- Schmidt, W.M.; Spiel, A.O.; Jilma, B.; Wolzt, M.; Muller, M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem. Biophys. Res. Commun. 2009, 380, 437–441. [Google Scholar] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stahler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar]
- Cabiati, M.; Federico, G.; Del Ry, S. Importance of Studying Non-Coding RNA in Children and Adolescents with Type 1 Diabetes. Biomedicines 2024, 12, 1988. [Google Scholar] [CrossRef]
- Roggli, E.; Gattesco, S.; Caille, D.; Briet, C.; Boitard, C.; Meda, P.; Regazzi, R. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 2012, 61, 1742–1751. [Google Scholar]
- Guay, C.; Kruit, J.K.; Rome, S.; Menoud, V.; Mulder, N.L.; Jurdzinski, A.; Mancarella, F.; Sebastiani, G.; Donda, A.; Gonzalez, B.J.; et al. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic beta Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab. 2019, 29, 348–361.e6. [Google Scholar]
- Sims, E.K.; Lakhter, A.J.; Anderson-Baucum, E.; Kono, T.; Tong, X.; Evans-Molina, C. MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia 2017, 60, 1057–1065. [Google Scholar]
- Tian, C.; Ouyang, X.; Lv, Q.; Zhang, Y.; Xie, W. Cross-talks between microRNAs and mRNAs in pancreatic tissues of streptozotocin-induced type 1 diabetic mice. Biomed. Rep. 2015, 3, 333–342. [Google Scholar]
- Mao, Y.; Mohan, R.; Zhang, S.; Tang, X. MicroRNAs as pharmacological targets in diabetes. Pharmacol. Res. 2013, 75, 37–47. [Google Scholar]
- Sahin, G.S.; Lee, H.; Engin, F. An accomplice more than a mere victim: The impact of beta-cell ER stress on type 1 diabetes pathogenesis. Mol. Metab. 2021, 54, 101365. [Google Scholar] [PubMed]
- La Sala, L.; Mrakic-Sposta, S.; Micheloni, S.; Prattichizzo, F.; Ceriello, A. Glucose-sensing microRNA-21 disrupts ROS homeostasis and impairs antioxidant responses in cellular glucose variability. Cardiovasc. Diabetol. 2018, 17, 105. [Google Scholar] [PubMed]
- Qadir, M.M.F.; Klein, D.; Alvarez-Cubela, S.; Dominguez-Bendala, J.; Pastori, R.L. The Role of MicroRNAs in Diabetes-Related Oxidative Stress. Int. J. Mol. Sci. 2019, 20, 5423. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Sun, Y.; Zhou, Y.; Zhang, Y.; Zhang, T.; Li, Y.; You, W.; Chang, X.; Yuan, L.; Han, X. MicroRNA-24 promotes pancreatic beta cells toward dedifferentiation to avoid endoplasmic reticulum stress-induced apoptosis. J. Mol. Cell Biol. 2019, 11, 747–760. [Google Scholar]
- Taheri, M.; Eghtedarian, R.; Dinger, M.E.; Ghafouri-Fard, S. Emerging roles of non-coding RNAs in the pathogenesis of type 1 diabetes mellitus. Biomed. Pharmacother. 2020, 129, 110509. [Google Scholar]
- Jankauskas, S.S.; Gambardella, J.; Sardu, C.; Lombardi, A.; Santulli, G. Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and Its Complications. Noncoding RNA 2021, 7, 39. [Google Scholar]
- Xue, L.; Xiong, C.; Li, J.; Ren, Y.; Zhang, L.; Jiao, K.; Chen, C.; Ding, P. miR-200-3p suppresses cell proliferation and reduces apoptosis in diabetic retinopathy via blocking the TGF-beta2/Smad pathway. Biosci. Rep. 2020, 40, BSR20201545. [Google Scholar] [CrossRef]
- Erener, S.; Mojibian, M.; Fox, J.K.; Denroche, H.C.; Kieffer, T.J. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology 2013, 154, 603–608. [Google Scholar]
- Coulson, D.J.; Bakhashab, S.; Latief, J.S.; Weaver, J.U. MiR-126, IL-7, CXCR1/2 receptors, inflammation and circulating endothelial progenitor cells: The study on targets for treatment pathways in a model of subclinical cardiovascular disease (type 1 diabetes mellitus). J. Transl. Med. 2021, 19, 140. [Google Scholar] [CrossRef]
- Nizam, R.; Malik, M.Z.; Jacob, S.; Alsmadi, O.; Koistinen, H.A.; Tuomilehto, J.; Alkandari, H.; Al-Mulla, F.; Thanaraj, T.A. Circulating hsa-miR-320a and its regulatory network in type 1 diabetes mellitus. Front. Immunol. 2024, 15, 1376416. [Google Scholar]
- Rasmi, Y.; Mohamed, Y.A.; Alipour, S.; Ahmed, S.; Abdelmajed, S.S. The role of miR-143/miR-145 in the development, diagnosis, and treatment of diabetes. J. Diabetes Metab. Disord. 2024, 23, 39–47. [Google Scholar] [PubMed]
- Swolin-Eide, D.; Forsander, G.; Pundziute Lycka, A.; Novak, D.; Grillari, J.; Diendorfer, A.B.; Hackl, M.; Magnusson, P. Circulating microRNAs in young individuals with long-duration type 1 diabetes in comparison with healthy controls. Sci. Rep. 2023, 13, 11634. [Google Scholar]
- Ding, L.B.; Li, Y.; Liu, G.Y.; Li, T.H.; Li, F.; Guan, J.; Wang, H.J. Long non-coding RNA PVT1, a molecular sponge of miR-26b, is involved in the progression of hyperglycemia-induced collagen degradation in human chondrocytes by targeting CTGF/TGF-beta signal ways. Innate Immun. 2020, 26, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yue, S.; Fang, J.; Zeng, J.; Chen, S.; Tian, J.; Nie, S.; Liu, X.; Ding, H. MicroRNA-10a/b inhibit TGF-beta/Smad-induced renal fibrosis by targeting TGF-beta receptor 1 in diabetic kidney disease. Mol. Ther. Nucleic Acids 2022, 28, 488–499. [Google Scholar]
- Daamouch, S.; Bluher, M.; Vazquez, D.C.; Hackl, M.; Hofbauer, L.C.; Rauner, M. MiR-144-5p and miR-21-5p do not drive bone disease in a mouse model of type 1 diabetes mellitus. JBMR Plus 2024, 8, ziae036. [Google Scholar]
- Douvris, A.; Vinas, J.; Burns, K.D. miRNA-486-5p: Signaling targets and role in non-malignant disease. Cell Mol. Life Sci. 2022, 79, 376. [Google Scholar]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Di Silvestre, D.; Prattichizzo, F.; Mozzillo, E.; Fattorusso, V.; La Sala, L.; Ceriello, A.; Puca, A.A.; et al. Plasma circulating miR-23~27~24 clusters correlate with the immunometabolic derangement and predict C-peptide loss in children with type 1 diabetes. Diabetologia 2020, 63, 2699–2712. [Google Scholar]
- Otmani, K.; Rouas, R.; Lagneaux, L.; Krayem, M.; Duvillier, H.; Berehab, M.; Lewalle, P. Acute myeloid leukemia-derived exosomes deliver miR-24-3p to hinder the T-cell immune response through DENN/MADD targeting in the NF-kappaB signaling pathways. Cell Commun. Signal 2023, 21, 253. [Google Scholar]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.L.; Coulson, D.J.; Yeoh, M.L.Y.; Tamara, A.; Latief, J.S.; Bakhashab, S.; Weaver, J.U. The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. Int. J. Mol. Sci. 2020, 21, 7217. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Yang, C.Y.; Rui, Z.L. MicroRNA-125b-5p improves pancreatic beta-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci. 2019, 224, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yan, J.; Xu, H.; Zhu, Y.; Liang, H.; Pan, W.; Yao, B.; Han, X.; Ye, J.; Weng, J. Two Novel MicroRNA Biomarkers Related to beta-Cell Damage and Their Potential Values for Early Diagnosis of Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2018, 103, 1320–1329. [Google Scholar] [CrossRef]
- Bagge, A.; Dahmcke, C.M.; Dalgaard, L.T. Syntaxin-1a is a direct target of miR-29a in insulin-producing beta-cells. Horm. Metab. Res. 2013, 45, 463–466. [Google Scholar]
- Jia, X.; Yu, L. Understanding Islet Autoantibodies in Prediction of Type 1 Diabetes. J. Endocr. Soc. 2023, 8, bvad160. [Google Scholar] [CrossRef]
- Arvan, P.; Pietropaolo, M.; Ostrov, D.; Rhodes, C.J. Islet autoantigens: Structure, function, localization, and regulation. Cold Spring Harb. Perspect. Med. 2012, 2, a007658. [Google Scholar] [CrossRef]
- Piganelli, J.D.; Mamula, M.J.; James, E.A. The Role of beta Cell Stress and Neo-Epitopes in the Immunopathology of Type 1 Diabetes. Front. Endocrinol. 2020, 11, 624590. [Google Scholar]
- Dwyer, A.J.; Ritz, J.M.; Mitchell, J.S.; Martinov, T.; Alkhatib, M.; Silva, N.; Tucker, C.G.; Fife, B.T. Enhanced CD4(+) and CD8(+) T cell infiltrate within convex hull defined pancreatic islet borders as autoimmune diabetes progresses. Sci. Rep. 2021, 11, 17142. [Google Scholar] [CrossRef]
- Snowhite, I.V.; Allende, G.; Sosenko, J.; Pastori, R.L.; Messinger Cayetano, S.; Pugliese, A. Association of serum microRNAs with islet autoimmunity, disease progression and metabolic impairment in relatives at risk of type 1 diabetes. Diabetologia 2017, 60, 1409–1422. [Google Scholar] [CrossRef]
- Atkinson, M.A.; Mirmira, R.G. The pathogenic “symphony” in type 1 diabetes: A disorder of the immune system, beta cells, and exocrine pancreas. Cell Metab. 2023, 35, 1500–1518. [Google Scholar] [PubMed]
- Clark, M.; Kroger, C.J.; Tisch, R.M. Type 1 Diabetes: A Chronic Anti-Self-Inflammatory Response. Front. Immunol. 2017, 8, 1898. [Google Scholar]
- Martino, M.; Galderisi, A.; Evans-Molina, C.; Dayan, C. Revisiting the Pattern of Loss of beta-Cell Function in Preclinical Type 1 Diabetes. Diabetes 2024, 73, 1769–1779. [Google Scholar] [PubMed]
- Burrack, A.L.; Martinov, T.; Fife, B.T. T Cell-Mediated Beta Cell Destruction: Autoimmunity and Alloimmunity in the Context of Type 1 Diabetes. Front. Endocrinol. 2017, 8, 343. [Google Scholar]
- Belgardt, B.F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015, 21, 619–627. [Google Scholar]
- Fayyad-Kazan, H.; Rouas, R.; Fayyad-Kazan, M.; Badran, R.; El Zein, N.; Lewalle, P.; Najar, M.; Hamade, E.; Jebbawi, F.; Merimi, M.; et al. MicroRNA profile of circulating CD4-positive regulatory T cells in human adults and impact of differentially expressed microRNAs on expression of two genes essential to their function. J. Biol. Chem. 2012, 287, 9910–9922. [Google Scholar]
- Wallberg, M.; Recino, A.; Phillips, J.; Howie, D.; Vienne, M.; Paluch, C.; Azuma, M.; Wong, F.S.; Waldmann, H.; Cooke, A. Anti-CD3 treatment up-regulates programmed cell death protein-1 expression on activated effector T cells and severely impairs their inflammatory capacity. Immunology 2017, 151, 248–260. [Google Scholar] [CrossRef]
- Benson, R.A.; Garcon, F.; Recino, A.; Ferdinand, J.R.; Clatworthy, M.R.; Waldmann, H.; Brewer, J.M.; Okkenhaug, K.; Cooke, A.; Garside, P.; et al. Non-Invasive Multiphoton Imaging of Islets Transplanted Into the Pinna of the NOD Mouse Ear Reveals the Immediate Effect of Anti-CD3 Treatment in Autoimmune Diabetes. Front. Immunol. 2018, 9, 1006. [Google Scholar] [CrossRef]
- Cao, Z.H.; Wu, Z.; Hu, C.; Zhang, M.; Wang, W.Z.; Hu, X.B. Endoplasmic reticulum stress and destruction of pancreatic beta cells in type 1 diabetes. Chin. Med. J. 2020, 133, 68–73. [Google Scholar] [CrossRef]
- Delmastro, M.M.; Piganelli, J.D. Oxidative stress and redox modulation potential in type 1 diabetes. Clin. Dev. Immunol. 2011, 2011, 593863. [Google Scholar]
- Padgett, L.E.; Broniowska, K.A.; Hansen, P.A.; Corbett, J.A.; Tse, H.M. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann. N. Y Acad. Sci. 2013, 1281, 16–35. [Google Scholar] [PubMed]
- Leenders, F.; Groen, N.; de Graaf, N.; Engelse, M.A.; Rabelink, T.J.; de Koning, E.J.P.; Carlotti, F. Oxidative Stress Leads to beta-Cell Dysfunction Through Loss of beta-Cell Identity. Front. Immunol. 2021, 12, 690379. [Google Scholar]
- Wei, J.; Zhang, Y.; Luo, Y.; Wang, Z.; Bi, S.; Song, D.; Dai, Y.; Wang, T.; Qiu, L.; Wen, L.; et al. Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1-Nrf2, Tgfbeta1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic. Biol. Med. 2014, 67, 91–102. [Google Scholar] [PubMed]
- Ghaffari, M.; Razi, S.; Zalpoor, H.; Nabi-Afjadi, M.; Mohebichamkhorami, F.; Zali, H. Association of MicroRNA-146a with Type 1 and 2 Diabetes and their Related Complications. J. Diabetes Res. 2023, 2023, 2587104. [Google Scholar]
- Velosa, A.P.; Teodoro, W.R.; dos Anjos, D.M.; Konno, R.; Oliveira, C.C.; Katayama, M.L.; Parra, E.R.; Capelozzi, V.L.; Yoshinari, N.H. Collagen V-induced nasal tolerance downregulates pulmonary collagen mRNA gene and TGF-beta expression in experimental systemic sclerosis. Respir. Res. 2010, 11, 1. [Google Scholar]
- Salas-Perez, F.; Codner, E.; Valencia, E.; Pizarro, C.; Carrasco, E.; Perez-Bravo, F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013, 218, 733–737. [Google Scholar]
- Eizirik, D.L.; Mandrup-Poulsen, T. A choice of death--the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia 2001, 44, 2115–2133. [Google Scholar]
- Tomita, T. Apoptosis of pancreatic beta-cells in Type 1 diabetes. Bosn. J. Basic. Med. Sci. 2017, 17, 183–193. [Google Scholar]
- Gurzov, E.N.; Eizirik, D.L. Bcl-2 proteins in diabetes: Mitochondrial pathways of beta-cell death and dysfunction. Trends Cell Biol. 2011, 21, 424–431. [Google Scholar] [CrossRef]
- Ortis, F.; Naamane, N.; Flamez, D.; Ladriere, L.; Moore, F.; Cunha, D.A.; Colli, M.L.; Thykjaer, T.; Thorsen, K.; Orntoft, T.F.; et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes 2010, 59, 358–374. [Google Scholar]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhao, S. miRNA-16-5p inhibits the apoptosis of high glucose-induced pancreatic beta cells via targeting of CXCL10: Potential biomarkers in type 1 diabetes mellitus. Endokrynol. Pol. 2020, 71, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Roggli, E.; Britan, A.; Gattesco, S.; Lin-Marq, N.; Abderrahmani, A.; Meda, P.; Regazzi, R. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010, 59, 978–986. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, W.; Kang, J.; Wang, X.; Sun, L. Role of the PI3K/AKT signalling pathway in apoptotic cell death in the cerebral cortex of streptozotocin-induced diabetic rats. Exp. Ther. Med. 2017, 13, 2417–2422. [Google Scholar] [CrossRef]
- Elmitwalli, O.; Darwish, R.; Al-Jabery, L.; Algahiny, A.; Roy, S.; Butler, A.E.; Hasan, A.S. The Emerging Role of p21 in Diabetes and Related Metabolic Disorders. Int. J. Mol. Sci. 2024, 25, 13209. [Google Scholar] [CrossRef]
- Singh, R.; Ha, S.E.; Wei, L.; Jin, B.; Zogg, H.; Poudrier, S.M.; Jorgensen, B.G.; Park, C.; Ronkon, C.F.; Bartlett, A.; et al. miR-10b-5p Rescues Diabetes and Gastrointestinal Dysmotility. Gastroenterology 2021, 160, 1662–1678.e18. [Google Scholar] [CrossRef]
- Lin, L.; Mahner, S.; Jeschke, U.; Hester, A. The Distinct Roles of Transcriptional Factor KLF11 in Normal Cell Growth Regulation and Cancer as a Mediator of TGF-beta Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 2928. [Google Scholar] [CrossRef]
- Pezzolesi, M.G.; Satake, E.; McDonnell, K.P.; Major, M.; Smiles, A.M.; Krolewski, A.S. Circulating TGF-beta1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 2015, 64, 3285–3293. [Google Scholar] [CrossRef]
- Pooja Rathan, V.; Bhuvaneshwari, K.; Nideesh Adit, G.; Kavyashree, S.; Thulasi, N.; Geetha, A.V.S.; Milan, K.L.; Ramkumar, K.M. Therapeutic potential of SMAD7 targeting miRNA in the pathogenesis of diabetic nephropathy. Arch. Biochem. Biophys. 2024, 764, 110265. [Google Scholar] [CrossRef]
- Qin, W.; Shi, Y.; Zhao, B.; Yao, C.; Jin, L.; Ma, J.; Jin, Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS ONE 2010, 5, e9429. [Google Scholar] [CrossRef]
- Chan, M.C.; Hilyard, A.C.; Wu, C.; Davis, B.N.; Hill, N.S.; Lal, A.; Lieberman, J.; Lagna, G.; Hata, A. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2010, 29, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Donath, M.Y.; Storling, J.; Maedler, K.; Mandrup-Poulsen, T. Inflammatory mediators and islet beta-cell failure: A link between type 1 and type 2 diabetes. J. Mol. Med. 2003, 81, 455–470. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Hu, Z.; Dong, J.; Wang, L.E.; Ma, H.; Liu, J.; Zhao, Y.; Tang, J.; Chen, X.; Dai, J.; Wei, Q.; et al. Serum microRNA profiling and breast cancer risk: The use of miR-484/191 as endogenous controls. Carcinogenesis 2012, 33, 828–834. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef]
- Li, L.M.; Hu, Z.B.; Zhou, Z.X.; Chen, X.; Liu, F.Y.; Zhang, J.F.; Shen, H.B.; Zhang, C.Y.; Zen, K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010, 70, 9798–9807. [Google Scholar] [CrossRef]
- Razumilava, N.; Bronk, S.F.; Smoot, R.L.; Fingas, C.D.; Werneburg, N.W.; Roberts, L.R.; Mott, J.L. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology 2012, 55, 465–475. [Google Scholar] [CrossRef]
- Zhang, H.; Zuo, Z.; Lu, X.; Wang, L.; Wang, H.; Zhu, Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 2012, 27, 594–598. [Google Scholar]
- Gonzalez-Martin, A.; Adams, B.D.; Lai, M.; Shepherd, J.; Salvador-Bernaldez, M.; Salvador, J.M.; Lu, J.; Nemazee, D.; Xiao, C. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat. Immunol. 2016, 17, 433–440. [Google Scholar] [CrossRef]
- Steele, C.; Hagopian, W.A.; Gitelman, S.; Masharani, U.; Cavaghan, M.; Rother, K.I.; Donaldson, D.; Harlan, D.M.; Bluestone, J.; Herold, K.C. Insulin secretion in type 1 diabetes. Diabetes 2004, 53, 426–433. [Google Scholar] [CrossRef]
- Cantley, J.; Ashcroft, F.M. Q&A: Insulin secretion and type 2 diabetes: Why do beta-cells fail? BMC Biol. 2015, 13, 33. [Google Scholar]
- Ha, S.E.; Singh, R.; Jin, B.; Baek, G.; Jorgensen, B.G.; Zogg, H.; Debnath, S.; Park, H.S.; Cho, H.; Watkins, C.M.; et al. miR-10a/b-5p-NCOR2 Regulates Insulin-Resistant Diabetes in Female Mice. Int. J. Mol. Sci. 2024, 25, 10147. [Google Scholar] [CrossRef]
- Barutta, F.; Corbetta, B.; Bellini, S.; Guarrera, S.; Matullo, G.; Scandella, M.; Schalkwijk, C.; Stehouwer, C.D.; Chaturvedi, N.; Soedamah-Muthu, S.S.; et al. MicroRNA 146a is associated with diabetic complications in type 1 diabetic patients from the EURODIAB PCS. J. Transl. Med. 2021, 19, 475. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [PubMed]
- Ellenrieder, V.; Buck, A.; Harth, A.; Jungert, K.; Buchholz, M.; Adler, G.; Urrutia, R.; Gress, T.M. KLF11 mediates a critical mechanism in TGF-beta signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology 2004, 127, 607–620. [Google Scholar]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar]
- Erener, S.; Ellis, C.E.; Ramzy, A.; Glavas, M.M.; O’Dwyer, S.; Pereira, S.; Wang, T.; Pang, J.; Bruin, J.E.; Riedel, M.J.; et al. Deletion of pancreas-specific miR-216a reduces beta-cell mass and inhibits pancreatic cancer progression in mice. Cell Rep. Med. 2021, 2, 100434. [Google Scholar]
- Wang, P.; Liu, Q.; Zhao, H.; Bishop, J.O.; Zhou, G.; Olson, L.K.; Moore, A. miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci. Rep. 2020, 10, 5302. [Google Scholar]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar]
- Tsukita, S.; Yamada, T.; Takahashi, K.; Munakata, Y.; Hosaka, S.; Takahashi, H.; Gao, J.; Shirai, Y.; Kodama, S.; Asai, Y.; et al. MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic beta-Cell Proliferation. EBioMedicine 2017, 15, 163–172. [Google Scholar]
- Scherm, M.G.; Serr, I.; Zahm, A.M.; Schug, J.; Bellusci, S.; Manfredini, R.; Salb, V.K.; Gerlach, K.; Weigmann, B.; Ziegler, A.G.; et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat. Commun. 2019, 10, 5697. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.; Kirchner, M.; Aharoni, R.; Ciolli Mattioli, C.; van den Bruck, D.; Gutkovitch, N.; Modepalli, V.; Selbach, M.; Moran, Y.; Chekulaeva, M. Conservation of miRNA-mediated silencing mechanisms across 600 million years of animal evolution. Nucleic Acids Res. 2017, 45, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Weber, M.J. New human and mouse microRNA genes found by homology search. FEBS J. 2005, 272, 59–73. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Gottmann, P.; Ouni, M.; Zellner, L.; Jahnert, M.; Rittig, K.; Walther, D.; Schurmann, A. Polymorphisms in miRNA binding sites involved in metabolic diseases in mice and humans. Sci. Rep. 2020, 10, 7202. [Google Scholar] [CrossRef]
- Brillante, S.; Volpe, M.; Indrieri, A. Advances in MicroRNA Therapeutics: From Preclinical to Clinical Studies. Hum. Gene Ther. 2024, 35, 628–648. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.; Ha, S.E.; Singh, R.; Kim, D.; Ro, S. microRNAs in Type 1 Diabetes: Roles, Pathological Mechanisms, and Therapeutic Potential. Int. J. Mol. Sci. 2025, 26, 3301. https://doi.org/10.3390/ijms26073301
Cho H, Ha SE, Singh R, Kim D, Ro S. microRNAs in Type 1 Diabetes: Roles, Pathological Mechanisms, and Therapeutic Potential. International Journal of Molecular Sciences. 2025; 26(7):3301. https://doi.org/10.3390/ijms26073301
Chicago/Turabian StyleCho, Hayeong, Se Eun Ha, Rajan Singh, David Kim, and Seungil Ro. 2025. "microRNAs in Type 1 Diabetes: Roles, Pathological Mechanisms, and Therapeutic Potential" International Journal of Molecular Sciences 26, no. 7: 3301. https://doi.org/10.3390/ijms26073301
APA StyleCho, H., Ha, S. E., Singh, R., Kim, D., & Ro, S. (2025). microRNAs in Type 1 Diabetes: Roles, Pathological Mechanisms, and Therapeutic Potential. International Journal of Molecular Sciences, 26(7), 3301. https://doi.org/10.3390/ijms26073301







