Integrated Analysis of mRNA and miRNA Associated with Reproduction in Female and Male Gonads in Abalone (Haliotis discus hannai)
Abstract
1. Introduction
2. Results
2.1. Overview of RNA-Seq Data
2.2. DEGs and Functional Enrichment Analyses
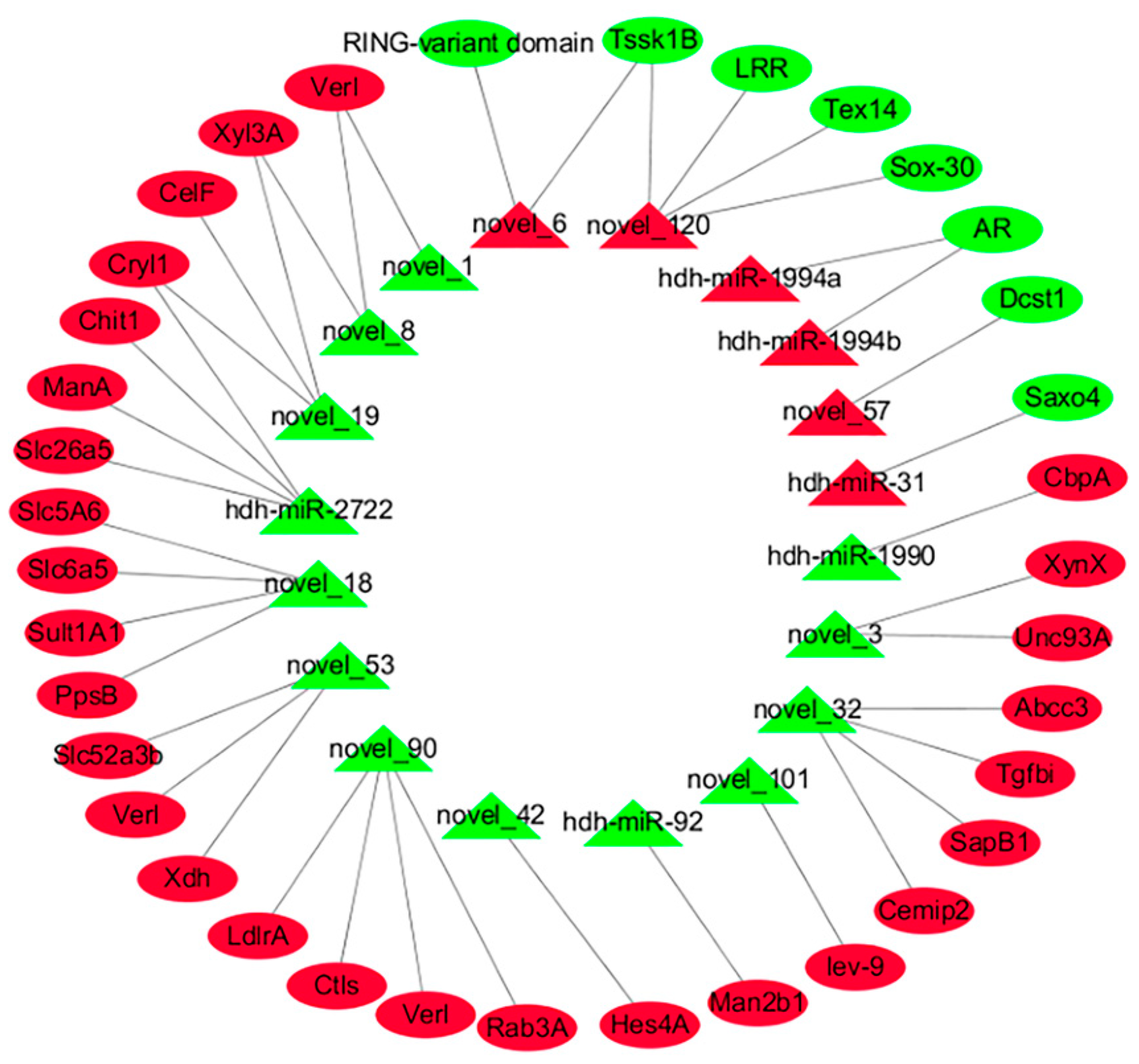
2.3. DEMs and Potential miRNA–mRNA Interaction Networks
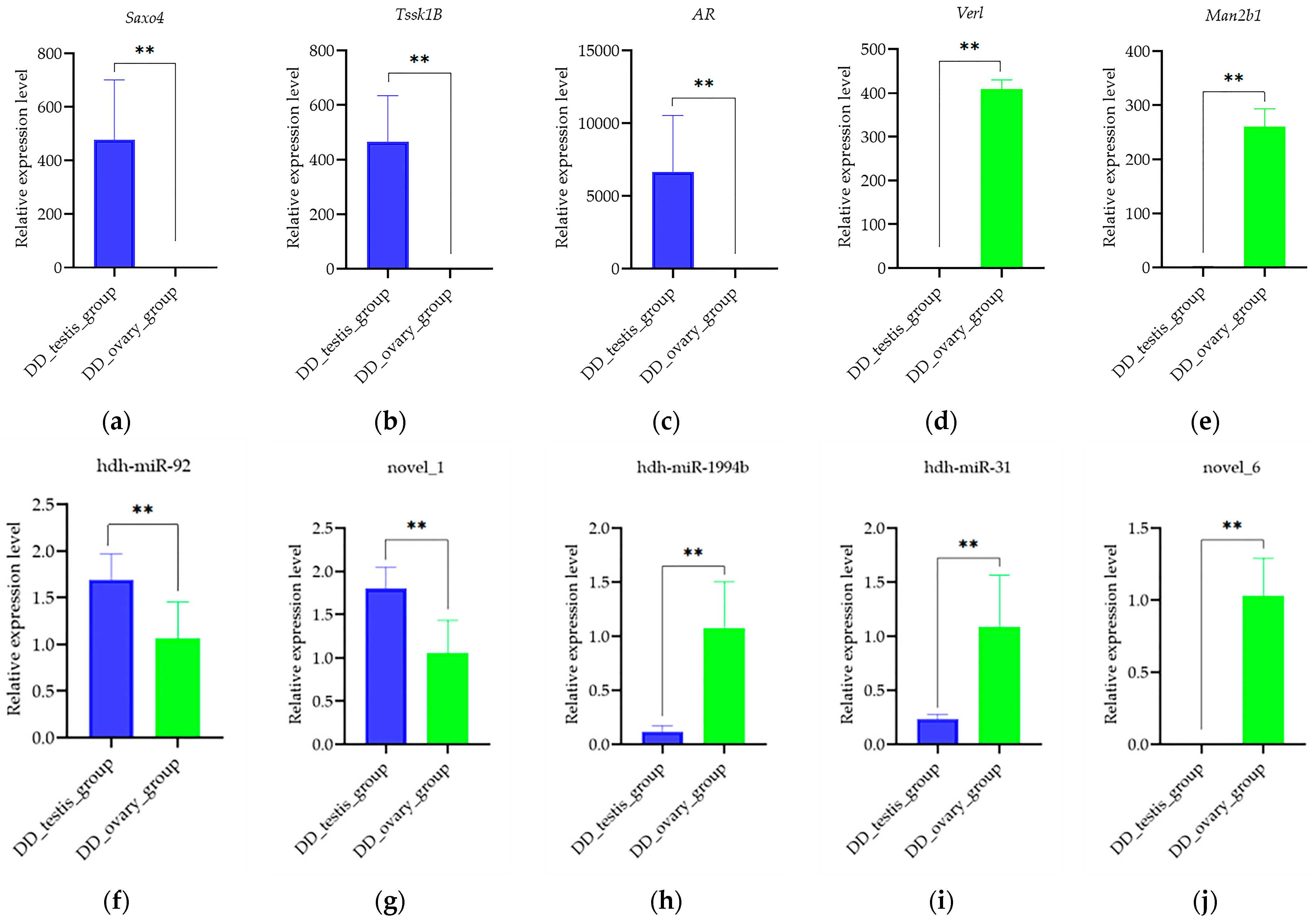
2.4. Verification Analysis of Identified DE-mRNA and DE-miRNA
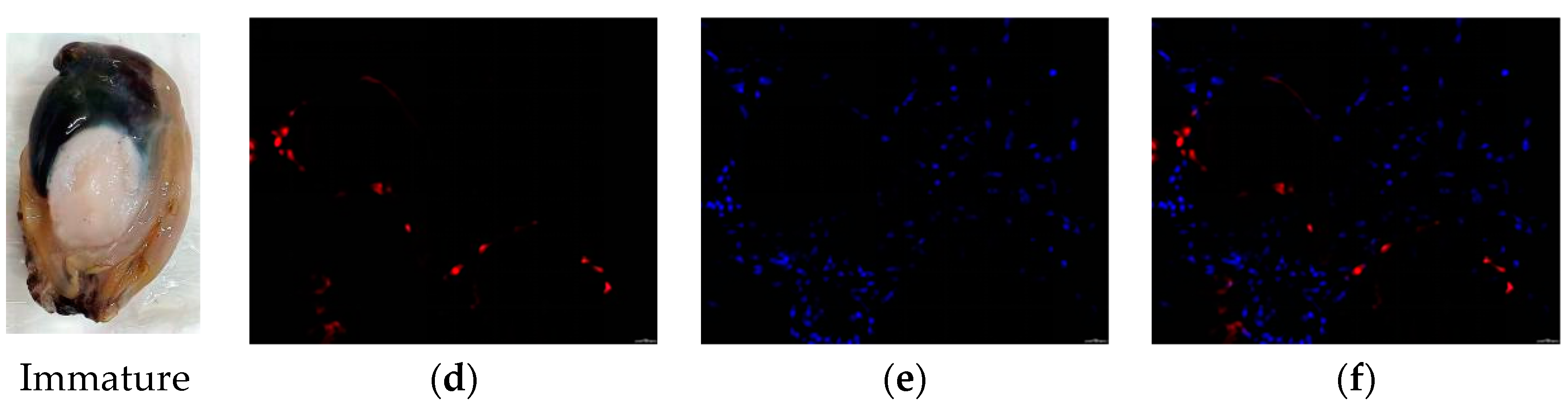
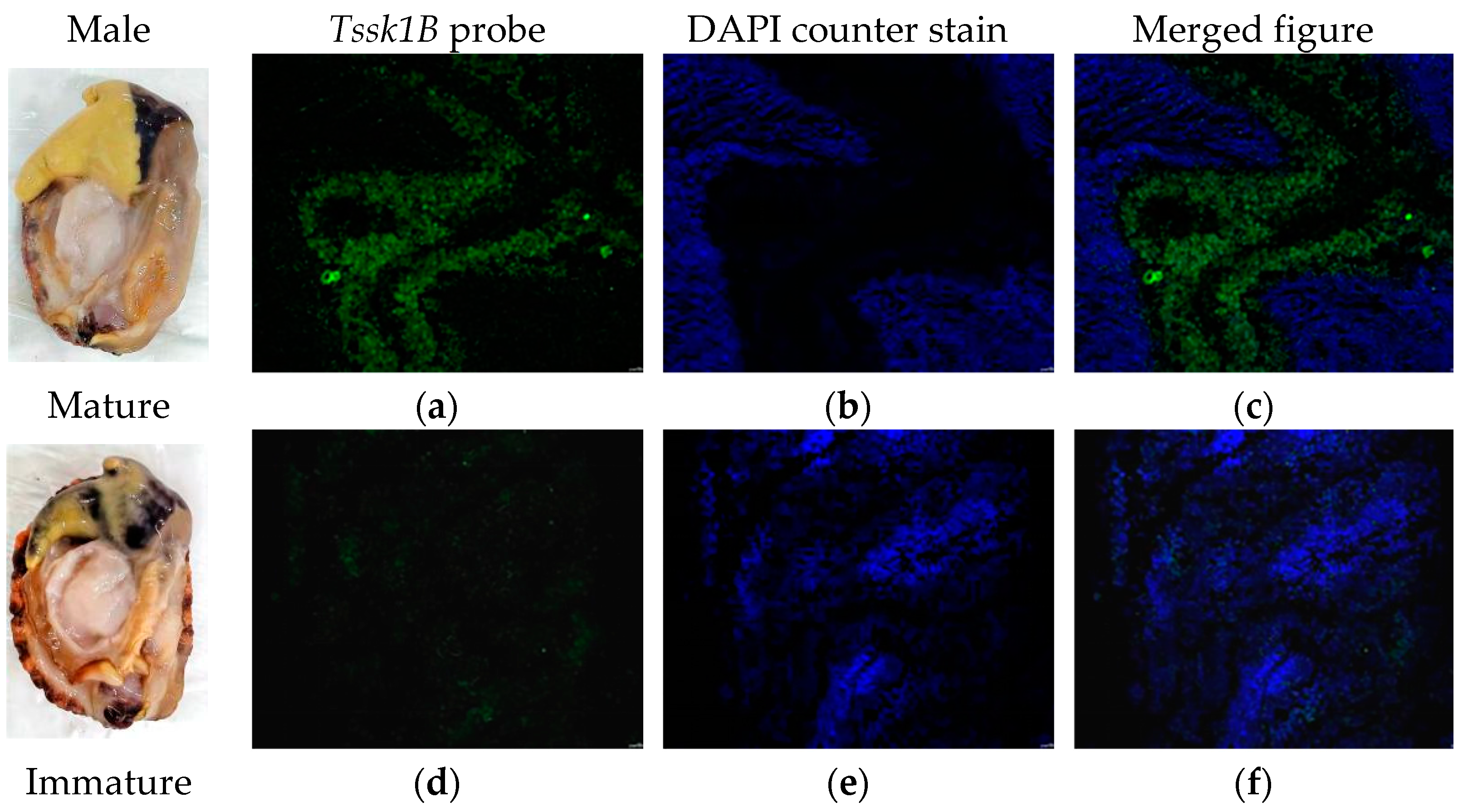
2.5. Localization of Verl and Tssk1B in the Gonad of H. discus hannai
3. Discussion
4. Materials and Methods
4.1. Abalones and RNA Preparation
4.2. Transcriptome Sequencing and Data Processing
4.3. sRNA Sequencing and Analysis
4.4. miRNA Target Prediction and Functional Analyses
4.5. Quantitative Real-Time PCR
4.6. Fluorescence In Situ Hybridization (FISH)
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| H. discus hannai | Haliotis discus hannai |
| DE | differentially expressed |
| Tssk | Testis-specific serine/threonine-protein kinase |
| Verl | Vitelline envelope sperm lysin receptor |
| FISH | fluorescence in situ hybridization |
| UTRs | untranslated regions |
| HTS | high-throughput sequencing |
| DEGs | differentially expressed genes |
| DEMs | differentially expressed miRNAs |
| brat | Brain tumor protein |
| Ppn | Papilin |
| Fut2 | Galactoside alpha-(1,2)-fucosyltransferase 2 |
| Tuba1A | Tubulin alpha-1A chain |
| Fasn | Fatty acid synthase |
| Inmt | Indolethylamine N-methyltransferase |
| Tpo | Thyroid peroxidase |
| hFat3 | Protocadherin Fat 3 |
| Zp-like | Zona pellucida-like domain |
| SOX-30 | Transcription factor SOX-30 |
| Tepp | Testis, prostate and placenta-expressed protein |
| HSF1 | Heat shock factor protein 1 |
| Saxo4 | Stabilizer of axonemal microtubules 4 |
| AR | Ankyrin repeats |
| Man2b1 | Lysosomal alpha-mannosidase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| MF | molecular functions |
| BP | biological processes |
| CC | cellular components |
| PPI | protein–protein interaction |
| qRT-PCR | Quantitative real-time PCR |
| SD | standard deviation |
| Abcc3 | ATP-binding cassette sub-family C member 3 |
| CbpA | Chitin binding Peritrophin-A domain |
| Chit1 | Chitotriosidase-1 |
| CelF | Endoglucanase F |
| XynX | Exoglucanase XynX |
| Cemip2 | Inactive cell surface hyaluronidase CEMIP2 |
| Cryl1 | Lambda-crystallin |
| Ctls | Lectin C-type domain |
| LdlrA | Low-density lipoprotein receptor domain class A |
| ManA | Mannan endo-1,4-beta-mannosidase |
| PpsB | Phenolphthiocerol/phthiocerol polyketide synthase subunit B |
| Slc26a5 | Prestin |
| lev-9 | Protein lev-9 |
| Unc93A | Protein unc-93 homolog A |
| Rab3A | Ras-related protein Rab-3A |
| SapB1 | Saposin-like type B, region 1 |
| Slc6a5 | Sodium- and chloride-dependent glycine transporter 2 |
| Sult1A1 | Sulfotransferase 1A1 |
| Hes4A | Transcription factor Hes4A |
| Tgfbi | Transforming growth factor-beta-induced protein ig-h3 |
| Xdh | Xanthine dehydrogenase |
| Xyl3A | Xylan 1,4-beta-xylosidase |
| Dcst1 | E3 ubiquitin-protein ligase Dcst1 |
| Tex14 | Inactive serine/threonine-protein kinase Tex14 |
| LRR | Leucine rich repeat |
References
- Qing, W. Preliminary Study on Gonadal Development and Mechanism of Oocyte Maturation of Hard Clam Mercenaria Mercenaria. Master’s Thesis, University of Chinese Academy of Sciences (Institute of Oceanography, Chinese Academy of Sciences), Qingdao, China, 2007. (In Chinese). [Google Scholar]
- Osada, M.; Matsumoto, T. Endocrine control of gametogenesis and spawning in bivalves. In Physiology of Molluscs, 1st ed.; Saleuddin, S., Mukai, S., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 180–220. [Google Scholar]
- Wang, W.; Wu, B.; Liu, Z.; Sun, X.; Zhou, L.; Xu, W.; Yu, T.; Zheng, Y.; Zhang, S. Comprehensive analysis on the regulation of differentially expressed of mRNA and ncRNA in different ovarian stages of ark shell Scapharca broughtonii. BMC Genom. 2023, 24, 563. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Song, Y.N.; Shi, L.L.; Liu, Z.Q.; Qiu, G.F. Global analysis of the ovarian microRNA transcriptome: Implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda). BMC Genom. 2014, 15, 547. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, G.S.; Yin, S.W.; Li, Z.C.; Wang, Q.T.; Chen, S.Q.; Zhou, G.Q. Integrated analysis of mRNA-seq and miRNA-seq reveals the potential roles of sexbiased miRNA-mRNA pairs in gonad tissue of dark sleeper (Odontobutis potamophila). BMC Genom. 2017, 18, 613. [Google Scholar] [CrossRef]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. Te drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef]
- He, Y.; Huang, C.X.; Chen, N.; Wu, M.; Huang, Y.; Liu, H.; Tang, R.; Wang, W.M.; Wang, H.L. The zebrafish miR-125c is induced under hypoxic stress via hypoxia-inducible factor 1 alpha and functions in cellular adaptations and embryogenesis. Oncotarget 2017, 8, 73846–73859. [Google Scholar] [CrossRef]
- Geiger, D.L. Distribution and biogeography of the Haliotidae (Gastropoda: Vetigastropoda) world-wide. Boll. Malacol. 2000, 35, 57–120. [Google Scholar]
- Luo, X.; Ke, C.; You, W. Estimates of correlations for shell morphological traits on body weight of interspecific hybrid abalone (Haliotis discus hannai and Haliotis gigantea). J. Shellfish Res. 2013, 32, 115–118. [Google Scholar] [CrossRef]
- Liu, X.Z.; Cu, L.F. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2023. [Google Scholar]
- Gnocchi, K.G.; Boldrini-França, J.; Gomes, L.C.; Chippari-Gomes, A.R. De novo assembly and annotation of the transcriptome of Astyanax lacustris liver unveil candidate genes to monitor response to environmental stress. Mar. Genom. 2020, 54, 100784. [Google Scholar] [CrossRef]
- Nazari, S.; Khoshkholgh, M.; Baeza, J.A. Comparative transcriptome sequencing analysis of the narrow-clawed crayfish Pontastacus leptodactylus (Eschscholtz, 1823) and discovery of candidate sex-related genes. Aquacult. Rep. 2022, 25, 101235. [Google Scholar] [CrossRef]
- Wang, H.; Tang, L.; Wei, H.; Lu, J.; Mu, C.; Wang, C. Transcriptomic analysis of adaptive mechanisms in response to sudden salinity drop in the mud crab, Scylla paramamosain. BMC Genom. 2018, 19, 421. [Google Scholar] [CrossRef]
- Lou, F.; Yang, T.; Han, Z.; Gao, T. Transcriptome analysis for identification of candidate genes related to sex determination and growth in Charybdis japonica. Gene 2018, 677, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, F.; Liu, S.; Zhao, H.; Lu, W.; Zhang, Q. Transcriptomic analysis reveals functional interaction of mRNA-lncRNA-miRNA in steroidogenesis and spermatogenesis of gynogenetic Japanese flounder (Paralichthys olivaceus). Biology 2022, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yang, Q.W.; Zhao, W.J.; Du, Q.Y.; Chang, Z.J. Effects of short-time exposure to atrazine on miRNA expression profiles in the gonad of common carp (Cyprinus carpio). BMC Genom. 2019, 20, 587. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Wei, P.; Chen, X.; Lin, Y.; Peng, J. Identification and characterization of microRNAs in the gonad of Trachinotus ovatus using Solexa sequencing. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 312–320. [Google Scholar] [CrossRef]
- Mendoza-Porras, O.; Botwright, N.A.; McWilliam, S.M.; Cook, M.T.; Harris, J.O.; Wijffels, G.; Colgrave, M.L. Exploiting genomic data to identify proteins involved in abalone reproduction. J. Proteom. 2014, 108, 337–353. [Google Scholar] [CrossRef]
- Mendoza-Porras, O.; Botwright, N.A.; Reverter, A.; Cook, M.T.; Harris, J.O.; Wijffels, G.; Colgrave, M.L. Identification of differentially expressed reproductive and metabolic proteins in the female abalone (Haliotis laevigata) gonad following artificial induction of spawning. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 127–138. [Google Scholar] [CrossRef]
- Kim, M.A.; Rhee, J.S.; Kim, T.H.; Lee, J.S.; Choi, A.Y.; Choi, B.S.; Choi, I.Y.; Sohn, Y.C. Alternative Splicing Profile and Sex-Preferential Gene Expression in the Female and Male Pacific Abalone Haliotis discus hannai. Genes 2017, 8, 99. [Google Scholar] [CrossRef]
- Yu, L.; Xu, D.; Ye, H.; Yue, H.; Ooka, S.; Kondo, H.; Yazawa, R.; Takeuchi, Y. Gonadal Transcriptome Analysis of Pacific Abalone Haliotis discus discus: Identification of Genes Involved in Germ Cell Development. Mar. Biotechnol. 2018, 20, 467–480. [Google Scholar]
- Kim, M.A.; Kim, T.H.; Lee, S.; Nam, B.H.; Lee, J.S.; Jang, W.; Sohn, Y.C. Ovarian transcriptome profiles associated with sexual maturation in Pacific abalone (Haliotis discus hannai). Genes Genom. 2020, 42, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xiao, J.; Jiang, Y.; Ke, Y.; Ke, C.; Cai, M. Mapping and marker identification for sex-determining in the Pacific abalone, Haliotis discus hannai Ino. Aquaculture 2021, 530, 735810. [Google Scholar] [CrossRef]
- Weng, X.; Xu, Y.; Dong, X.; Luo, X.; You, W.; Ke, C.; Cai, M. Sex-specific markers developed by next-generation sequencing confirmed a male heterogametic sex determination in small abalone, Haliotis diversicolor. Aquaculture 2022, 555, 738256. [Google Scholar] [CrossRef]
- Wilburn, D.B.; Tuttle, L.M.; Klevit, R.E.; Swanson, W.J. Solution structure of sperm lysin yields novel insights into molecular dynamics of rapid protein evolution. Proc. Natl. Acad. Sci. USA 2018, 115, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Litscher, E.S.; Wassarman, P.M. Zona Pellucida Proteins, Fibrils, and Matrix. Annu. Rev. Biochem. 2020, 89, 695–715. [Google Scholar] [CrossRef] [PubMed]
- Salicioni, A.M.; Gervasi, M.G.; Sosnik, J.; Tourzani, D.A.; Nayyab, S.; Caraballo, D.A.; Visconti, P.E. Testis-specific serine kinase protein family in male fertility and as targets for non-hormonal male contraception. Biol. Reprod. 2020, 103, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Neila-Montero, M.; Alvarez, M.; Riesco, M.F.; Montes-Garrido, R.; Palacin-Martinez, C.; Silva-Rodríguez, A.; Martín-Cano, F.E.; Peña, F.J.; de Paz, P.; Anel, L.; et al. Ovine fertility by artificial insemination in the breeding season could be affected by intraseasonal variations in ram sperm proteomic profile. Theriogenology 2023, 208, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Ford, W. Glycolysis and sperm motility: Does a spoonful of sugar help the flagellum go round? Hum. Reprod. Update 2006, 12, 269–274. [Google Scholar] [CrossRef]
- Marco, A. Sex-biased expression of microRNAs in Drosophila melanogaster. Open Biol. 2014, 4, 140024. [Google Scholar] [CrossRef]
- Tonelli, F.M.P.; Lacerda, S.M.S.N.; Procópio, M.S.; Lemos, B.L.S.; de Franca, L.R.; Resende, R.R. Gene delivery to Nile tilapia cells for transgenesis and the role of PI3K-c2α in angiogenesis. Sci. Rep. 2017, 7, 44317. [Google Scholar] [CrossRef]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [PubMed]
- Sun, L.; Lu, S.; Bai, M.; Xiang, L.; Li, J.; Jia, C.; Jiang, H. Integrative microRNA-mRNA analysis of muscle tissues in Qianhua Mutton Merino and Small Tail Han sheep reveals key roles for oar-miR-655-3p and oar-miR-381-5p. DNA Cell Biol. 2019, 38, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Abdalla, B.A.; Zheng, M.; He, X.; Cai, B.; Han, P.; Ouyang, H.; Chen, B.; Nie, Q.; Zhang, X. Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. Mol. Genet. Genom. 2018, 293, 69–80. [Google Scholar]
- Zhao, W.; Xiao, W.; Sun, J.; Chen, M.; Ma, M.; Cao, Y.; Cen, W.; Li, R.; Luo, J. An integration of microRNA and transcriptome sequencing analysis reveal regulatory roles of miRNAs in response to chilling stress in wild rice. Plants 2022, 11, 977. [Google Scholar] [CrossRef]
- Waiho, K.; Fazhan, H.; Zhang, Y.; Zhang, Y.; Li, S.; Zheng, H.; Liu, W.; Ikhwanuddin, M.; Ma, H. Gonadal microRNA Expression Profiles and Their Potential Role in Sex Differentiation and Gonadal Maturation of Mud Crab Scylla paramamosain. Mar. Biotechnol. 2019, 21, 320–334. [Google Scholar]
- Hanif, M.A.; Hossen, S.; Cho, Y.; Sukhan, Z.P.; Choi, C.Y.; Kho, K.H. Characterization and Expression Analysis of Mollusk-like Growth Factor: A Secreted Protein Involved in Pacific Abalone Embryonic and Larval Development. Biology 2022, 11, 1445. [Google Scholar] [CrossRef] [PubMed]
- Hossen, S.; Sukhan, Z.P.; Kim, S.C.; Hanif, M.A.; Kong, I.K.; Kho, K.H. Molecular Cloning and Functional Characterization of Catalase in Stress Physiology, Innate Immunity, Testicular Development, Metamorphosis, and Cryopreserved Sperm of Pacific Abalone. Antioxidants 2023, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Uki, N. Technical study on artificial spawning of abalone, Genus Haliotis I. Relation between water temperature and advancing sexual maturity of Haliotis discus hannai Ino. Bull. Tohoku Reg. Fish. Res. Lab. 1974, 33, 69–78. [Google Scholar]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Choi, M.J.; Kim, G.D.; Kim, J.M.; Lim, H.K. Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2015, 16, 27520–27534. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with highthroughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef]
- Zhong, L.; Zhang, F.; Zhai, Y.; Cao, Y.H.; Zhang, S.; Chang, Y.Q. Identifcation and comparative analysis of complement C3-associated microRNAs in immune response of Apostichopus japonicus by high-throughput sequencing. Sci. Rep. 2015, 5, 17763. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An Integrative microRNA Evolutionary Analysis Platform for Next-generation Sequencing Experiments. BMC Bioinform. 2010, 13, 140. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. R Bioconduct. 2012. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [PubMed]
- Zheng, Z.; Huang, R.; Tian, R.; Jiao, Y.; Du, X. Pm-miR-133 hosting in one potential lncRNA regulates RhoA expression in pearl oyster Pinctada martensii. Gene 2016, 591, 484–489. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–428. [Google Scholar]

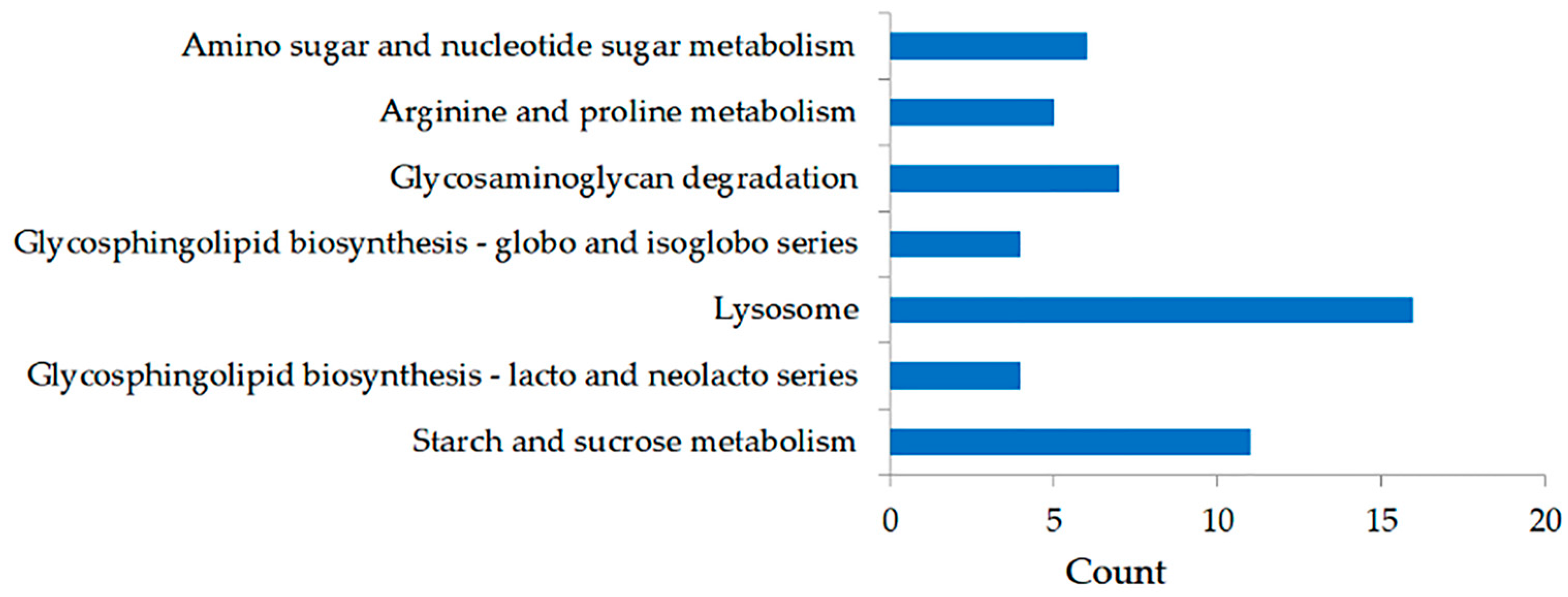



| Sample Name | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error Rate | Q20 (%) | Q30 (%) | GC pct (%) |
|---|---|---|---|---|---|---|---|---|
| DD_ovary_1 | 44,600,466 | 6.69G | 43,341,220 | 6.5G | 0.03 | 94.87 | 87.52 | 45.16 |
| DD_ovary_2 | 47,513,192 | 7.13G | 46,428,612 | 6.96G | 0.03 | 95.07 | 87.84 | 45.29 |
| DD_ovary_3 | 44,400,154 | 6.66G | 43,435,352 | 6.52G | 0.03 | 94.53 | 86.61 | 43.46 |
| DD_testis_1 | 41,592,936 | 6.24G | 40,138,628 | 6.02G | 0.03 | 97.25 | 92.19 | 43.31 |
| DD_testis_2 | 45,197,534 | 6.78G | 44,052,766 | 6.61G | 0.03 | 97.06 | 91.82 | 43.38 |
| DD_testis_3 | 44,737,000 | 6.71G | 42,770,608 | 6.42G | 0.03 | 96.86 | 91.44 | 43.87 |
| Sample | Total_Reads | N% > 10% | Low Quality | 5_adapter_ contamine | 3_adapter_null or insert_null | With ployA/T/G/C | Clean Reads |
|---|---|---|---|---|---|---|---|
| DD_ovary_1 | 12,604,189 (100.00%) | 1 (0.00%) | 0 (0.00%) | 2907 (0.02%) | 263,476 (2.09%) | 3627 (0.03%) | 12,334,178 (97.86%) |
| DD_ovary_2 | 11,763,492 (100.00%) | 319 (0.00%) | 0 (0.00%) | 11,695 (0.10%) | 91,539 (0.78%) | 35,964 (0.31%) | 11,623,975 (98.81%) |
| DD_ovary_3 | 11,148,631 (100.00%) | 270 (0.00%) | 0 (0.00%) | 11,784 (0.11%) | 134,483 (1.21%) | 30,690 (0.28%) | 10,971,404 (98.41%) |
| DD_testis_1 | 11,558,139 (100.00%) | 0 (0.00%) | 0 (0.00%) | 2243 (0.02%) | 359,076 (3.11%) | 2194 (0.02%) | 11,194,626 (96.85%) |
| DD_testis_2 | 11,598,714 (100.00%) | 0 (0.00%) | 0 (0.00%) | 7447 (0.06%) | 247,111 (2.13%) | 8378 (0.07%) | 11,335,778 (97.73%) |
| DD_testis_3 | 11,563,092 (100.00%) | 0 (0.00%) | 0 (0.00%) | 2606 (0.02%) | 580,785 (5.02%) | 2072 (0.02%) | 10,977,629 (94.94%) |
| Target DEG id | Target DEG Name | log2Fold Change | padj | Related miRNA |
|---|---|---|---|---|
| HDH_G17212 | Alpha-amylase | 11.5793044 | 8.34E-14 | hdh-miR-133-3p |
| HDH_G10591 | ATP-binding cassette sub-family C member 3 (Abcc3) | 10.69052989 | 3.88E-14 | novel_32 |
| HDH_G16571 | Chitin binding Peritrophin-A domain (CbpA) | 10.02494546 | 1.08E-09 | hdh-miR-1990 |
| HDH_G27689 | Chitotriosidase-1 (Chit1) | 12.04039088 | 1.27E-14 | hdh-miR-2722 |
| HDH_G29537 | Endoglucanase F (CelF) | 13.61183447 | 2.04E-18 | novel_19 |
| HDH_G17091 | Exoglucanase XynX (XynX) | 10.07419645 | 2.56E-08 | novel_3 |
| HDH_G14591 | Inactive cell surface hyaluronidase CEMIP2 (Cemip2) | 14.81063531 | 3.02E-25 | novel_32 |
| HDH_G05585 | Lambda-crystallin (Cryl1) | 10.20412277 | 1.31E-09 | hdh-miR-2722, novel_19 |
| HDH_G13593 | Lectin C-type domain (Ctls) | 10.28631776 | 2.36E-09 | novel_90 |
| HDH_G08135 | Low-density lipoprotein receptor domain class A (LdlrA) | 14.26186176 | 5.70E-19 | novel_90 |
| HDH_G26485 | Lysosomal alpha-mannosidase (Man2b1) | 12.22683725 | 4.35E-17 | hdh-miR-92 |
| HDH_G25248 | Mannan endo-1,4-beta-mannosidase (ManA) | 12.05066365 | 2.40E-14 | hdh-miR-2722 |
| HDH_G06159 | Phenolphthiocerol/phthiocerol polyketide synthase subunit B (PpsB) | 11.98933126 | 4.70E-19 | novel_18 |
| HDH_G14448 | Prestin (Slc26a5) | 10.15083509 | 1.76E-12 | hdh-miR-2722 |
| HDH_G06399 | Protein lev-9 (lev-9) | 18.16934184 | 1.96E-35 | novel_101 |
| HDH_G15576 | Protein unc-93 homolog A (Unc93A) | 10.02044907 | 1.97E-11 | novel_3 |
| HDH_G15755 | Ras-related protein Rab-3A (Rab3A) | 10.03697922 | 1.02E-12 | novel_90 |
| HDH_G27130 | Saposin-like type B, region 1 (SapB1) | 12.75468774 | 2.94E-15 | novel_32 |
| HDH_G25000 | Sodium- and chloride-dependent glycine transporter 2 (Slc6a5) | 11.09950664 | 6.36E-15 | novel_18 |
| HDH_G21292 | Slc5a6 | 10.49241993 | 1.19E-13 | novel_18 |
| HDH_G21270 | Scl52a3B | 10.96700804 | 8.10E-15 | novel_53 |
| HDH_G16512 | Sulfotransferase 1A1 (Sult1A1) | 10.29513478 | 3.53E-10 | novel_18 |
| HDH_G29406 | Transcription factor Hes4A (Hes4A) | 11.25346065 | 2.90E-16 | novel_42 |
| HDH_G24939 | Transforming growth factor-beta-induced protein ig-h3 (Tgfbi) | 11.38049792 | 2.95E-13 | novel_32 |
| HDH_G12983 | Vitelline envelope sperm lysin receptor (Verl) | 15.48186129 | 1.00E-33 | novel_90 |
| HDH_G17385 | Verl | 19.59400277 | 8.33E-54 | novel_53 |
| HDH_G15719 | Verl | 18.78354843 | 9.64E-49 | novel_1, novel_8 |
| HDH_G14936 | Xanthine dehydrogenase (Xdh) | 10.06294149 | 1.01E-09 | novel_53 |
| HDH_G06239 | Xylan 1,4-beta-xylosidase (Xyl3A) | 11.28666914 | 2.85E-11 | novel_19, novel_8 |
| HDH_G31217 | Ankyrin repeats (AR) | −13.11878319 | 9.27E-25 | hdh-miR-1994a, hdh-miR-1994b |
| HDH_G04117 | E3 ubiquitin-protein ligase Dcst1 (Dcst1) | −10.29220265 | 5.34E-15 | novel_57 |
| HDH_G27469 | Inactive serine/threonine-protein kinase Tex14 (Tex14) | −12.05306941 | 3.31E-137 | novel_120 |
| HDH_G02229 | Leucine rich repeat (LRR) | −13.50459197 | 1.33E-26 | novel_120 |
| HDH_G08545 | RING-variant domain | −14.2614608 | 5.53E-29 | novel_6 |
| HDH_G05810 | Stabilizer of axonemal microtubules 4 (Saxo4) | −10.25719851 | 4.38E-55 | hdh-miR-31 |
| HDH_G26330 | Testis-specific serine/threonine-protein kinase 1 (Tssk1B) | −10.66140224 | 1.63E-58 | novel_120, novel_6 |
| HDH_G12498 | Transcription factor Sox-30 (Sox-30) | −13.81154742 | 5.52E-27 | novel_120 |
| Primer | Sequence (5′-3′) |
|---|---|
| Saxo4-qF | GTTCAAGGGTCTTCGAGGCA |
| Saxo4-qR | CGTGAAATAACCGGGCTGCT |
| Tssk1B-qF | CCACCATTCTGACCATCCCT |
| Tssk1B-qR | CCTCCTTCTTCTTCCTCTCGG |
| AR-qF | GAAAATGGGATCCTCGGCTG |
| AR-qR | TTACCCCTCACCGCTTGAAT |
| Verl-qF | GACTTCCGGGCCATCTGTAA |
| Verl-qR | ACGTTGGAGTTCTGTCTCCT |
| Man2b1-qF | GGAGGCTAAAGCGCTCATCA |
| Man2b1-qR | CCGAACTTTTGCTCAGCGTT |
| β-actin-qF | GGTATCCTCACCCTCAAGT |
| β-actin-qR | GGGTCATCTTTTCACGGTTG |
| hdh-miR-92 | AATTGCACTTGTCCCGGCCTGC |
| novel_1 | AATTGCACTCGTCCCGGCCTGCAA |
| hdh-miR-1994b | TGAGACAGTGTGTCCTCCCTCA |
| hdh-miR-31 | AGGCAAGATGTTGGCATAGCT |
| novel_6 | TCGAGGAAGTAGAAGACCTTGACGT |
| Probe Name | Digestive Condition | Probe Sequence (5′-3′) | Repair Condition | Probe Concentration | Hybridization Temperature | Name of the Corresponding Signal Probe |
|---|---|---|---|---|---|---|
| Verl | Proteinase K was digested at 40 °C for 10 min. | TATGCAGGTAATGGTGCCGTCAAG /GTAATCGACCCTTCCGGTTCCAAG /TTGCGAATCTTGTGTTCGTCCTCG | Tissue sections were kept in a citric acid solution (pH 6.0) repair box and incubated in a water bath at 90 °C for 48 min. | 500 nM | 40 °C | CY3 |
| Tssk1B | Proteinase K was digested at 40 °C for 10 min. | CTCGTGAAGGACCAGCAGAGAAGC /CATAGGACCCACAGAAGGTCTCCATC /GTTCTCGGCAGTCTTTAGACACCTGC | Tissue sections were kept in a citric acid solution (pH 6.0) repair box and incubated in a water bath at 90 °C for 48 min. | 500 nM | 40 °C | FAM (488) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhou, M.; She, Z.; Chen, J.; Ke, C. Integrated Analysis of mRNA and miRNA Associated with Reproduction in Female and Male Gonads in Abalone (Haliotis discus hannai). Int. J. Mol. Sci. 2025, 26, 3235. https://doi.org/10.3390/ijms26073235
Huang J, Zhou M, She Z, Chen J, Ke C. Integrated Analysis of mRNA and miRNA Associated with Reproduction in Female and Male Gonads in Abalone (Haliotis discus hannai). International Journal of Molecular Sciences. 2025; 26(7):3235. https://doi.org/10.3390/ijms26073235
Chicago/Turabian StyleHuang, Jianfang, Mingcan Zhou, Zhenghan She, Jianming Chen, and Caihuan Ke. 2025. "Integrated Analysis of mRNA and miRNA Associated with Reproduction in Female and Male Gonads in Abalone (Haliotis discus hannai)" International Journal of Molecular Sciences 26, no. 7: 3235. https://doi.org/10.3390/ijms26073235
APA StyleHuang, J., Zhou, M., She, Z., Chen, J., & Ke, C. (2025). Integrated Analysis of mRNA and miRNA Associated with Reproduction in Female and Male Gonads in Abalone (Haliotis discus hannai). International Journal of Molecular Sciences, 26(7), 3235. https://doi.org/10.3390/ijms26073235





