Genome-Wide Identification and Characterization of the Growth-Regulating Factor Gene Family Responsive to Abiotic Stresses and Phytohormone Treatments in Populus ussuriensis
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Discovery of GRFs in P. ussuriensis
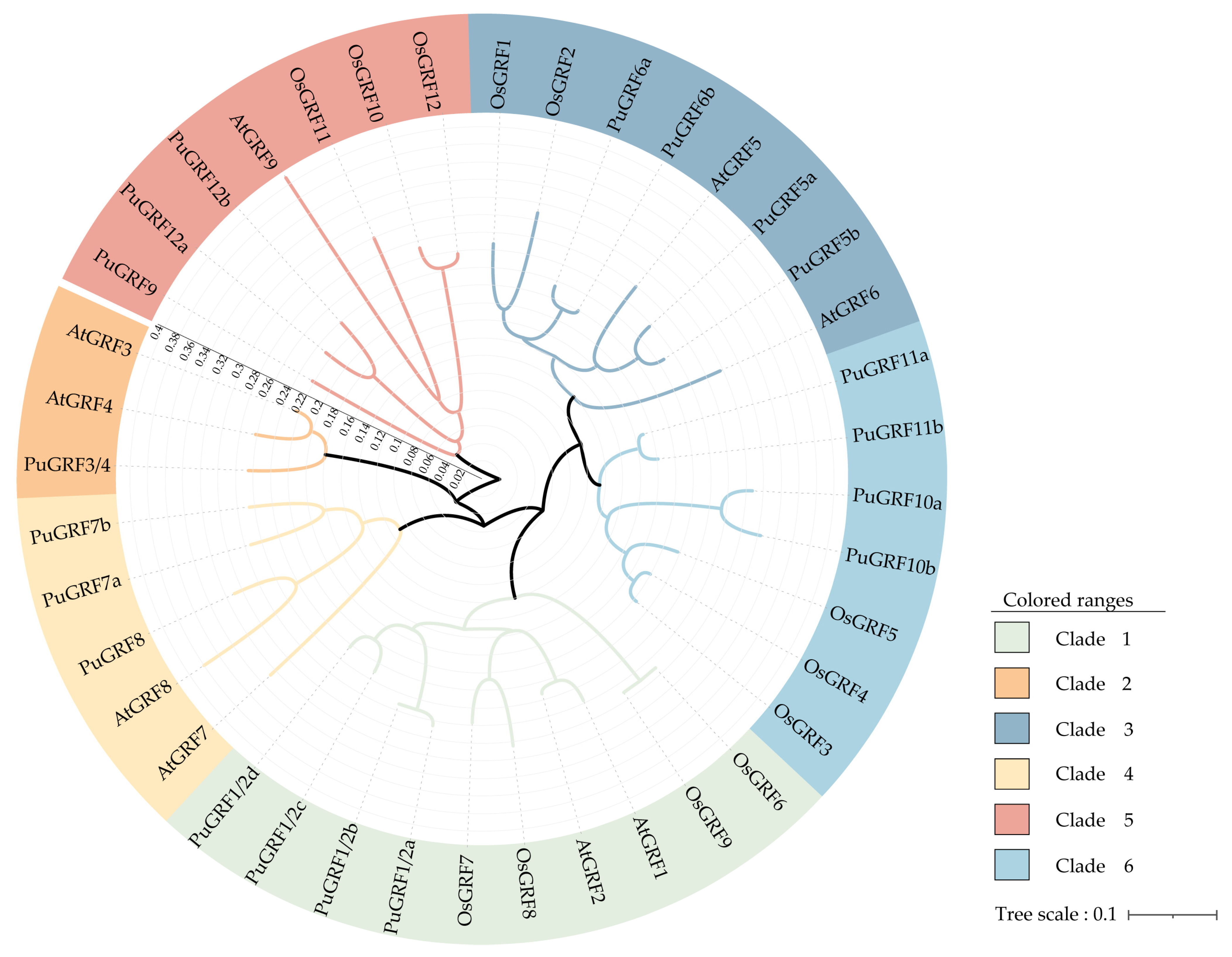
2.2. Protein Evolution of PuGRFs
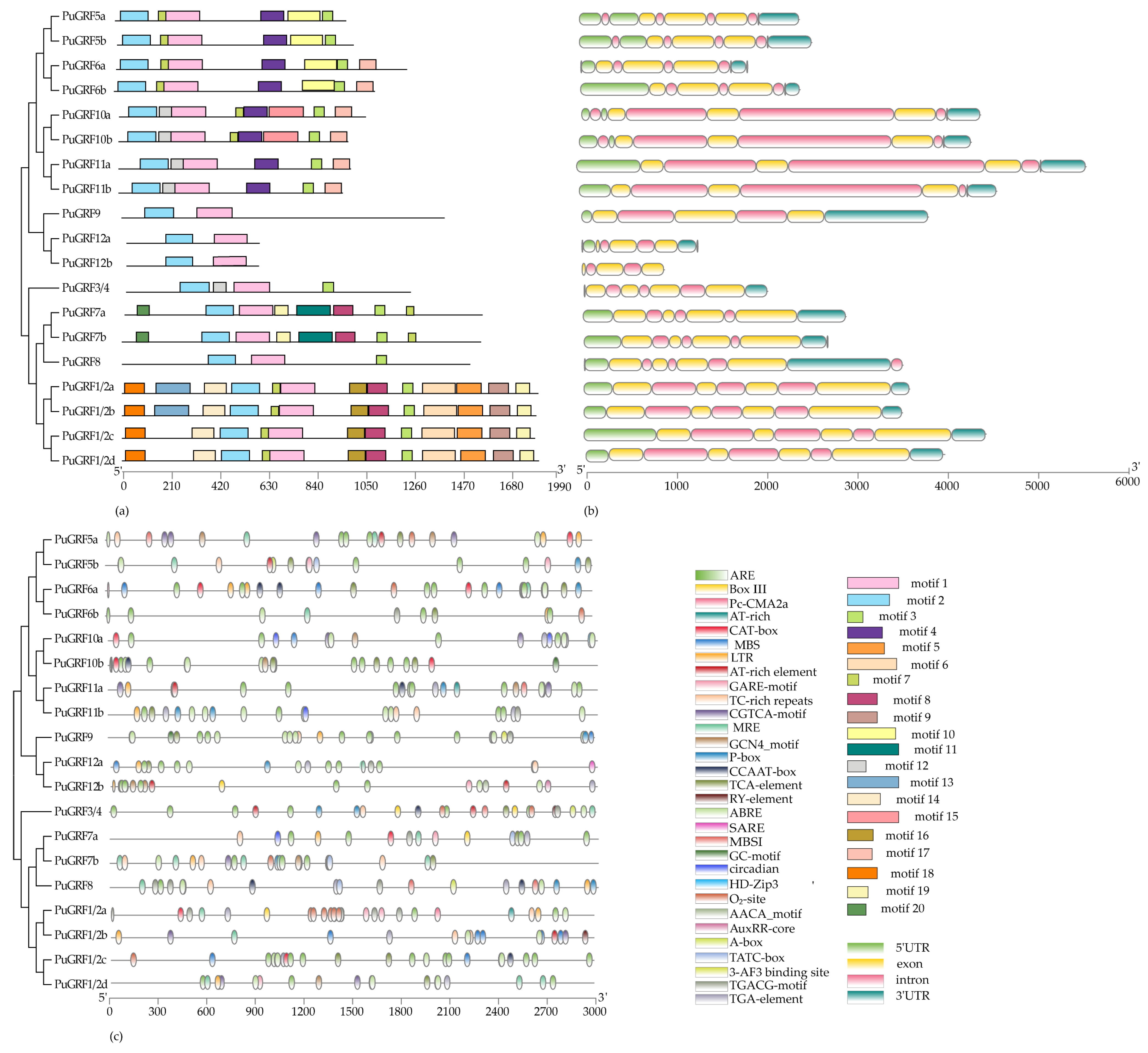
2.3. The Protein Motif, Structure, and Cis-Element Analysis of PuGRFs
2.4. Expression Pattern of PuGRFs
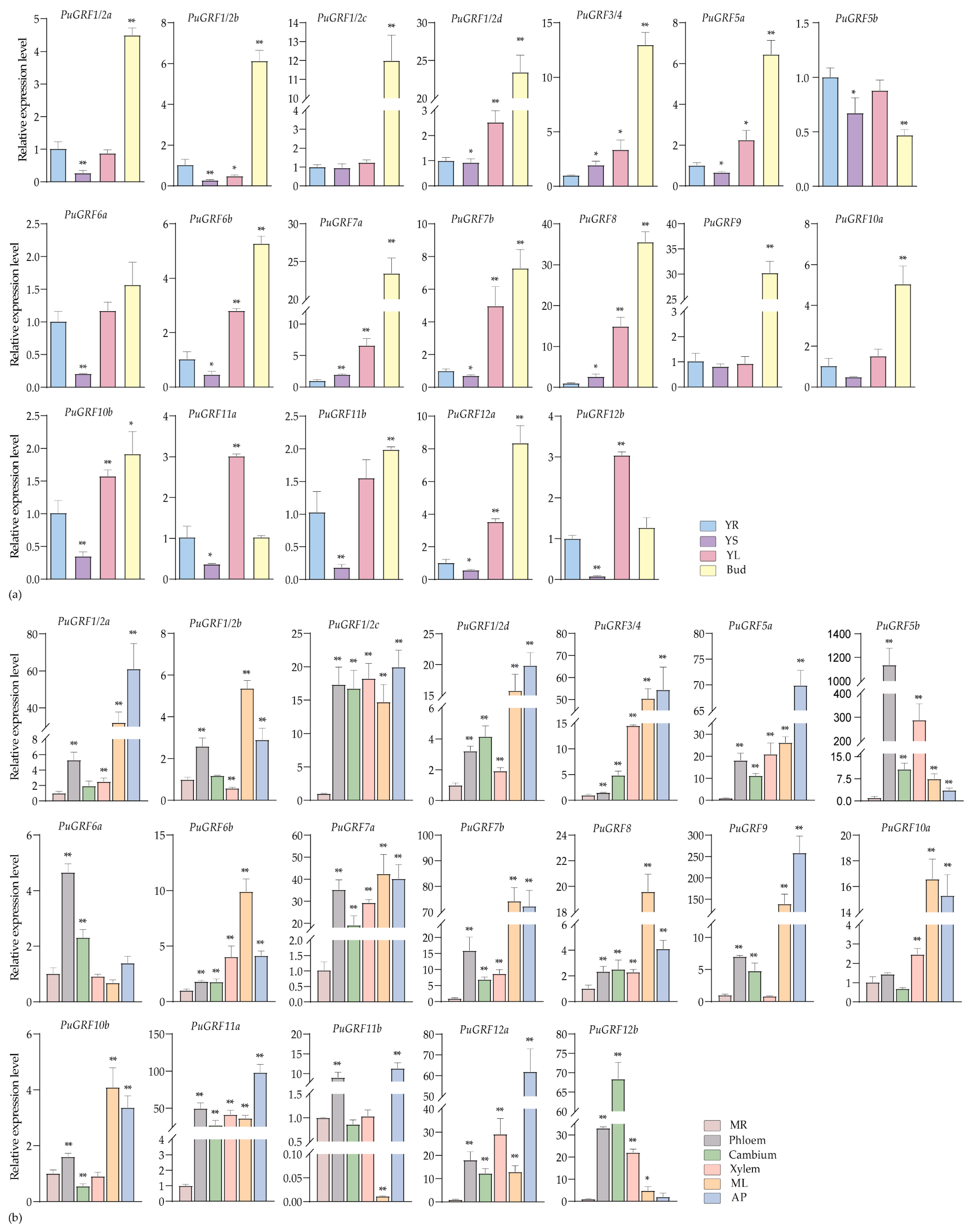
2.4.1. Expression Analysis of PuGRFs During Growth and Development of P. ussuriensis
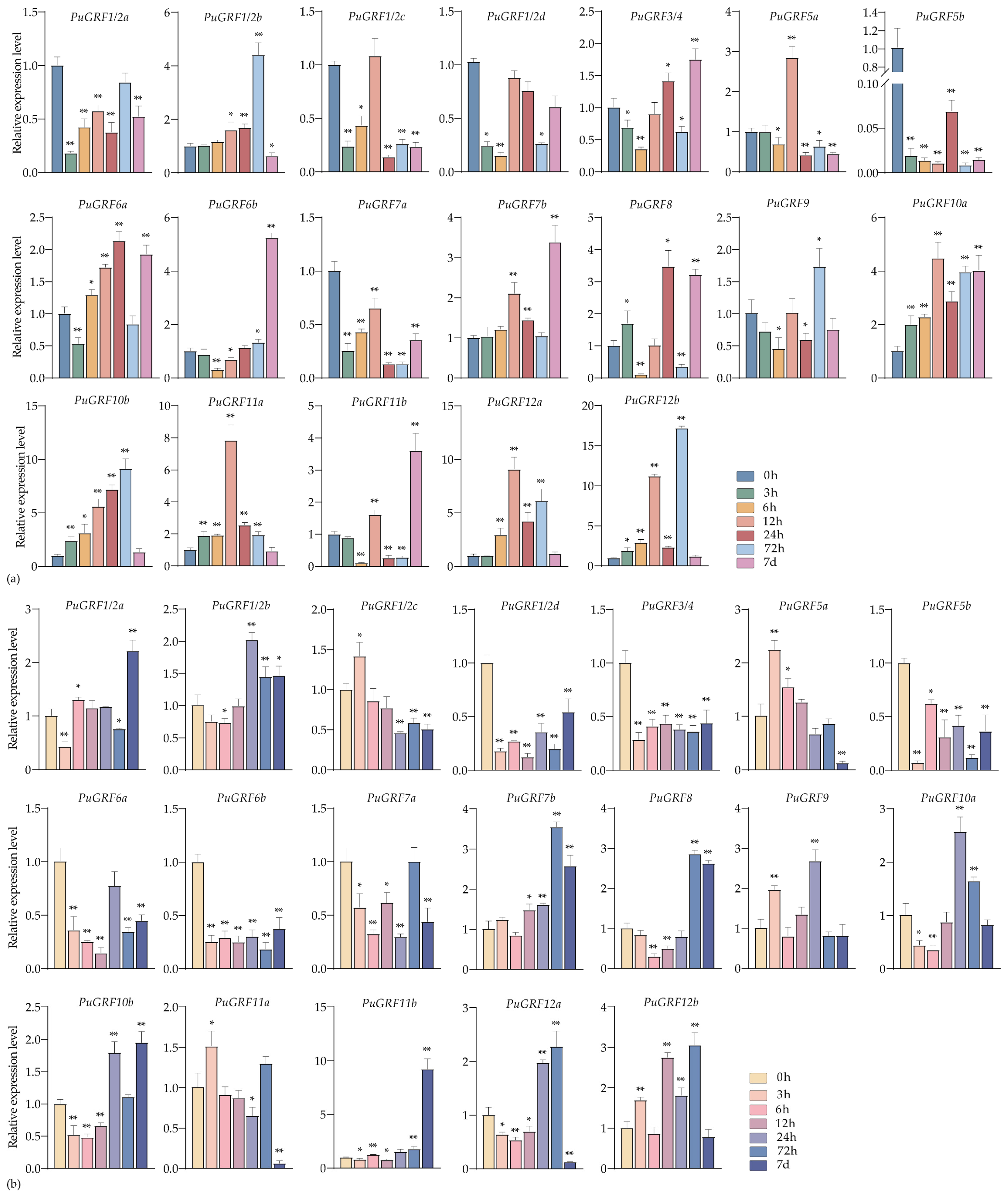
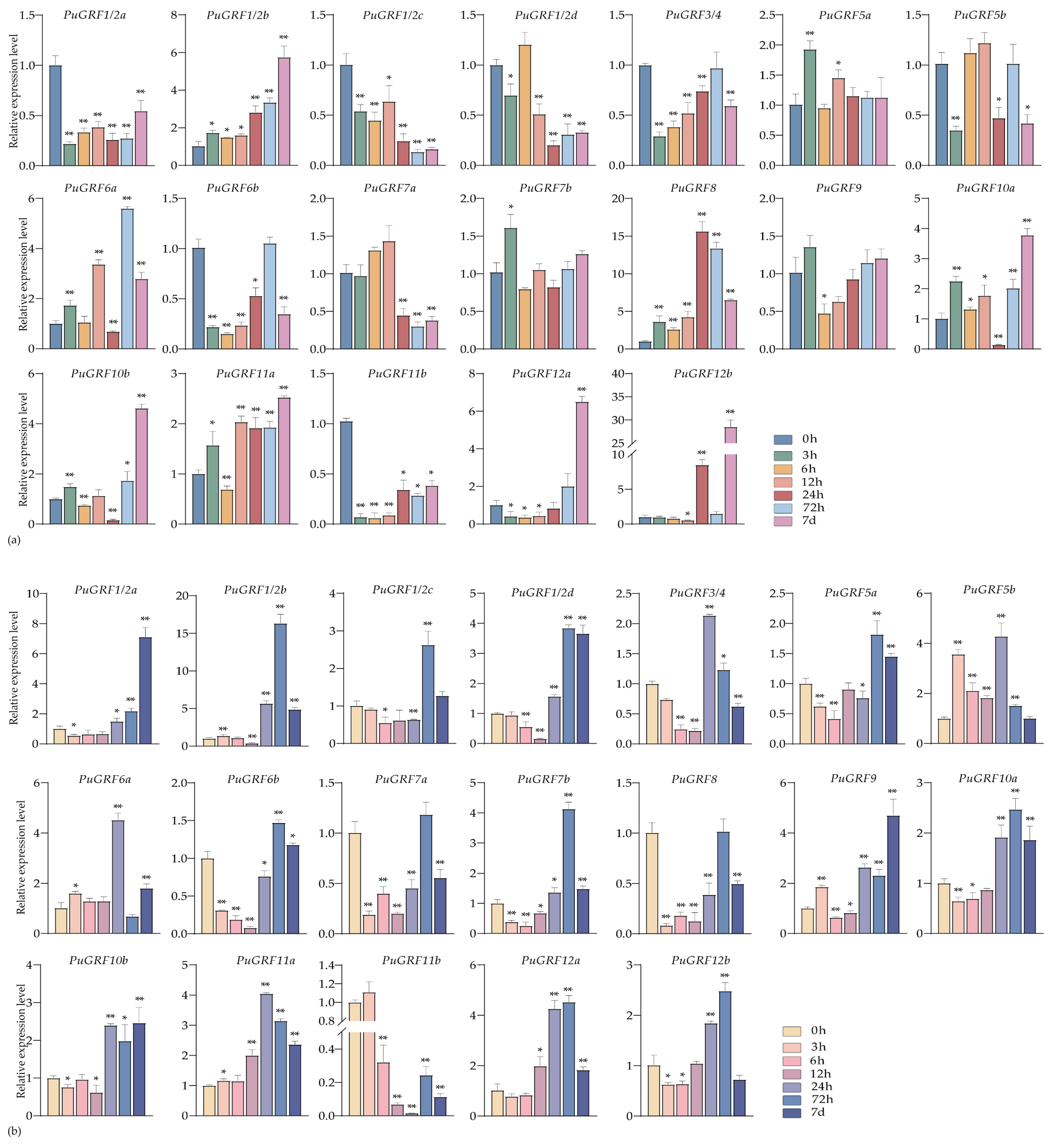
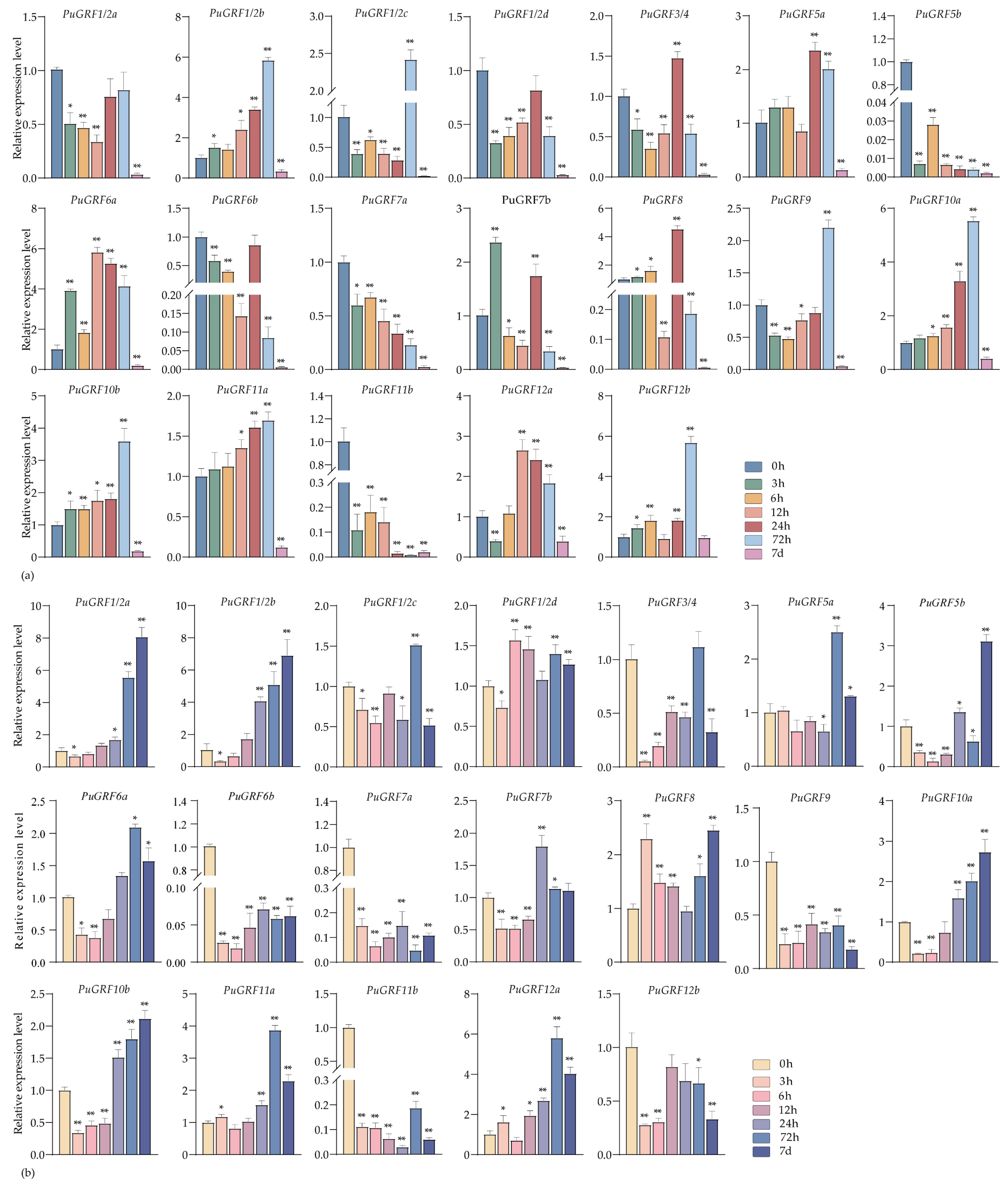
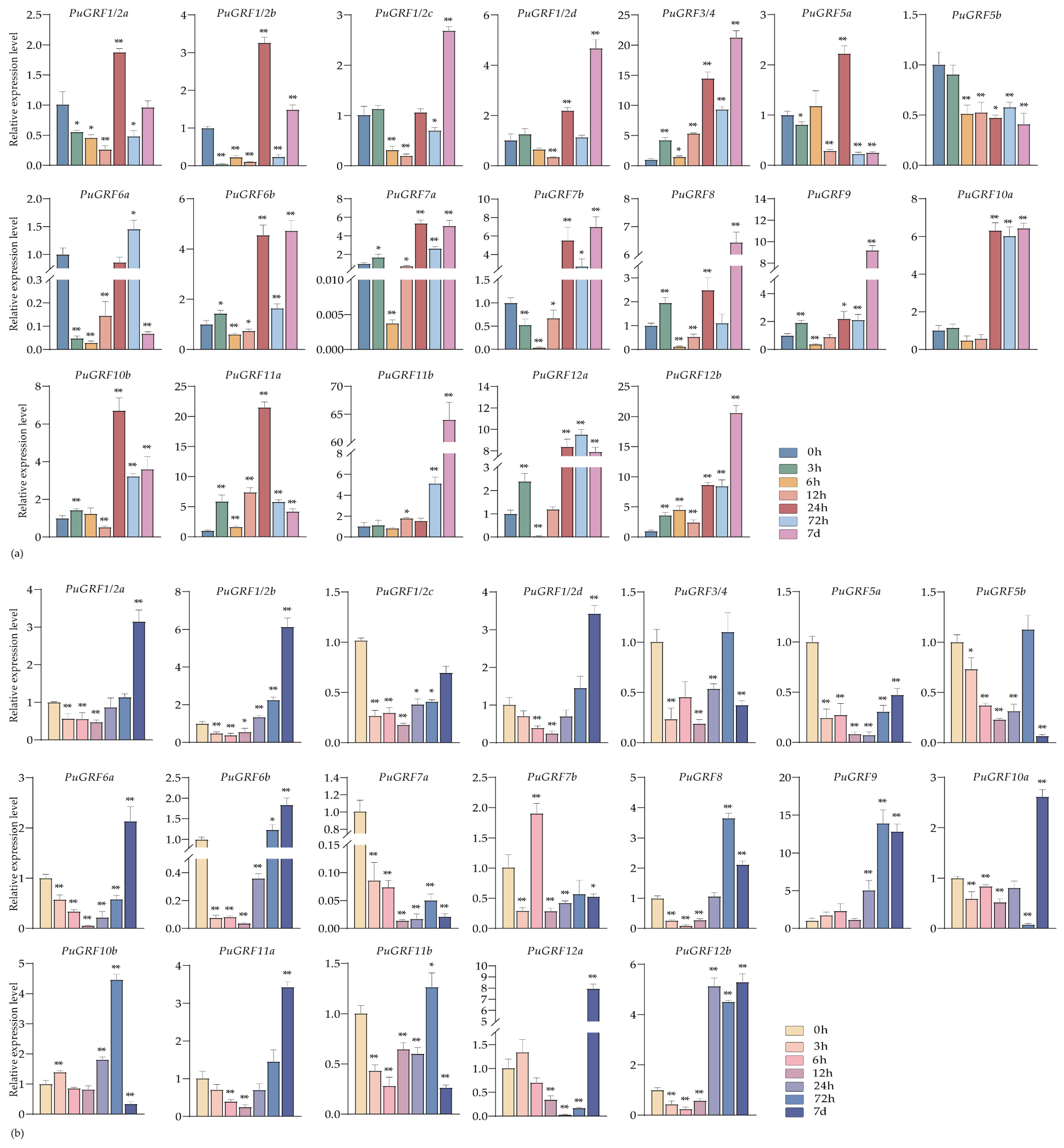
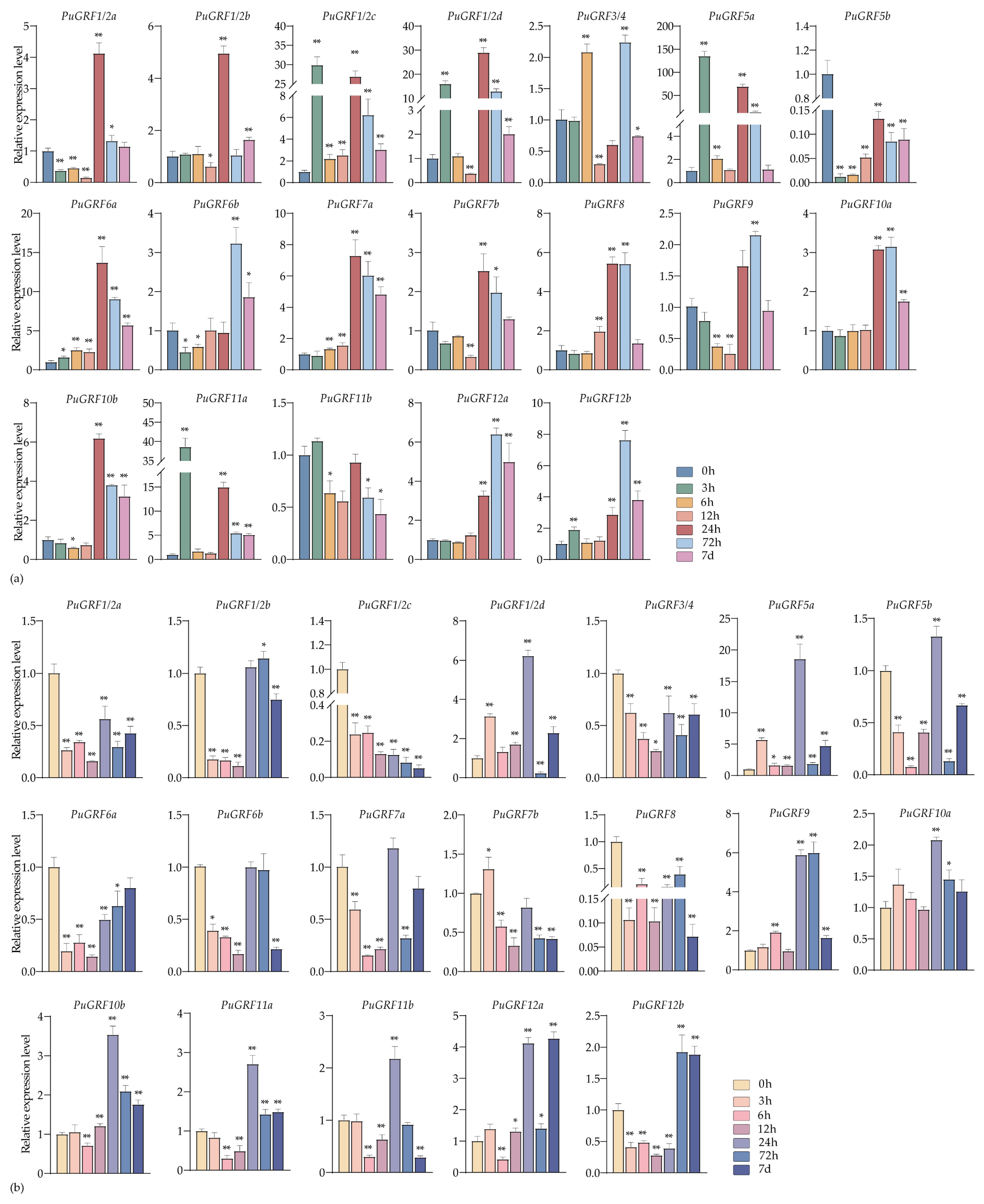
2.4.2. Expression Analysis of PuGRFs Under Salt and Osmotic Stresses
2.4.3. Expression Analysis of PuGRFs Under ABA, MeJA and SA Treatment
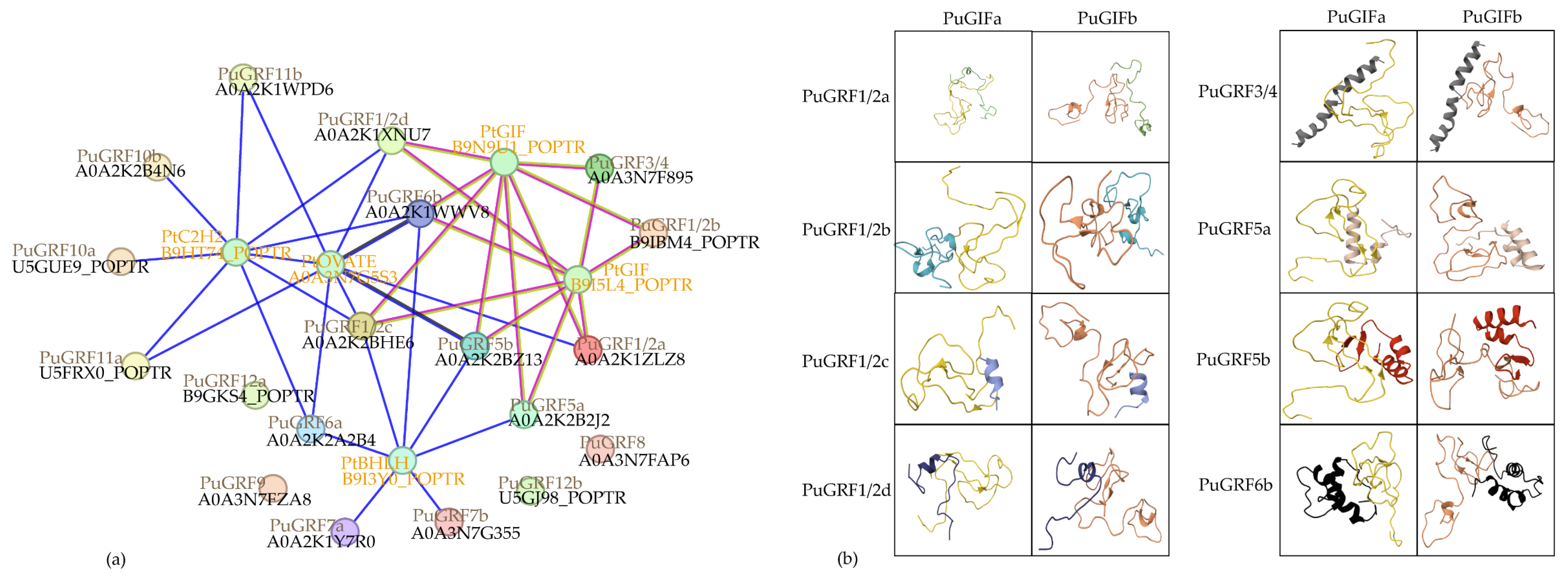
2.5. Predict and Analyze the Interacting Proteins of PuGRFs
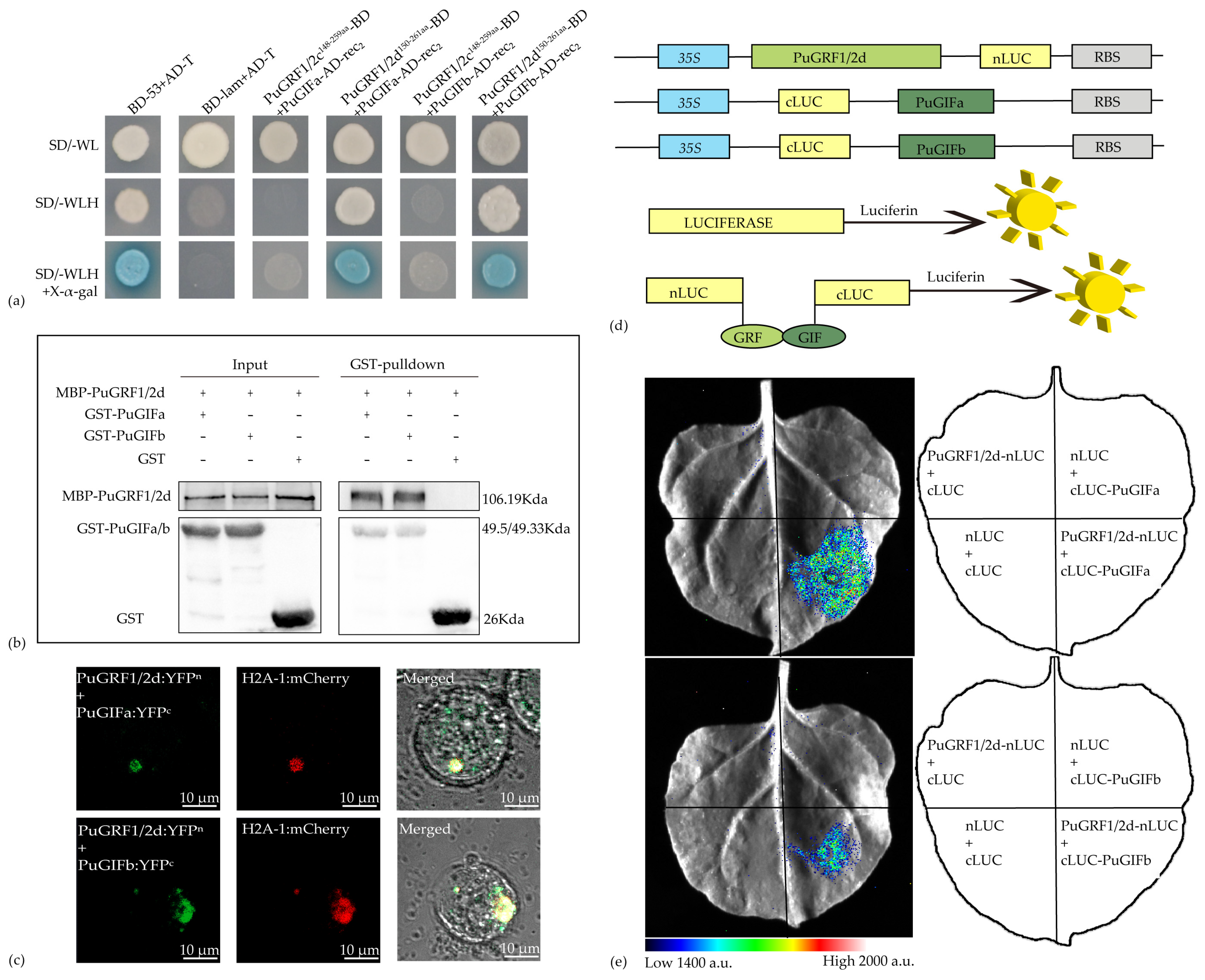
2.6. Verification of the Interaction Relationship Between PuGRFs and PuGIFa/b
3. Discussion
4. Materials and Methods
4.1. Identifcation of PuGRFs in P. ussuriensis
4.2. Phylogenetic Investigation of the PuGRFs
4.3. Analysis of Conserved Motifs, Gene Structure, and Promoter Regions in PuGRFs
4.4. Plant Materials and Treatments
4.5. RNA Isolation and RT-qPCR Analysis
4.6. Protein-Protein Interaction Studies of PuGRFs
4.7. Y2H Assays
4.8. BiFC
4.9. LCI Assays
4.10. In Vitro Pull-Down Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kooke, R.; Keurentjes, J.J.B. Multi-dimensional regulation of metabolic networks shaping plant development and performance. J. Exp. Bot. 2011, 63, 3353–3365. [Google Scholar] [CrossRef] [PubMed]
- Van der Knapp, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar]
- Wang, J.; Zhou, H.; Zhao, Y.; Sun, P.; Tang, F.; Song, X.; Lu, M.-Z. Characterization of poplar growth-regulating factors and analysis of their function in leaf size control. BMC Plant Biol. 2020, 20, 509. [Google Scholar] [CrossRef]
- Khisti, M.; Avuthu, T.; Yogendra, K.; Valluri, V.K.; Kudapa, H.; Reddy, P.S.; Tyagi, W. Genome-wide identification and expression profiling of growth-regulating factor (GRF) and GRF-interacting factor (GIF) gene families in chickpea and pigeonpea. Sci. Rep. 2024, 14, 17178. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cheng, W.; Li, S.; Li, Y.; Wang, X.; Xie, J.; He, Y.; Wang, Y.; Niu, Y.; Bao, X.; et al. Genome-wide identification and expression analysis of the growth regulating factor (GRF) family in Jatropha curcas. PLoS ONE 2021, 16, e0254711. [Google Scholar]
- Fu, M.-K.; He, Y.-N.; Yang, X.-Y.; Tang, X.; Wang, M.; Dai, W.-S. Genome-wide identification of the GRF family in sweet orange (Citrus sinensis) and functional analysis of the CsGRF04 in response to multiple abiotic stresses. BMC Genom. 2024, 25, 37. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef]
- Kim, J.H. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 2019, 52, 227–238. [Google Scholar] [CrossRef]
- Lee, B.H.; Ko, J.-H.; Lee, S.; Lee, Y.; Pak, J.-H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR Gene Family Performs an Overlapping Function in Determining Organ Size as Well as Multiple Developmental Properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef]
- Hoe, J.K.; Tsukaya, H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar]
- Cheng, Z.; Wen, S.; Wu, Y.; Shang, L.; Wu, L.; Lyu, D.; Yu, H.; Wang, J.; Jian, H. Comparatively Evolution and Expression Analysis of GRF Transcription Factor Genes in Seven Plant Species. Plants 2023, 12, 2790. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xie, Q.; Yan, J.; Chen, J.; Chen, Q. Genome-Wide Identification and Characterization of the Abiotic-Stress-Responsive GRF Gene Family in Diploid Woodland Strawberry (Fragaria vesca). Plants 2021, 10, 1916. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.-F.; Huang, J.-Q.; Liu, X.; Huang, C.-C.; Zheng, Z.-S.; Zhang, X.-F.; Shangguan, X.-X.; Wang, L.-J.; Zhang, Y.-G.; Wendel, J.F.; et al. Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genom. 2020, 21, 575. [Google Scholar] [CrossRef]
- Horiguchi, G.; Kim, G.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef]
- Jathar, V.; Saini, K.; Chauhan, A.; Rani, R.; Ichihashi, Y.; Ranjan, A. Spatial control of cell division by GA-OsGRF7/8 module in a leaf explaining the leaf length variation between cultivated and wild rice. New Phytol. 2022, 234, 867–883. [Google Scholar]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid-and osmotic stress–responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar]
- Liu, Z.-Y.; Han, Y.-T.; Wang, C.-Y.; Lei, X.-J.; Wang, Y.-Y.; Dong, W.-F.; Xie, Q.-J.; Fu, Y.-J.; Gao, C.-Q. The growth-regulating factor PdbGRF1 positively regulates the salt stress response in Populus davidiana × P. bolleana. Plant Sci. 2022, 326, 111502. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Rodriguez, E.R.; Mecchia, M.A.; Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Zhao, S.; Li, D.; Wei, M.; Li, C. PuHSFA4a Enhances Tolerance To Excess Zinc by Regulating Reactive Oxygen Species Production and Root Development in Populus. Plant Physiol. 2019, 180, 2254–2271. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Liu, Q.; Wang, Z.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Nie, J.; Liu, B.; Lv, K.; et al. PuHox52-mediated hierarchical multilayered gene regulatory network promotes adventitious root formation in Populus ussuriensis. New Phytol. 2020, 228, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Zhang, M.; Sun, J.; Zhao, Y.; Pak, S.; Ma, M.; Chen, Y.; Lu, H.; Yang, J.; Wei, H.; et al. PuHox52 promotes coordinated uptake of nitrate, phosphate, and iron under nitrogen deficiency in Populus ussuriensis. J. Integr. Plant Biol. 2022, 65, 791–809. [Google Scholar]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2021, 34, 718–741. [Google Scholar] [CrossRef]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.-L.; Chen, Y.-P.; Li, G.-B.; Zhu, Y.; Yang, X.-M.; Zhang, L.-L.; Zhao, Z.-X.; et al. miR396-OsGRFs Module Balances Growth and Rice Blast Disease-Resistance. Front. Plant Sci. 2019, 9, 1999. [Google Scholar] [CrossRef]
- Rikiishi, K.; Matsuura, T.; Maekawa, M. TaABF1, ABA response element binding factor 1, is related to seed dormancy and ABA sensitivity in wheat (Triticum aestivum L.) seeds. J. Cereal Sci. 2010, 52, 236–238. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Choi, D.; Kim, J.H.; Kende, H. Whole Genome Analysis of the OsGRF Gene Family Encoding Plant-Specific Putative Transcription Activators in Rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Wang, Q.; Lv, K.; Du, K.; Zhang, W.; Li, Q.; Kang, X.; Wei, H. Growth-regulating factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021, 230, 612–628. [Google Scholar] [CrossRef]
- Beltramino, M.; Ercoli, M.F.; Debernardi, J.M.; Goldy, C.; Rojas, A.M.L.; Nota, F.; Alvarez, M.E.; Vercruyssen, L.; Inzé, D.; Palatnik, J.F.; et al. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar] [CrossRef]
- Zhou, H.; Wei, K.; Jiang, C.; Zhao, J.Q.; Song, X.Q.; Liu, M.Z. Negative negulation of GRF1/2d on the formation and development of adventitious roots in Populus alba × P. glandulosa “84K”. Sci. Silvae Sin. 2017, 53, 33–39. [Google Scholar]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Manuel Debernardi, J.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.-J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Kirolinko, C.; Hobecker, K.; Wen, J.; Mysore, K.S.; Niebel, A.; Blanco, F.A.; Zanetti, M.E. Auxin Response Factor 2 (ARF2), ARF3, and ARF4 Mediate Both Lateral Root and Nitrogen Fixing Nodule Development in Medicago truncatula. Front. Plant Sci. 2021, 12, 659061. [Google Scholar] [CrossRef]
- Poethig, R.S.; Omidbakhshfard, M.A.; Olas, J.J.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar]
- Wang, J.; Zhou, H.; Zhao, Y.; Jiang, C.; Li, J.; Tang, F.; Liu, Y.; Zhao, S.; Hu, J.; Song, X.; et al. PagGRF12a interacts with PagGIF1b to regulate secondary xylem development through modulating PagXND1a expression in Populus alba × P. glandulosa. J. Integr. Plant Biol. 2021, 63, 1683–1694. [Google Scholar]
- Tian, Y.; Zhao, Y.; Sun, Y.; El-Kassaby, Y.A.; Song, G.; Mi, Y.; Han, J.; Li, Y. PagGRF11 Overexpression Promotes Stem Development and Dwarfing in Populus. Int. J. Mol. Sci. 2022, 23, 7858. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef]
- Kim, G.; Ryu, H.; Sung, J. Hormonal Crosstalk and Root Suberization for Drought Stress Tolerance in Plants. Biomolecules 2022, 12, 811. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Lv, Q.; Wang, L.; Han, S.; Wang, J.; Chen, Y.; Zhu, W.; Zhang, X.; Bao, F.; Hu, Y.; et al. Abscisic acid–mediated autoregulation of the MYB41-BRAHMA module enhances drought tolerance in Arabidopsis. Plant Physiol. 2024, 196, 1608–1626. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Yin, Q.; Chen, Y.; Sun, M.; Yuan, X.; Ding, Z.; Kong, X. ERF114/115/109 are essential for jasmonate-repressed non-canonical JAZ8 activity in JA signaling. Cell Rep. 2025, 44, 115222. [Google Scholar] [CrossRef]
- Kim, Y.; Takahashi, S.; Miyao, M. Relationship between reduction in rice (Nipponbare) leaf blade size under elevated CO2 and miR396-GRF module. Plant Signal. Behav. 2022, 17, 2041280. [Google Scholar]
- Pan, W.; Cheng, Z.; Han, Z.; Yang, H.; Zhang, W.; Zhang, H. Efficient transformation and genome editing of watermelon assisted by genes that enco dedevelopmental regulators. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lu, Y.; Zeng, J.; Liu, Q. The Rice miR396-GRF-GIF-SWI/SNF Module: A Player in GA Signaling. Front. Plant Sci. 2022, 12, 786641. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, M.; Zhang, X.; Gu, L.; Li, J.; Ming, F.; Wang, M.; Wang, Z. MIR396-GRF/GIF enhances in planta shoot regeneration of Dendrobium catenatum. BMC Genom. 2024, 25, 543. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.; Wang, S.; Cai, H.; Yang, M.; Dong, Y. Genome-wide molecular evolution analysis of the GRF and GIF gene families in Plantae (Archaeplastida). BMC Genom. 2024, 25, 74. [Google Scholar] [CrossRef]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. miR396a-Mediated Basic Helix–Loop–Helix Transcription Factor bHLH74 Repression Acts as a Regulator for Root Growth in Arabidopsis Seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef]
- Zhang, D.-F.; Li, B.; Jia, G.-Q.; Zhang, T.-F.; Dai, J.-R.; Li, J.-S.; Wang, S.-C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Watanabe, S.; Kojima, K.; Ide, Y.; Sasaki, S. Establishment of a tissue culture system of Populus euphratica Oliv. Bull. Tokyo Univ. For. 1999, 107, 87–92. [Google Scholar]
- Gaur, A.; Kumar, P.; Thakur, A.K.; Srivastava, D.K. REVIEW: In vitro plant regeneration studies and their potential applications in Populus spp.: A review. Isr. J. Plant Sci. 2016, 63, 77–84. [Google Scholar] [CrossRef]
- Wang, H.; Leng, X.; Xu, X.; Li, C. Comprehensive Analysis of the TIFY Gene Family and Its Expression Profiles under Phytohormone Treatment and Abiotic Stresses in Roots of Populus trichocarpa. Forests 2020, 11, 315. [Google Scholar] [CrossRef]
- Wang, H.; Leng, X.; Yang, J.; Zhang, M.; Zeng, M.; Xu, X.; Wang, F.; Li, C. Comprehensive analysis of AHL gene family and their expression under drought stress and ABA treatment in Populus trichocarpa. PeerJ 2021, 9, e10932. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Q.; Wang, H.; Zhang, H.; Xu, X.; Li, C.; Yang, C. Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci. Rep. 2016, 6, 36274. [Google Scholar] [CrossRef]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2011, 9, 173–175. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Li, W.; Chen, H.; Li, Q.; Sun, Y.-H.; Shi, R.; Lin, C.-Y.; Wang, J.P.; Chen, H.-C.; Chuang, L.; et al. A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protoc. 2014, 9, 2194–2205. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.-M. Firefly Luciferase Complementation Imaging Assay for Protein-Protein Interactions in Plants. Plant Physiol. 2007, 146, 323–324. [Google Scholar] [CrossRef]
- Zhang, D.; Tan, W.; Yang, F.; Han, Q.; Deng, X.; Guo, H.; Liu, B.; Yin, Y.; Lin, H. A BIN2-GLK1 Signaling Module Integrates Brassinosteroid and Light Signaling to Repress Chloroplast Development in the Dark. Dev. Cell 2021, 56, 310–324.e7. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID of the Homologous Genes in P. trichocarpa | Full (bp) | Protein Length (aa) | Theoretical (pI) | Molecular Weight (Da) | Predicted Location(s) | Predicted microRNAs (ptc-miR) |
|---|---|---|---|---|---|---|---|
| PuGRF1/2a | Potri.007G007100 | 1833 | 611 | 7.28 | 66,518.92 | Nucleus | 396a-396g, 7822, 159c, 169y, 6439a, 6454, 7835 |
| PuGRF1/2b | Potri.014G007200 | 1824 | 607 | 6.44 | 65,962.14 | Nucleus | 396a-396g, 6421, 7829 |
| PuGRF1/2c | Potri.002G115100 | 1818 | 605 | 7.22 | 65,496.67 | Nucleus | 396a-396g, 169b, 164a-164f, 399d, 6426a-6426b |
| PuGRF1/2d | Potri.014G012800 | 1836 | 611 | 8.61 | 66,179.14 | Nucleus | 396a-396g, 164a-164f, 397a, 6444, 6466, 6473 |
| PuGRF3/4 | Potri.006G115200 | 1203 | 400 | 7.33 | 43,882.40 | Nucleus | 396a-396g, 390a-d, 482d, 7813, 7824 |
| PuGRF5a | Potri.003G065000 | 1023 | 340 | 7.25 | 37,883.77 | Nucleus | 396a-396g, 2111a-2111b, 399a-399d, 399j, 478e, 395b-395k, 475d |
| PuGRF5b | Potri.001G169100 | 1047 | 348 | 7.74 | 39,163.48 | Nucleus | 6426a-6426b, 396a-396g, 399f-399i, 399d, 6444, 159c, 169n, 393a-363c, 481d |
| PuGRF6a | Potri.006G143200 | 1287 | 428 | 8.88 | 48,392.25 | Nucleus | 396a-396g, 476b, 7814, 156k-156l, 395a, 169s, 172a-172f, 390a-390d, 476a, 6442 |
| PuGRF6b | Potri.018G065400 | 1155 | 384 | 8.64 | 43,491.38 | Nucleus | 396a-396g, 156a-156l, 390a-390d, 7814, 156a-156f, 474a-474b, 7817b |
| PuGRF7a | Potri.012G022600 | 1608 | 535 | 7.71 | 58,256.64 | Nucleus | 396a-396g, 172b, 172d-172e, 172g-172i, 475a-475b, 475d, 1444a, 6461, 6475, 7835, 7841 |
| PuGRF7b | Potri.015G006200 | 1614 | 537 | 8.37 | 68,981.47 | Nucleus | 396a-396g, 1444a-1444c, 172a-172i, 6425a-6425e, 6447 |
| PuGRF8 | Potri.001G082700 | 1572 | 523 | 6.96 | 56,896.02 | Nucleus | 396a-396g, 472b, 482d, 390a-390d, 6471 |
| PuGRF9 | Potri.014G071800 | 1377 | 458 | 9.11 | 50,349.58 | Nucleus | 396a-396g, 6470, 6427 |
| PuGRF10a | Potri.001G132600 | 1086 | 361 | 9.3 | 40,379.25 | Nucleus | 396a-396g, 172b, 172g, 6445a-6445b, 6472, 7828 |
| PuGRF10b | Potri.003G100800 | 1011 | 336 | 9.1 | 37,297.52 | Nucleus | 396a-396g, 6445a-6445b, 171l, 7815 |
| PuGRF11a | Potri.013G077500 | 1023 | 340 | 7.72 | 37,402.25 | Nucleus | 396a-396g, 164a-164f, 475d, 7822 |
| PuGRF11b | Potri.019G042300 | 987 | 328 | 8.96 | 36,233.07 | Nucleus | 396a-396g, 390a-390d, 408, 164f, 6423, 7813 |
| PuGRF12a | Potri.001G114000 | 609 | 202 | 10.06 | 22,477.85 | Nucleus | 396a-396g |
| PuGRF12b | Potri.003G118100 | 606 | 201 | 9.94 | 22,313.44 | Mitochondrion; Nucleus | 396a-396g, 6480 |
| Name | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dock | Confidence | Dock | Confidence | Dock | Confidence | Dock | Confidence | Dock | Confidence | ||
| PuGRF1/2a | PuGIFa | −171.37 | 0.6053 | −161.52 | 0.5573 | −160.81 | 0.5538 | −157 | 0.5349 | −154.84 | 0.5242 |
| PuGIFb | −185.91 | 0.6722 | −164.85 | 0.5737 | −160.97 | 0.5546 | −160.28 | 0.5512 | −154.92 | 0.5246 | |
| PuGRF1/2b | PuGIFa | −235.84 | 0.8477 | −234.64 | 0.8446 | −209.29 | 0.766 | −193.59 | 0.7051 | −190.97 | 0.6941 |
| PuGIFb | −203.77 | 0.7456 | −195.5 | 0.713 | −190.32 | 0.6913 | −183.81 | 0.6629 | −181.4 | 0.652 | |
| PuGRF1/2c | PuGIFa | −144.31 | 0.4716 | −138.13 | 0.4409 | −136.26 | 0.4317 | −129.22 | 0.3976 | −127.87 | 0.3911 |
| PuGIFb | −145.95 | 0.4798 | −134.19 | 0.4216 | −131.63 | 0.4092 | −130.46 | 0.4035 | −128.24 | 0.3929 | |
| PuGRF1/2d | PuGIFa | −190.73 | 0.6931 | −177.09 | 0.6322 | −171.51 | 0.6059 | −170.98 | 0.6034 | −170.94 | 0.6032 |
| PuGIFb | −184.81 | 0.6673 | −164.6 | 0.5725 | −159.5 | 0.5474 | −157.81 | 0.539 | −156.01 | 0.53 | |
| PuGRF3/4 | PuGIFa | −196.47 | 0.717 | −194.87 | 0.7104 | −187.22 | 0.678 | −185.67 | 0.6712 | −184.51 | 0.666 |
| PuGIFb | −191.07 | 0.6945 | −185.84 | 0.6719 | −180.97 | 0.6501 | −179.17 | 0.6418 | −166.01 | 0.5794 | |
| PuGRF5a | PuGIFa | −189.6 | 0.6886 | −185.4 | 0.67 | −173.42 | 0.615 | −170.67 | 0.6019 | −170.67 | 0.6019 |
| PuGIFb | −202.75 | 0.7417 | −189.25 | 0.6868 | −179.49 | 0.6433 | −178.57 | 0.6391 | −177.39 | 0.6336 | |
| PuGRF5b | PuGIFa | −176.72 | 0.6305 | −174.26 | 0.619 | −167.51 | 0.5867 | −165.87 | 0.5787 | −165.76 | 0.5782 |
| PuGIFb | −190.4 | 0.6917 | −177.21 | 0.6328 | −176.9 | 0.6313 | −176.57 | 0.6298 | −176.51 | 0.6295 | |
| PuGRF6b | PuGIFa | −248.48 | 0.8776 | −218.49 | 0.7973 | −212.27 | 0.7765 | −209.91 | 0.7682 | −204.96 | 0.7501 |
| PuGIFb | −203.15 | 0.7433 | −202.32 | 0.7401 | −200.22 | 0.7319 | −196.61 | 0.7175 | −195.06 | 0.7112 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Liu, Y.; Chai, Y.; Zhang, H.; Wei, M.; Li, C. Genome-Wide Identification and Characterization of the Growth-Regulating Factor Gene Family Responsive to Abiotic Stresses and Phytohormone Treatments in Populus ussuriensis. Int. J. Mol. Sci. 2025, 26, 3288. https://doi.org/10.3390/ijms26073288
Zhao Y, Liu Y, Chai Y, Zhang H, Wei M, Li C. Genome-Wide Identification and Characterization of the Growth-Regulating Factor Gene Family Responsive to Abiotic Stresses and Phytohormone Treatments in Populus ussuriensis. International Journal of Molecular Sciences. 2025; 26(7):3288. https://doi.org/10.3390/ijms26073288
Chicago/Turabian StyleZhao, Ying, Yuqi Liu, Yuan Chai, Hedan Zhang, Ming Wei, and Chenghao Li. 2025. "Genome-Wide Identification and Characterization of the Growth-Regulating Factor Gene Family Responsive to Abiotic Stresses and Phytohormone Treatments in Populus ussuriensis" International Journal of Molecular Sciences 26, no. 7: 3288. https://doi.org/10.3390/ijms26073288
APA StyleZhao, Y., Liu, Y., Chai, Y., Zhang, H., Wei, M., & Li, C. (2025). Genome-Wide Identification and Characterization of the Growth-Regulating Factor Gene Family Responsive to Abiotic Stresses and Phytohormone Treatments in Populus ussuriensis. International Journal of Molecular Sciences, 26(7), 3288. https://doi.org/10.3390/ijms26073288






