Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule
Abstract
1. Introduction
2. Results
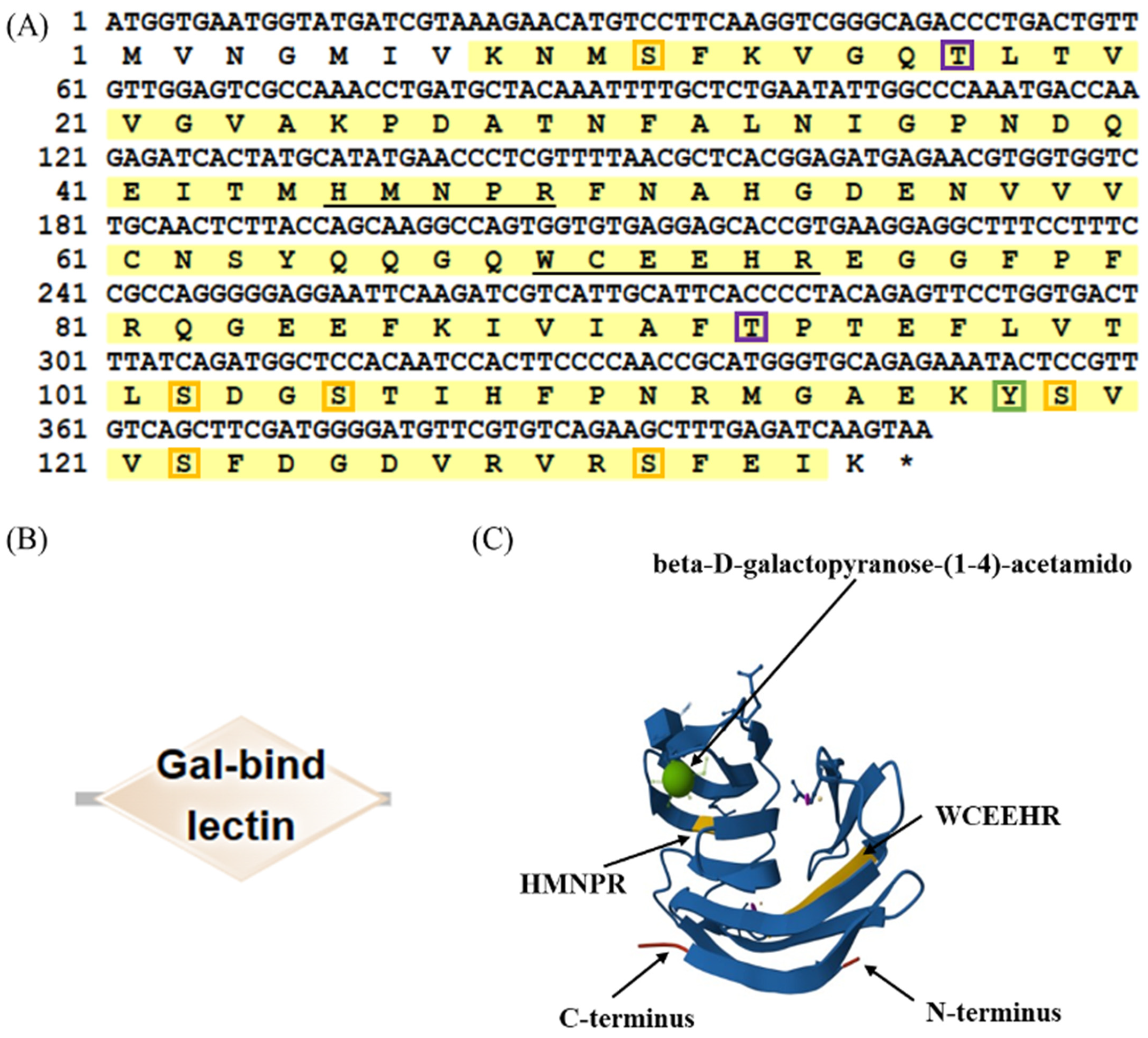
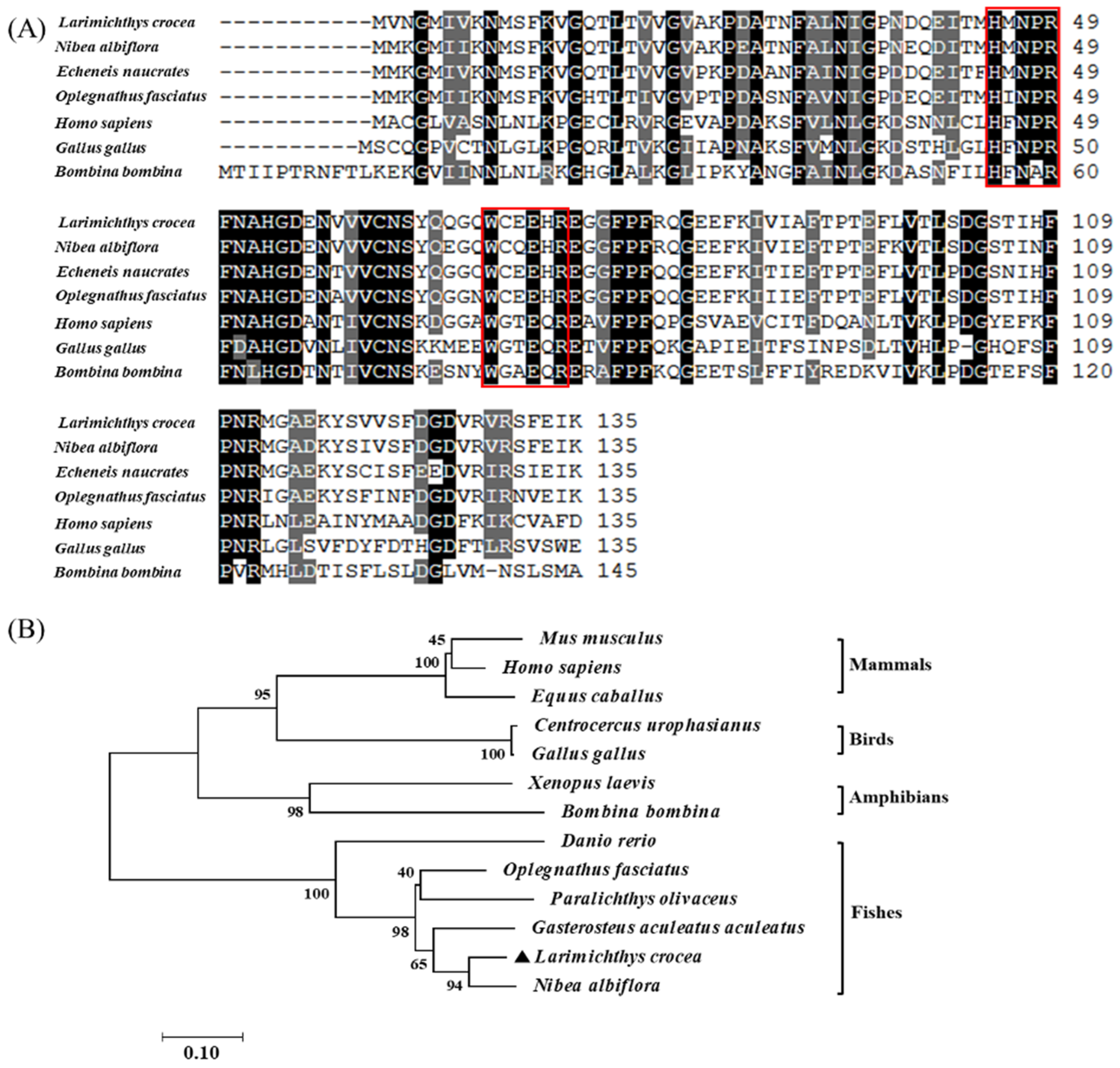
2.1. Sequence Analysis and Molecular Characteristics of LcβLectin
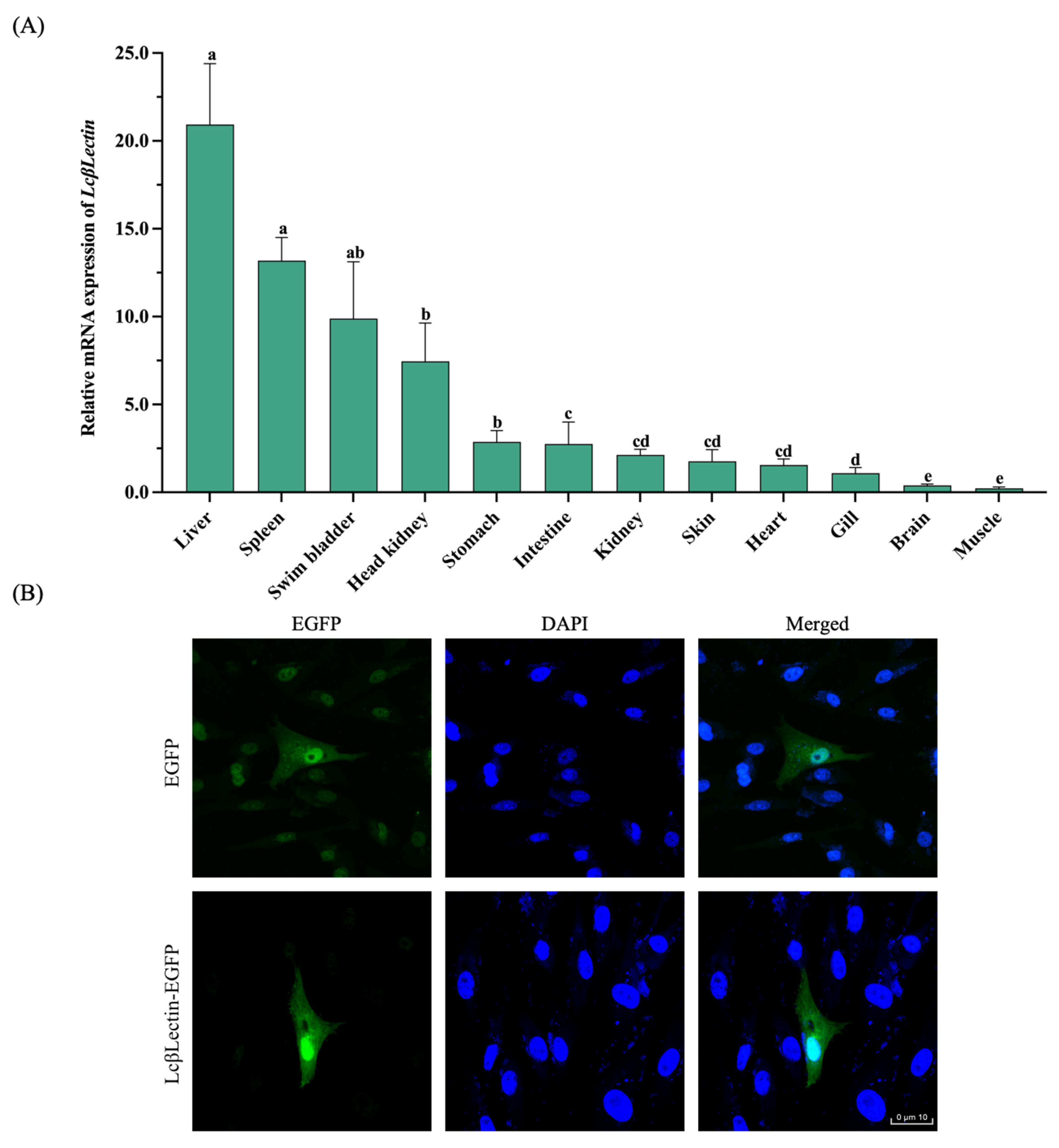
2.2. Tissue Expression Pattern and Cellular Distribution
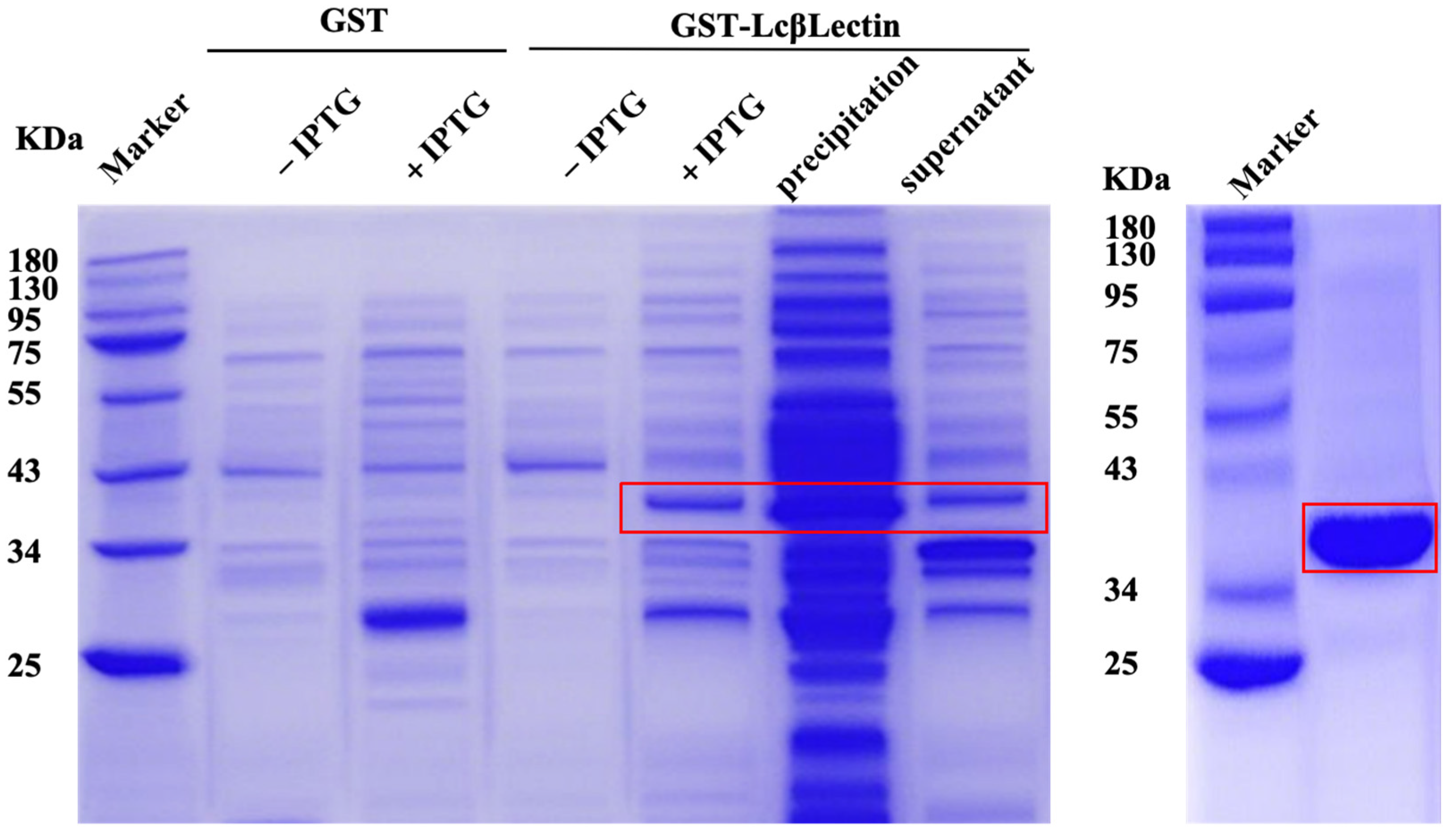
2.3. Prokaryotic Expression and Purification of LcβLectin Protein
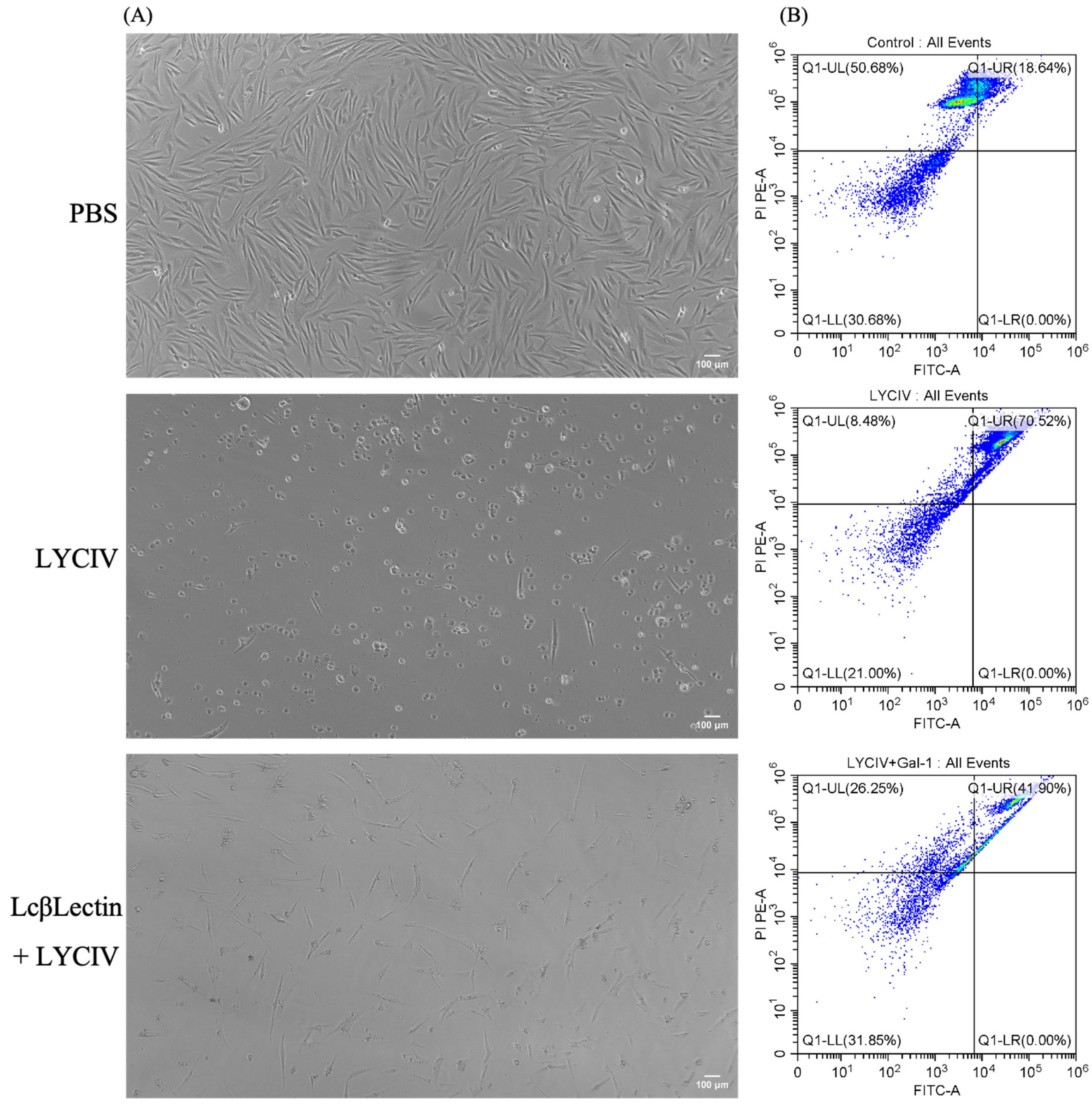
2.4. Neutralizing Virus Activity of Recombinant LcβLectin
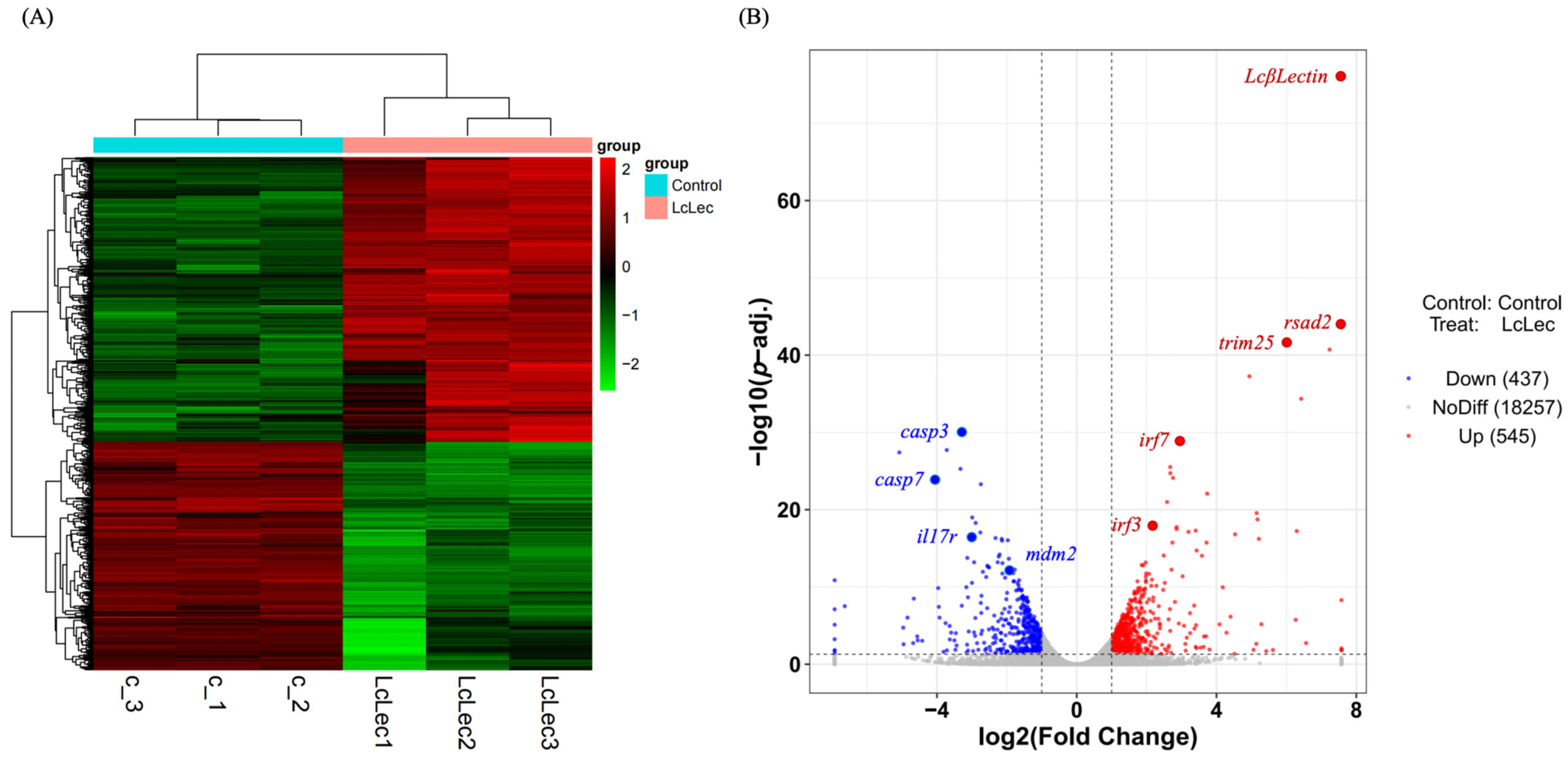
2.5. Transcriptome Data Analysis and Identification of DEGs
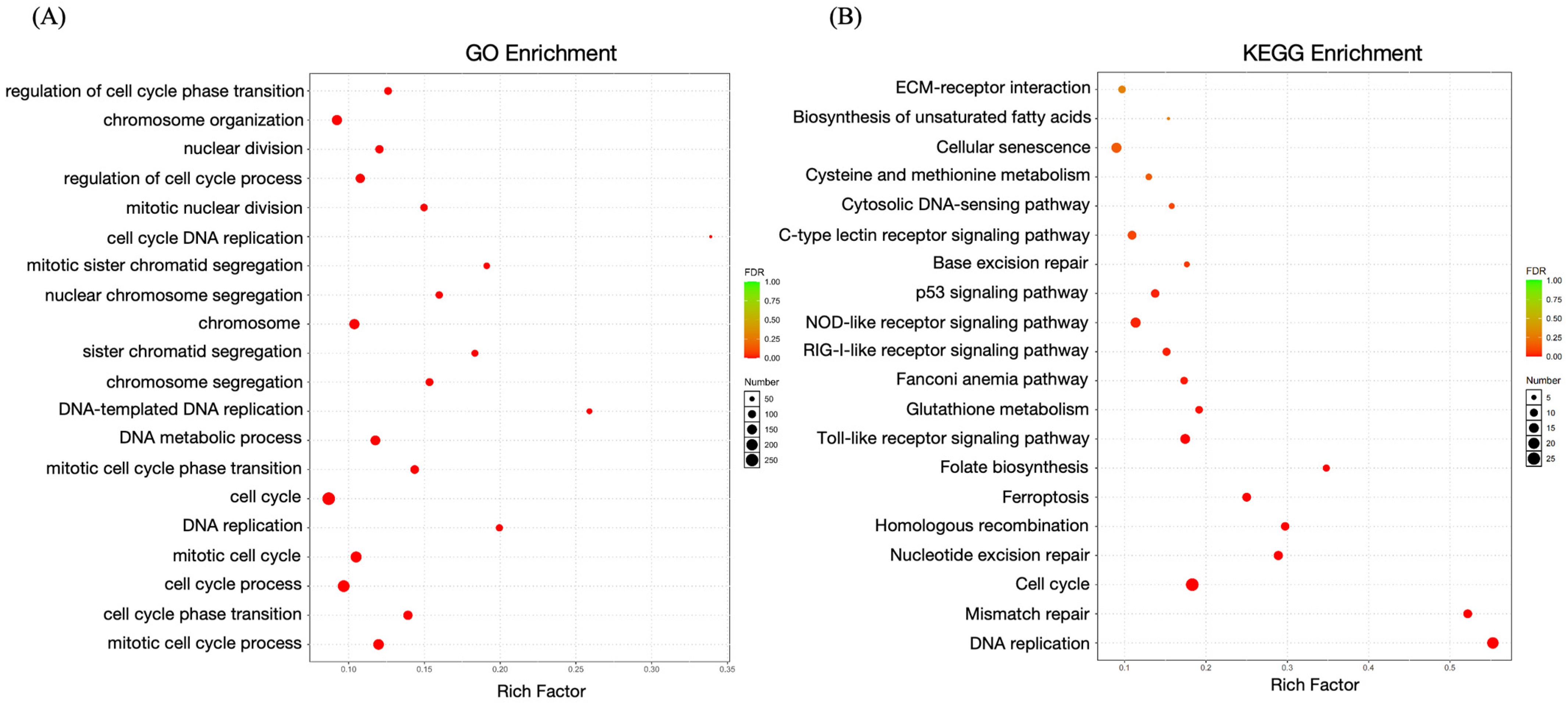
2.6. Enrichment Analysis of DEGs
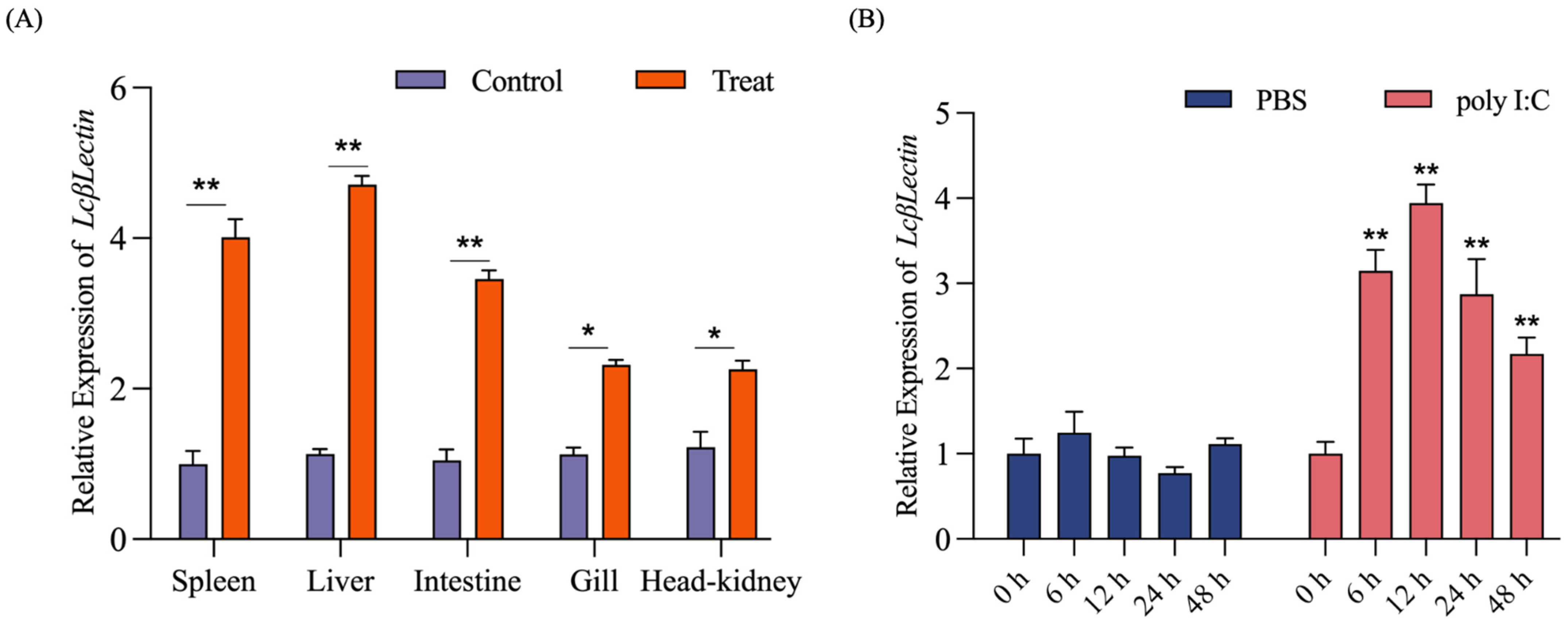
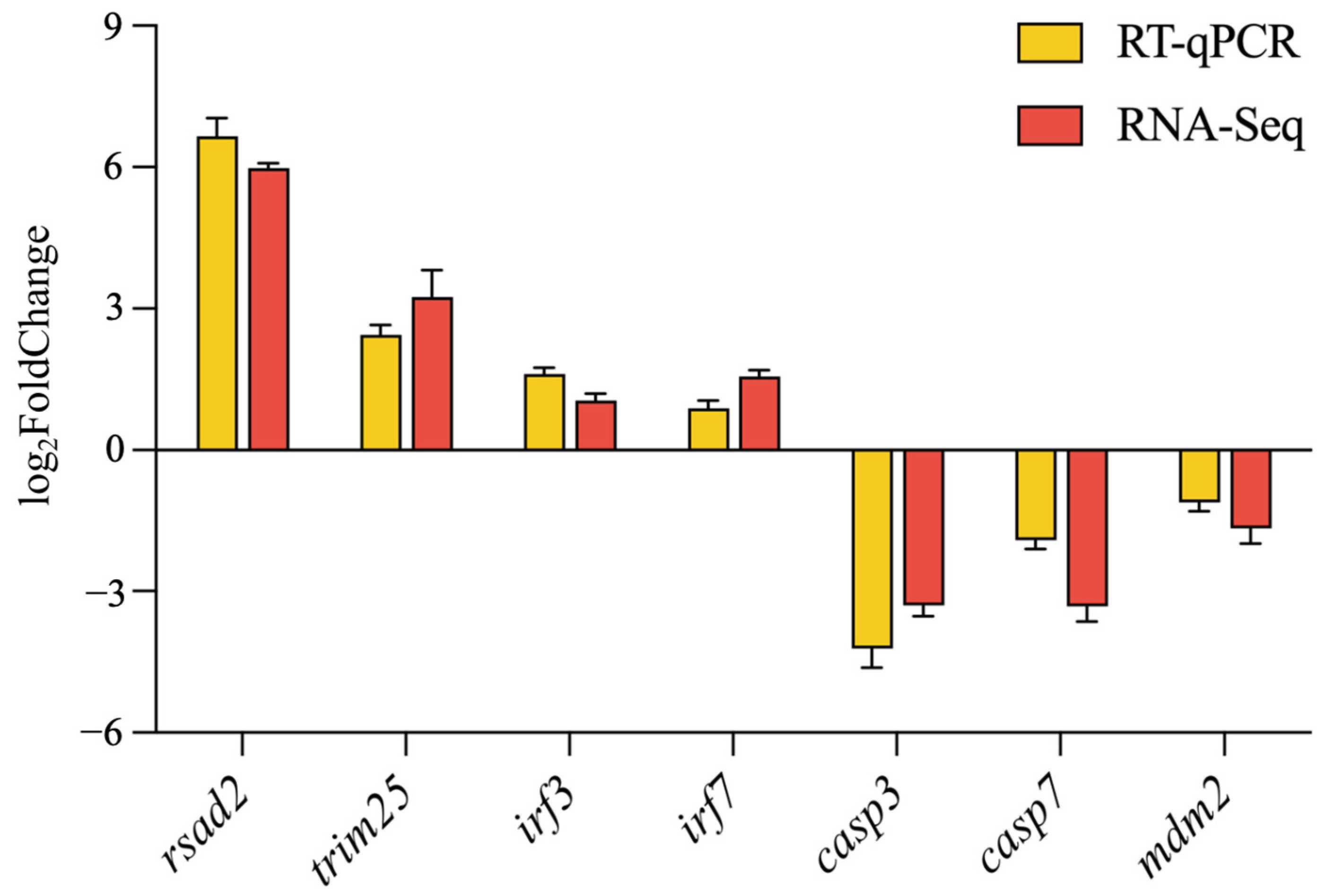
2.7. RT-qPCR Verification of DEGs
3. Discussion
4. Materials and Methods
4.1. Sample Collection, RNA Extraction, and cDNA Synthesization
4.2. Cell Culture
4.3. Bioinformatics Analysis and Plasmid Construction
4.4. Real-Time Quantitative PCR (RT-qPCR)
4.5. LcβLectin Subcellular Localization
4.6. Prokaryotic Expression and Purification of LcβLectin
4.7. Annexin V-FITC/PI Flow Cytometer Assay
4.8. LcβLectin Overexpression
4.9. Library Construction, Sequencing, and Transcriptomic Analysis
4.10. RT-qPCR Verification
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Litman, G.W.; Cannon, J.P.; Dishaw, L.J. Reconstructing Immune Phylogeny: New Perspectives. Nat. Rev. Immunol. 2005, 5, 866–879. [Google Scholar] [CrossRef]
- Reis E Sousa, C.; Yamasaki, S.; Brown, G.D. Myeloid C-Type Lectin Receptors in Innate Immune Recognition. Immunity 2024, 57, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.J.; Sancho, D.; Slack, E.C.; LeibundGut-Landmann, S.; Sousa, C.R.E. Myeloid C-Type Lectins in Innate Immunity. Nat. Immunol. 2006, 7, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Li, Q.; Yang, Y.; Gao, H.; Han, F. Antimicrobial Functions of Galectins from Fish, Mollusks, and Crustaceans: A Review. J. Agric. Food Chem. 2024, 72, 24895–24907. [Google Scholar] [CrossRef] [PubMed]
- Bänfer, S.; Jacob, R. Galectins. Curr. Biol. 2022, 32, R406–R408. [Google Scholar] [CrossRef]
- Liu, F.-T.; Stowell, S.R. The Role of Galectins in Immunity and Infection. Nat. Rev. Immunol. 2023, 23, 479–494. [Google Scholar] [CrossRef]
- Blazev, R.; Ashwood, C.; Abrahams, J.L.; Chung, L.H.; Francis, D.; Yang, P.; Watt, K.I.; Qian, H.; Quaife-Ryan, G.A.; Hudson, J.E.; et al. Integrated Glycoproteomics Identifies a Role of N-Glycosylation and Galectin-1 on Myogenesis and Muscle Development. Mol. Cell. Proteom. 2021, 20, 100030. [Google Scholar] [CrossRef]
- Eckardt, V.; Miller, M.C.; Blanchet, X.; Duan, R.; Leberzammer, J.; Duchene, J.; Soehnlein, O.; Megens, R.T.; Ludwig, A.; Dregni, A.; et al. Chemokines and Galectins Form Heterodimers to Modulate Inflammation. EMBO Rep. 2020, 21, e47852. [Google Scholar] [CrossRef]
- Ayona, D.; Fournier, P.-E.; Henrissat, B.; Desnues, B. Utilization of Galectins by Pathogens for Infection. Front. Immunol. 2020, 11, 1877. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Huang, K.; Yuan, L.; Ma, R.; Zhou, S.; Jiang, J.; Wang, Y.; Xie, J. Identification and Functional Characterization of Galectin-3 in Silver Pomfret (Pampus argenteus). Aquaculture 2024, 592, 741241. [Google Scholar] [CrossRef]
- Gabius, H.-J. The Magic of the Sugar Code. Trends Biochem. Sci. 2015, 40, 341. [Google Scholar] [CrossRef]
- Gabius, H.-J.; Kaltner, H.; Kopitz, J.; André, S. The Glycobiology of the CD System: A Dictionary for Translating Marker Designations into Glycan/Lectin Structure and Function. Trends Biochem. Sci. 2015, 40, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Banerjee, A.; Amzel, L.M.; Vasta, G.R.; Bianchet, M.A. Structure of the Zebrafish Galectin-1-L2 and Model of Its Interaction with the Infectious Hematopoietic Necrosis Virus (IHNV) Envelope Glycoprotein. Glycobiology 2019, 29, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Poisa-Beiro, L.; Dios, S.; Ahmed, H.; Vasta, G.R.; Martínez-López, A.; Estepa, A.; Alonso-Gutiérrez, J.; Figueras, A.; Novoa, B. Nodavirus Infection of Sea Bass (Dicentrarchus labrax) Induces up-Regulation of Galectin-1 Expression with Potential Anti-Inflammatory Activity. J. Immunol. 2009, 183, 6600–6611. [Google Scholar] [CrossRef] [PubMed]
- Natnan, M.; Mayalvanan, Y.; Jazamuddin, F.; Aizat, W.; Low, C.-F.; Goh, H.-H.; Azizan, K.; Bunawan, H.; Baharum, S. Omics Strategies in Current Advancements of Infectious Fish Disease Management. Biology 2021, 10, 1086. [Google Scholar] [CrossRef]
- Saura, M.; Carabaño, M.J.; Fernández, A.; Cabaleiro, S.; Doeschl-Wilson, A.B.; Anacleto, O.; Maroso, F.; Millán, A.; Hermida, M.; Fernández, C.; et al. Disentangling Genetic Variation for Resistance and Endurance to Scuticociliatosis in Turbot Using Pedigree and Genomic Information. Front. Genet. 2019, 10, 539. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Z.; Li, M.; Zhang, X.; Shi, Q.; Xu, Z. Fish Genomics and Its Application in Disease-Resistance Breeding. Rev. Aquac. 2024, 17, e12973. [Google Scholar] [CrossRef]
- Chen, X.H.; Lin, K.B.; Wang, X.W. Outbreaks of an Iridovirus Disease in Maricultured Large Yellow Croaker, Larimichthys crocea (Richardson), in China. J. Fish Dis. 2003, 26, 615–619. [Google Scholar] [CrossRef]
- Zheng, Z.; Chi, H.; Liu, X.; Yang, X.; Chen, X.; Pan, Y.; Gong, H. A New Embryonic Cell Line YCE1 from Large Yellow Croaker (Larimichthys crocea) and Its Susceptibility to Large Yellow Croaker Iridovirus. Aquaculture 2023, 565, 739079. [Google Scholar] [CrossRef]
- Wang, G.; Luan, Y.; Wei, J.; Li, Y.; Shi, H.; Cheng, H.; Bai, A.; Xie, J.; Xu, W.; Qin, P. Genetic and Pathogenic Characterization of a New Iridovirus Isolated from Cage-Cultured Large Yellow Croaker (Larimichthys crocea) in China. Viruses 2022, 14, 208. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, B.; Zou, W.; Han, F. Unveiling the Molecular Characteristics and Antibacterial Activity of Tandem-Repeat-Type Galectin-8 in Large Yellow Croaker (Larimichthys crocea). Fish Shellfish Immunol. 2024, 153, 109849. [Google Scholar] [CrossRef]
- Yang, R.-Y.; Rabinovich, G.A.; Liu, F.-T. Galectins: Structure, Function and Therapeutic Potential. Expert Rev. Mol. Med. 2008, 10, e17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, W.; Mu, C.; Li, R.; Song, W.; Ye, Y.; Shi, C.; Liu, L.; Wang, H.; Wang, C. Identification and Characterization of a Novel Galectin from the Mud Crab Scylla Paramamosain. Fish Shellfish Immunol. 2020, 98, 699–709. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Huang, M.; Yi, Q.; Guo, Y.; Gai, Y.; Wang, H.; Zhang, H.; Song, L. A Galectin from Eriocheir Sinensis Functions as Pattern Recognition Receptor Enhancing Microbe Agglutination and Haemocytes Encapsulation. Fish Shellfish Immunol. 2016, 55, 10–20. [Google Scholar] [CrossRef]
- Zhang, C.; Xue, Z.; Yu, Z.; Wang, H.; Liu, Y.; Li, H.; Wang, L.; Li, C.; Song, L. A Tandem-Repeat Galectin-1 from Apostichopus Japonicus with Broad PAMP Recognition Pattern and Antibacterial Activity. Fish Shellfish Immunol. 2020, 99, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, Q.; Li, W.; Luo, S.; Han, F.; Wang, Z. Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora). Int. J. Mol. Sci. 2023, 24, 3298. [Google Scholar] [CrossRef]
- Yang, M.-L.; Chen, Y.-H.; Wang, S.-W.; Huang, Y.-J.; Leu, C.-H.; Yeh, N.-C.; Chu, C.-Y.; Lin, C.-C.; Shieh, G.-S.; Chen, Y.-L.; et al. Galectin-1 Binds to Influenza Virus and Ameliorates Influenza Virus Pathogenesis. J. Virol. 2011, 85, 10010–10020. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.; Yin, X.; Yang, Y.; Wu, L.; Wu, H.; Li, B.; Guo, Z.; Ye, J. Functional Characterization of a Mannose-Binding Lectin (MBL) from Nile Tilapia (Oreochromis niloticus) in Non-Specific Cell Immunity and Apoptosis in Monocytes/Macrophages. Fish Shellfish Immunol. 2019, 87, 265–274. [Google Scholar] [CrossRef]
- Calandra, T.; Roger, T. Macrophage Migration Inhibitory Factor: A Regulator of Innate Immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef]
- Lee, E.H.; Rikihisa, Y. Absence of Tumor Necrosis Factor Alpha, Interleukin-6 (IL-6), and Granulocyte-Macrophage Colony-Stimulating Factor Expression but Presence of IL-1beta, IL-8, and IL-10 Expression in Human Monocytes Exposed to Viable or Killed Ehrlichia Chaffeensis. Infect. Immun. 1996, 64, 4211–4219. [Google Scholar] [CrossRef]
- Thulasitha, W.S.; Umasuthan, N.; Whang, I.; Nam, B.-H.; Lee, J. Antimicrobial Response of Galectin-1 from Rock Bream Oplegnathus Fasciatus: Molecular, Transcriptional, and Biological Characterization. Fish Shellfish Immunol. 2016, 50, 66–78. [Google Scholar] [CrossRef]
- Niu, J.; Huang, Y.; Liu, X.; Luo, G.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Functional Characterization of Galectin-3 from Nile Tilapia (Oreochromis niloticus) and Its Regulatory Role on Monocytes/Macrophages. Fish Shellfish Immunol. 2019, 95, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef]
- Fernandes-Santos, C.; Azeredo, E.L.D. Innate Immune Response to Dengue Virus: Toll-like Receptors and Antiviral Response. Viruses 2022, 14, 992. [Google Scholar] [CrossRef]
- Arad, U.; Madar-Balakirski, N.; Angel-Korman, A.; Amir, S.; Tzadok, S.; Segal, O.; Menachem, A.; Gold, A.; Elkayam, O.; Caspi, D. Galectin-3 Is a Sensor-Regulator of Toll-like Receptor Pathways in Synovial Fibroblasts. Cytokine 2015, 73, 30–35. [Google Scholar] [CrossRef]
- Chou, W.; Tsai, K.; Hsieh, P.; Wu, C.; Jou, I.; Tu, Y.; Ma, C. Galectin-3 Facilitates Inflammation and Apoptosis in Chondrocytes through Upregulation of the TLR -4-mediated Oxidative Stress Pathway in TC28A2 Human Chondrocyte Cells. Environ. Toxicol. 2022, 37, 478–488. [Google Scholar] [CrossRef]
- Panda, S.K.; Facchinetti, V.; Voynova, E.; Hanabuchi, S.; Karnell, J.L.; Hanna, R.N.; Kolbeck, R.; Sanjuan, M.A.; Ettinger, R.; Liu, Y.-J. Galectin-9 Inhibits TLR7-Mediated Autoimmunity in Murine Lupus Models. J. Clin. Investig. 2018, 128, 1873–1887. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A Diverse Range of Gene Products Are Effectors of the Type I Interferon Antiviral Response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef]
- Liu, D.; Zheng, H.; Li, Y.; Zhou, P.; Jin, H.; Luo, R. Molecular Cloning and Functional Characterization of Duck Janus Kinase 1. Mol. Immunol. 2020, 117, 29–36. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, L.; Fang, Q.; Hui, W.H.; Zhou, Z.H. 3.3 Å Cryo-EM Structure of a Nonenveloped Virus Reveals a Priming Mechanism for Cell Entry. Cell 2010, 141, 472–482. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.-H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-Finger E3 Ubiquitin Ligase Is Essential for RIG-I-Mediated Antiviral Activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Mocarski, E.S.; Upton, J.W.; Kaiser, W.J. Viral Infection and the Evolution of Caspase 8-Regulated Apoptotic and Necrotic Death Pathways. Nat. Rev. Immunol. 2012, 12, 79–88. [Google Scholar] [CrossRef]
- Porter, A.G.; Jänicke, R.U. Emerging Roles of Caspase-3 in Apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, Y. A P53 Amino-Terminal Nuclear Export Signal Inhibited by DNA Damage-Induced Phosphorylation. Science 2001, 292, 1910–1915. [Google Scholar] [CrossRef]
- Cui, K.; Li, Q.; Xu, D.; Zhang, J.; Gao, S.; Xu, W.; Mai, K.; Ai, Q. Establishment and Characterization of Two Head Kidney Macrophage Cell Lines from Large Yellow Croaker (Larimichthys crocea). Dev. Comp. Immunol. 2020, 102, 103477. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, B.; Yang, Y.; Li, W.; Han, F. Cloning, Subcellular Localization and Antibacterial Functional Analysis of NK-Lysin in Yellow Drum (Nibea albiflora). Fish Shellfish Immunol. 2023, 141, 109061. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Latin Name | Foreign Name | Genbank Accession | Identity (%) |
|---|---|---|---|
| Larimichthys crocea | Large yellow croaker | XP_010736086.1 | 100.00 |
| Nibea albiflora | Yellow drum | WEU52361.1 | 90.37 |
| Echeneis naucrates | Live sharksucker | XP_029364677.1 | 84.44 |
| Gasterosteus aculeatus | Three-spined stickleback | XP_040046198.1 | 82.96 |
| Pungitius pungitius | Ninespine stickleback | XP_037331494.1 | 82.22 |
| Oplegnathus fasciatus | Rock bream | ADV35589.1 | 81.48 |
| Homo sapiens | Human | NP_002296.1 | 40.46 |
| Equus caballus | Horse | AQX42328.1 | 40.00 |
| Mus musculus | Mouse | AAH99479.1 | 39.23 |
| Xenopus laevis | Frog | AAK11514.1 | 39.20 |
| Gallus gallus | Chicken | NP_990826.1 | 39.06 |
| Centrocercus urophasianus | Greater sage-grouse | XP_042693815.1 | 39.06 |
| Bombina bombina | Fire-bellied toad | XP_053577272.1 | 38.71 |
| Group | Raw Reads (M) | Clean Reads (M) | Clean Bases (Gb) | Q30 (%) | Uniquely Mapping Gene Ratio (%) |
|---|---|---|---|---|---|
| c1 | 41.37 | 40.78 | 6.14 | 96.98 | 94.67 |
| c2 | 51.29 | 50.55 | 7.60 | 97.05 | 94.59 |
| c3 | 43.54 | 42.96 | 6.46 | 97.12 | 94.60 |
| LcLec-1 | 53.64 | 52.90 | 7.95 | 97.13 | 94.10 |
| LcLec-2 | 58.58 | 57.81 | 8.70 | 97.16 | 94.32 |
| LcLec-3 | 70.95 | 70.01 | 10.54 | 97.73 | 94.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wu, H.; Huang, Y.; Yang, Y.; Yekefenhazi, D.; Zou, W.; Han, F. Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule. Int. J. Mol. Sci. 2025, 26, 3251. https://doi.org/10.3390/ijms26073251
Zhang J, Wu H, Huang Y, Yang Y, Yekefenhazi D, Zou W, Han F. Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule. International Journal of Molecular Sciences. 2025; 26(7):3251. https://doi.org/10.3390/ijms26073251
Chicago/Turabian StyleZhang, Jiawei, Hongling Wu, Ying Huang, Yao Yang, Dinaer Yekefenhazi, Wenzheng Zou, and Fang Han. 2025. "Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule" International Journal of Molecular Sciences 26, no. 7: 3251. https://doi.org/10.3390/ijms26073251
APA StyleZhang, J., Wu, H., Huang, Y., Yang, Y., Yekefenhazi, D., Zou, W., & Han, F. (2025). Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule. International Journal of Molecular Sciences, 26(7), 3251. https://doi.org/10.3390/ijms26073251





