Identification of Long-Distance Mobile mRNAs Responding to Drought Stress in Heterografted Tomato Plants
Abstract
:1. Introduction
2. Results
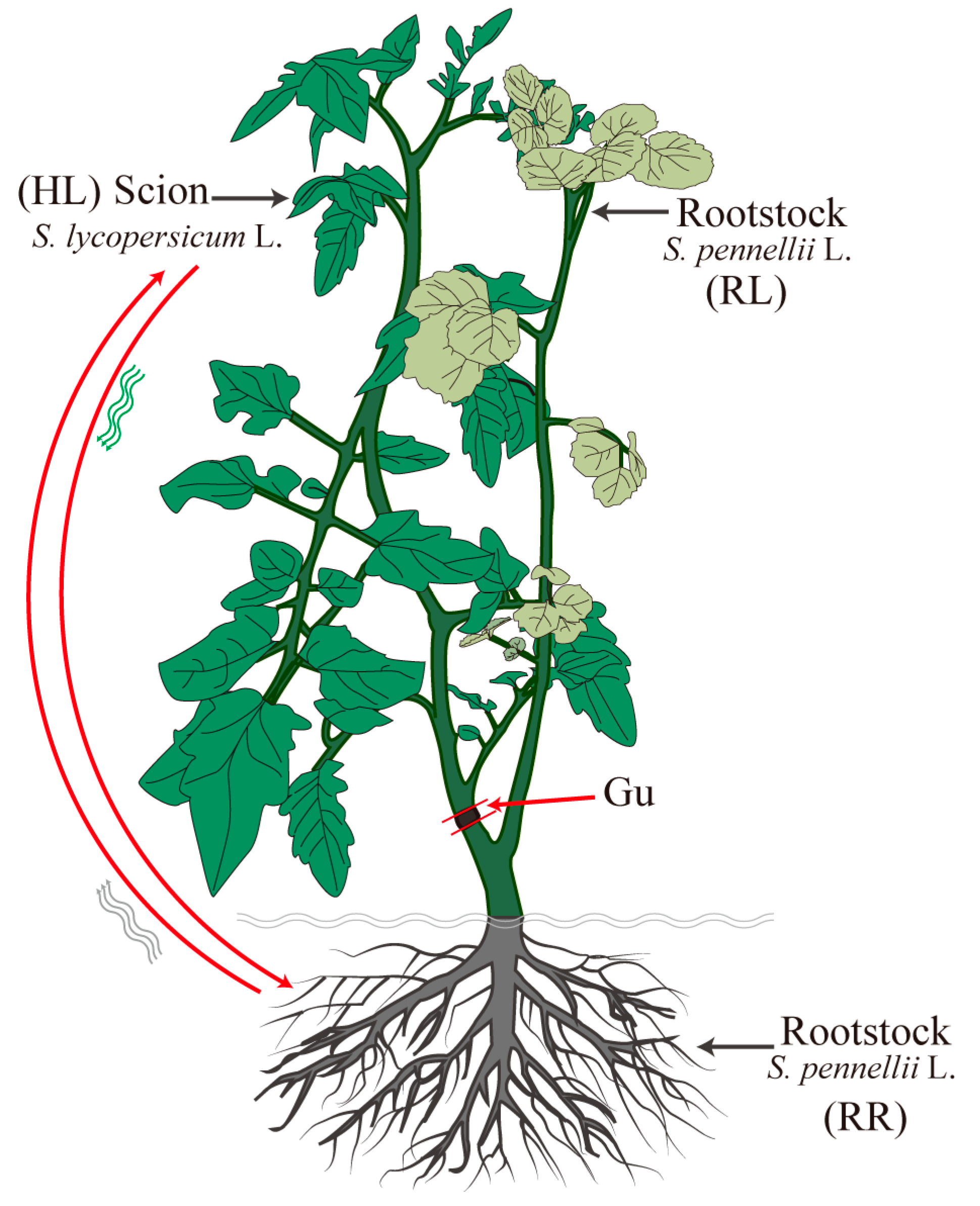
2.1. A Heterograft System for Identifying the Long-Distance Mobile mRNAs
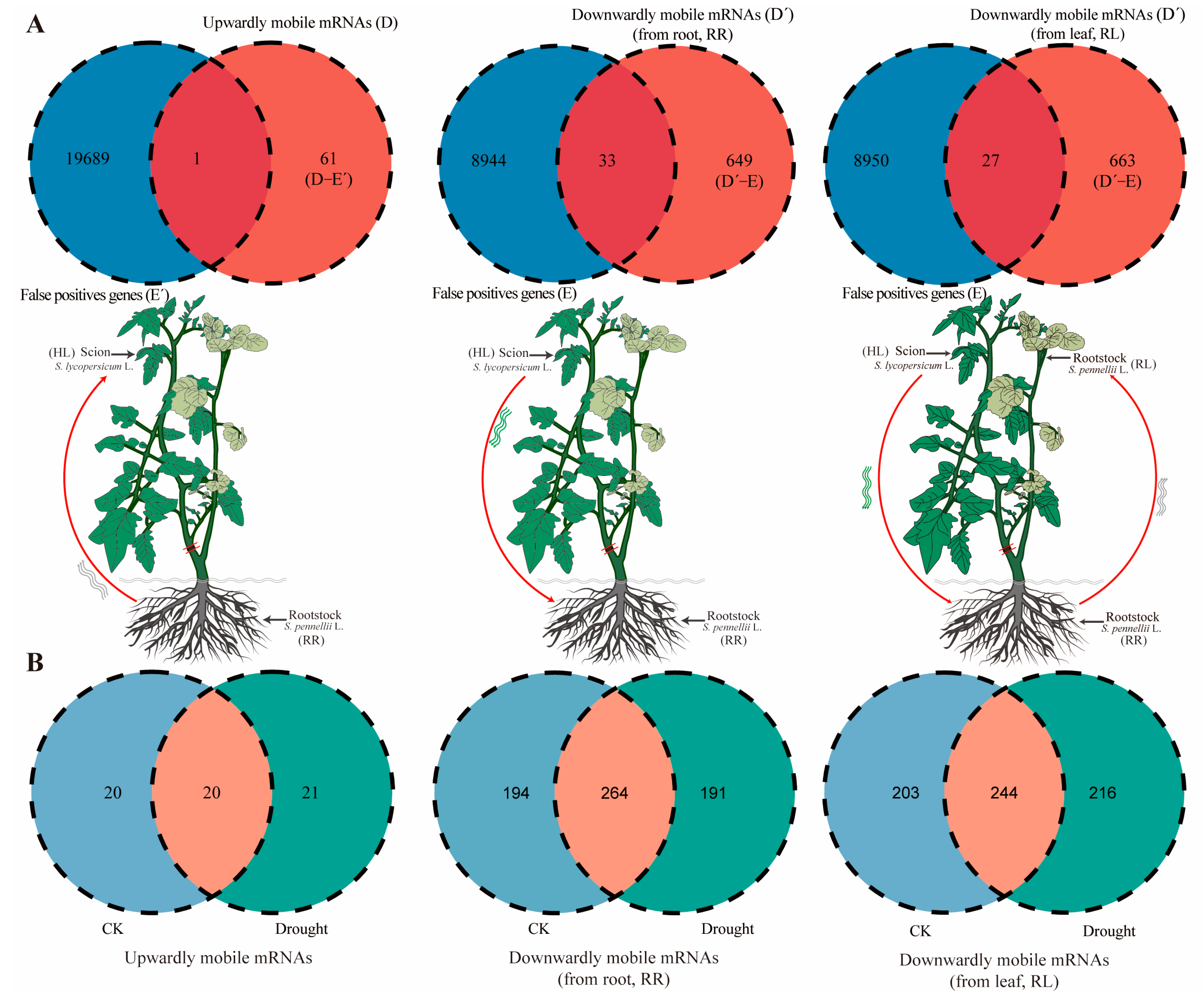
2.2. Long-Distance Movement of mRNAs in Scion to Rootstock
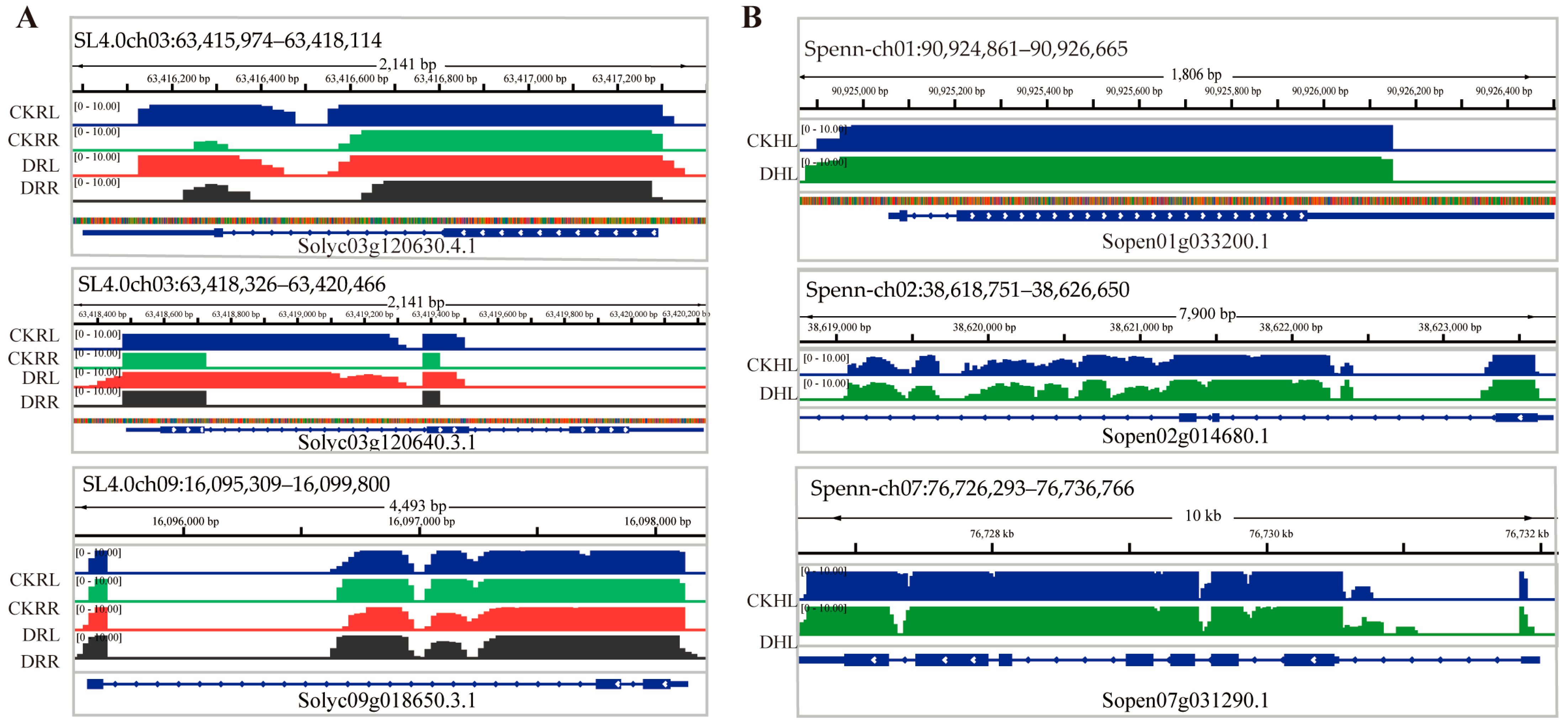
2.3. mRNAs Move from Leaf (Scion) to Root to Leaf (Rootstock)
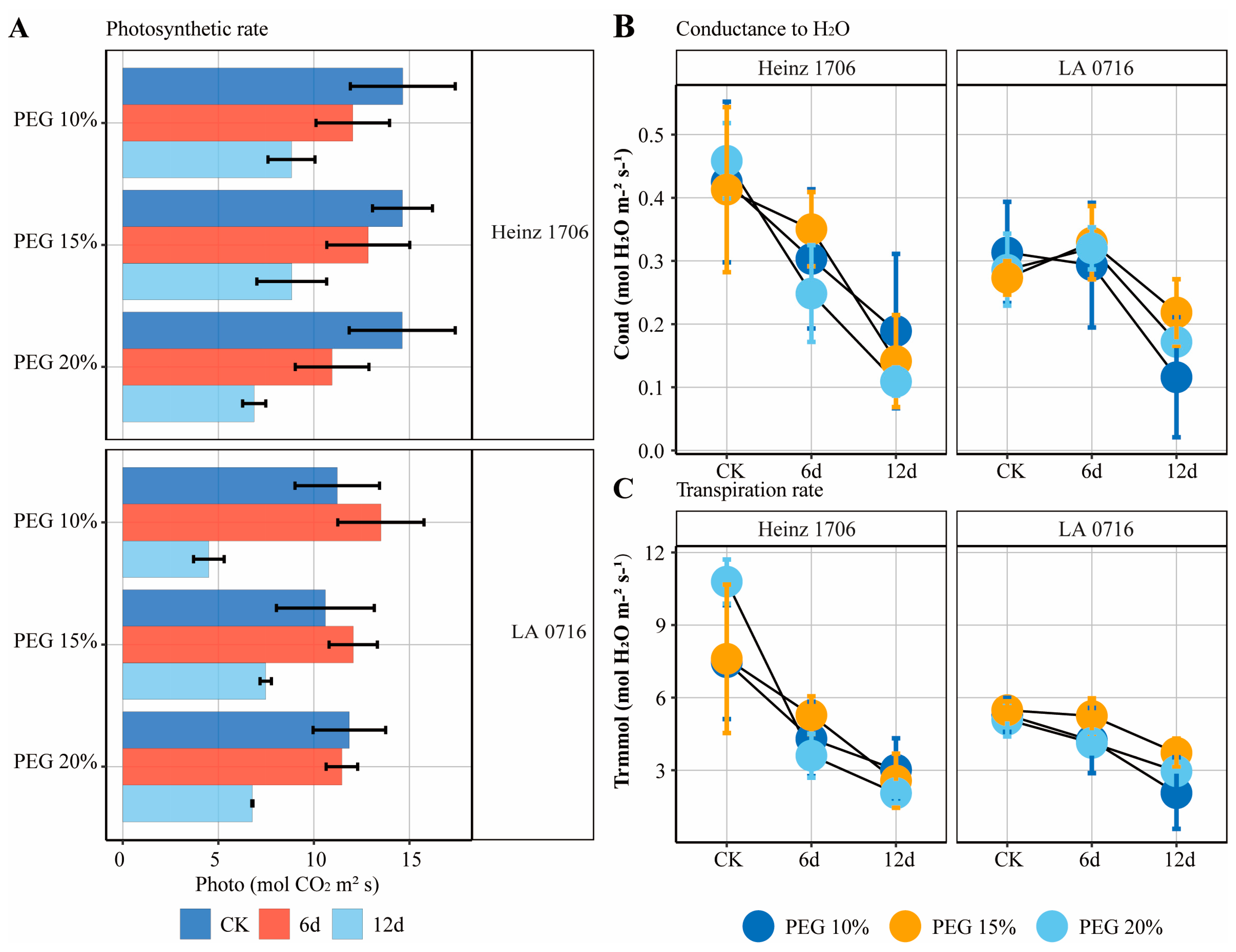
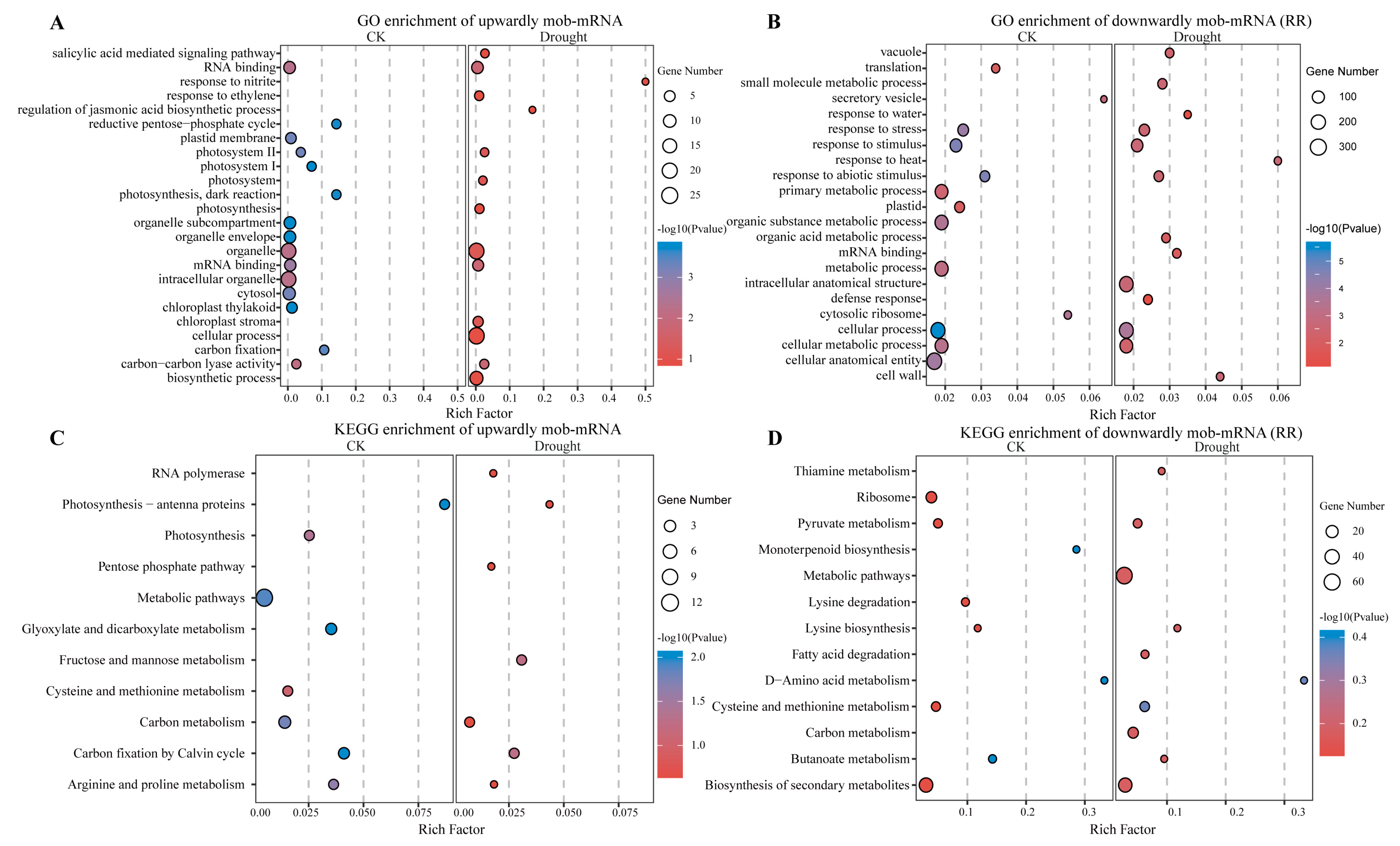
2.4. Effects of mRNAs Migration Under Drought Stress
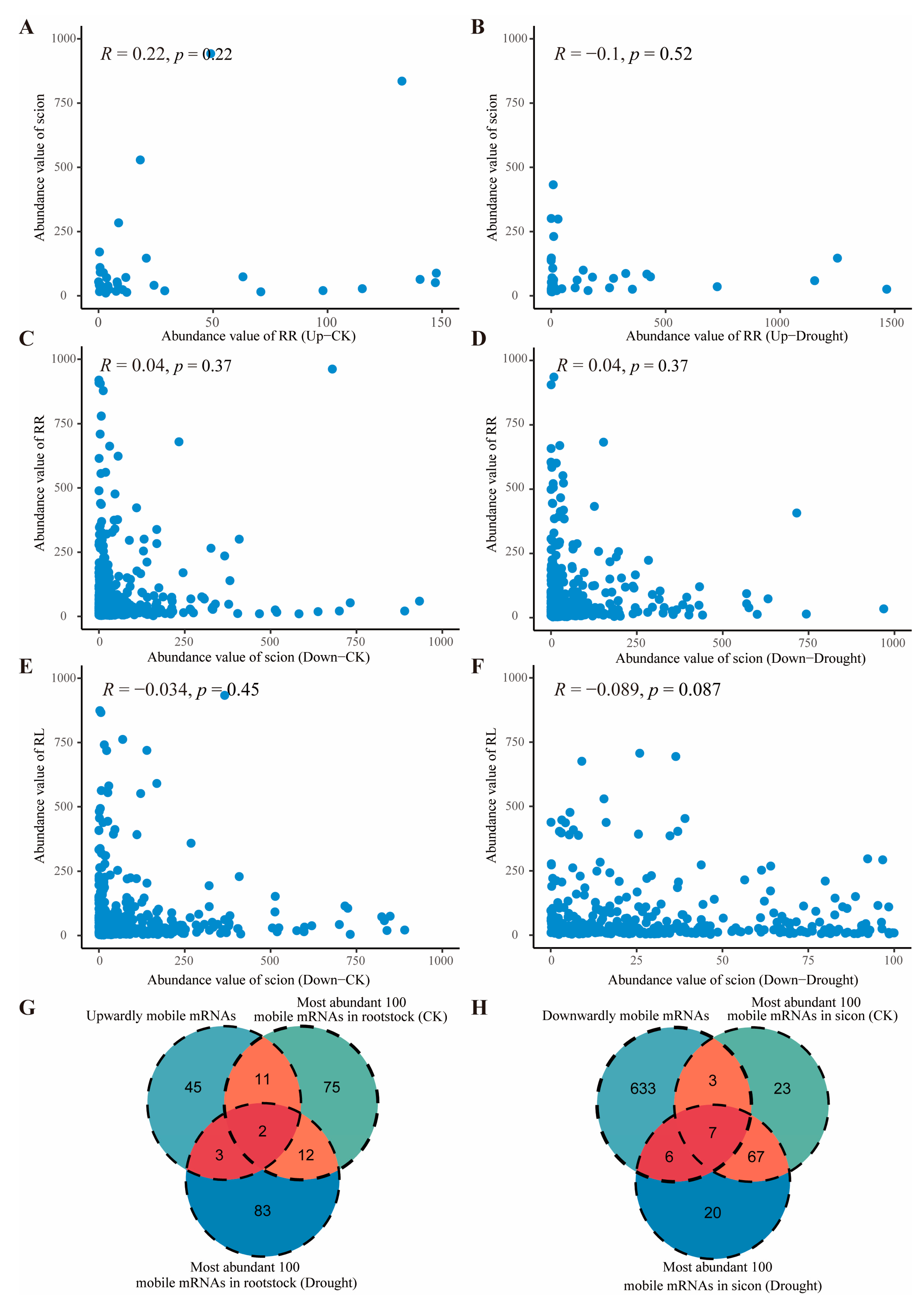
2.5. Movement of mob-mRNAs Is Independent of Abundance in Source to Sink Tissue
2.6. RNA-Binding Proteins Are Conserved
3. Discussion
4. Materials and Methods
4.1. Plant Grafting and Drought Stress Treatment
4.2. Photosynthetic Rate Determination
4.3. Sample Collection
4.4. Total RNA Extraction, RNA-Seq Library Construction and Analysis
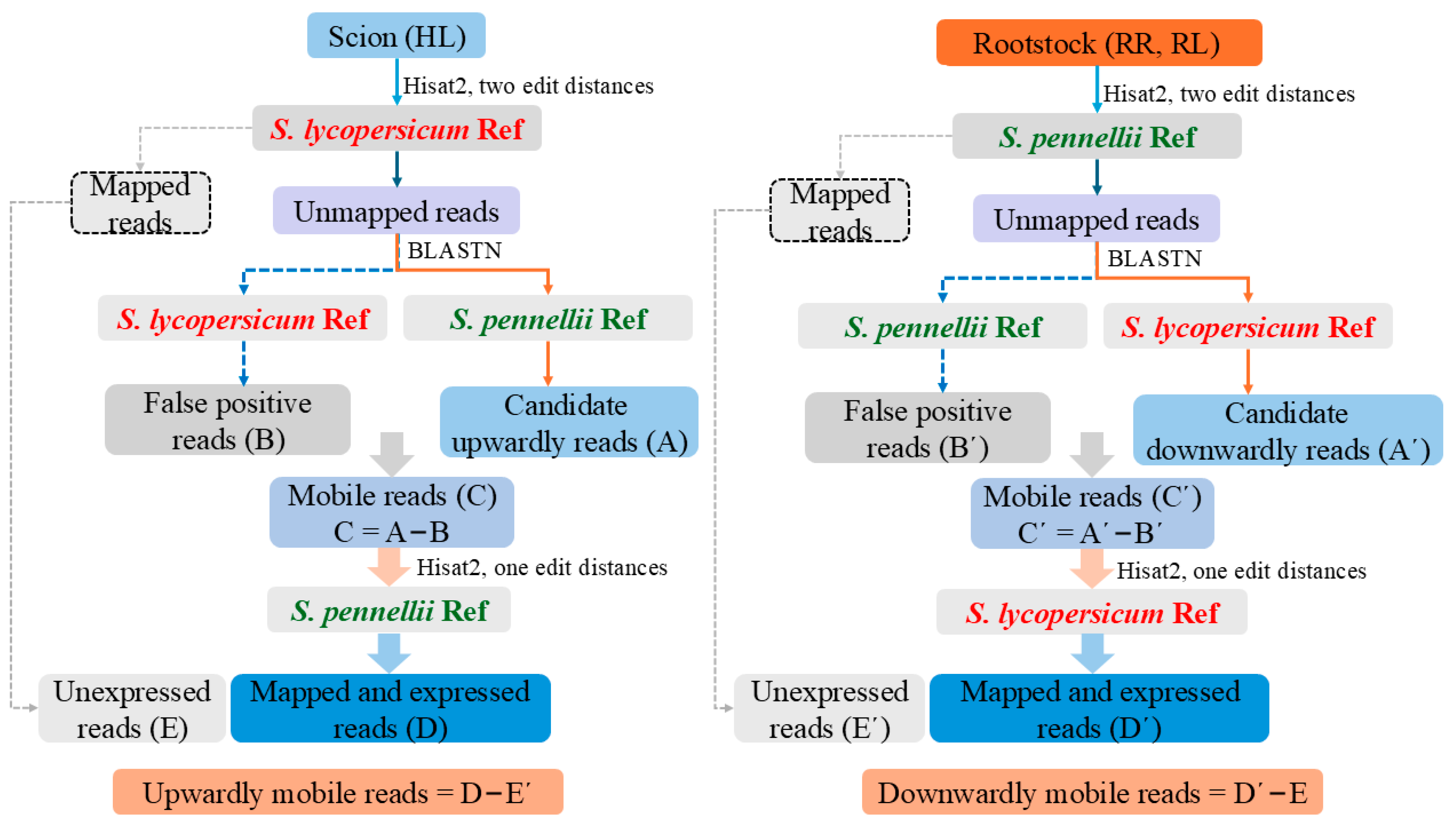
4.5. Identification of Mobile mRNAs
4.6. Functional Enrichment Analysis of Mobile mRNAs
4.7. Analysis of RBPs Structural Domain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar]
- Klee, H.J.; Tieman, D.M. The genetics of fruit flavour preferences. Nat. Rev. Genet. 2018, 19, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Imtiaz, M.; Kong, Q.; Cheng, F.; Ahmed, W.; Huang, Y.; Bie, Z. Grafting: A technique to modify Ion accumulation in horticultural crops. Front. Plant. Sci. 2016, 7, 1457. [Google Scholar] [CrossRef] [PubMed]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; von Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Gaion, L.A.; Braz, L.T.; Carvalho, R.F. Grafting in vegetable crops: A great technique for agriculture. Int. J. Veg. Sci. 2018, 24, 85–102. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 2018, 4, 869–878. [Google Scholar] [CrossRef]
- Feng, M.; Augstein, F.; Kareem, A.; Melnyk, C.W. Plant grafting: Molecular mechanisms and applications. Mol. Plant 2024, 17, 75–91. [Google Scholar] [CrossRef]
- Albacete, A.; Martínez-Andújar, C.; Martínez-Pérez, A.; Thompson, A.J.; Dodd, I.C.; Pérez-Alfocea, F. Unravelling rootstock×scion interactions to improve food security. J. Exp. Bot. 2015, 66, 2211–2226. [Google Scholar] [CrossRef]
- Miao, C.; Zhang, Y.; Cui, J.; Zhang, H.; Wang, H.; Jin, H.; Lu, P.; He, L.; Zhou, Q.; Yu, J.; et al. An enhanced interaction of graft and exogenous SA on photosynthesis, phytohormone, and transcriptome analysis in yomato under salinity stress. Int. J. Mol. Sci. 2024, 25, 10799. [Google Scholar] [CrossRef]
- Aydin, A. Effects of grafting with wild tomato (Solanum pimpinellifolium and Solanum habrochaites) rootstocks on growth and leaf mineral accumulation in salt stress. Hortic. Environ. Biotechnol. 2024, 65, 785–801. [Google Scholar] [CrossRef]
- Wang, R.; Yang, Y.; Xu, K.; Wang, T.; Elsadek, M.A.; Yuan, L.; Hu, Z.; Lv, Y.; Yuan, X.; Chen, X. Multi-omics analysis reveals improvement of tomato quality by grafting on goji rootstock. Food Qual. Saf. 2024, 8, fyae023. [Google Scholar]
- Westwood, J.H. RNA transport: Delivering the message. Nat. Plants 2015, 1, 15038. [Google Scholar] [PubMed]
- Kehr, J.; Kragler, F. Long distance RNA movement. New Phytol. 2018, 218, 29–40. [Google Scholar]
- Reagan, B.C.; Ganusova, E.E.; Fernandez, J.C.; McCray, T.N.; Burch-Smith, T.M. RNA on the move: The plasmodesmata perspective. Plant Sci. 2018, 275, 1–10. [Google Scholar] [PubMed]
- Maizel, A.; Markmann, K.; Timmermans, M.; Wachter, A. To move or not to move: Roles and specificity of plant RNA mobility. Curr. Opin. Plant Biol. 2020, 57, 52–60. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Nogueira, F.T.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C. Pattern formation via small RNA mobility. Genes Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, L.; Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009, 150, 378–387. [Google Scholar]
- Kim, G.; LeBlanc, M.L.; Wafula, E.K.; dePamphilis, C.W.; Westwood, J.H. Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 2014, 345, 808–811. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, Y.; Ham, B.K.; Chen, J.; Yoshida, A.; Kochian, L.V.; Fei, Z.; Lucas, W.J. Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants 2016, 2, 16033. [Google Scholar]
- Xia, C.; Zheng, Y.; Huang, J.; Zhou, X.; Li, R.; Zha, M.; Wang, S.; Huang, Z.; Lan, H.; Turgeon, R.; et al. Elucidation of the mechanisms of long-distance mRNA movement in a Nicotiana Benthamiana/Tomato heterograft system. Plant Physiol. 2018, 177, 745–758. [Google Scholar] [CrossRef]
- Xia, C.; Huang, J.; Lan, H.; Zhang, C. Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts. Plants 2020, 9, 876. [Google Scholar] [CrossRef] [PubMed]
- Notaguchi, M.; Higashiyama, T.; Suzuki, T. Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 2015, 56, 311–321. [Google Scholar] [CrossRef]
- Davoudi, M.; Song, M.; Zhang, M.; Chen, J.; Lou, Q. Long-distance control of pumpkin rootstock over cucumber scion under drought stress as revealed by transcriptome sequencing and mobile mRNAs identifications. Hortic. Res. 2022, 9, uhab033. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Xing, N.; Wu, X.; Wu, X.; Wang, B.; Lu, Z.; Xu, P.; Tao, Y.; Li, G.; et al. A universal pipeline for mobile mRNA detection and insights into heterografting advantages under chilling stress. Hortic. Res. 2020, 7, 13. [Google Scholar] [CrossRef]
- Roney, J.K.; Khatibi, P.A.; Westwood, J.H. Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol. 2007, 143, 1037–1043. [Google Scholar] [CrossRef]
- Westwood, J.H.; Roney, J.K.; Khatibi, P.A.; Stromberg, V.K. RNA translocation between parasitic plants and their hosts. Pest. Manag Sci. 2009, 65, 533–539. [Google Scholar] [CrossRef]
- LeBlanc, M.; Kim, G.; Patel, B.; Stromberg, V.; Westwood, J. Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant cuscuta pentagona. New Phytol. 2013, 200, 1225–1233. [Google Scholar] [CrossRef]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Xu, Z.; Ma, H.; Hao, Y.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; Han, Z.; Wu, T. Characteristics of long-distance mobile mRNAs from shoot to root in grafted plant species. Hortic. Plant J. 2024, 10, 25–37. [Google Scholar] [CrossRef]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 727. [Google Scholar] [CrossRef]
- Stahl, Y.; Faulkner, C. Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci. 2016, 21, 450–459. [Google Scholar] [PubMed]
- Kitagawa, M.; Jackson, D. Plasmodesmata-mediated Cell-to-Cell communication in the shoot apical meristem: How stem cells talk. Plants 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Doering-Saad, C.; Newbury, H.J.; Bale, J.S.; Pritchard, J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 2002, 53, 631–637. [Google Scholar]
- Ivashikina, N.; Deeken, R.; Ache, P.; Kranz, E.; Pommerrenig, B.; Sauer, N.; Hedrich, R. Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J. 2003, 36, 931–945. [Google Scholar] [CrossRef]
- Omid, A.; Keilin, T.; Glass, A.; Leshkowitz, D.; Wolf, S. Characterization of phloem-sap transcription profile in melon plants. J. Exp. Bot. 2007, 58, 3645–3656. [Google Scholar] [CrossRef] [PubMed]
- Kanehira, A.; Yamada, K.; Iwaya, T.; Tsuwamoto, R.; Kasai, A.; Nakazono, M.; Harada, T. Apple phloem cells contain some mRNAs transported over long distances. Tree Genet. Genomes 2010, 6, 635–642. [Google Scholar] [CrossRef]
- Luo, K.R.; Huang, N.C.; Yu, T.S. Selective Targeting of Mobile mRNAs to Plasmodesmata for Cell-to-Cell Movement. Plant Physiol. 2018, 177, 604–614. [Google Scholar]
- Zhang, C.; Yu, X.; Ayre, B.G.; Turgeon, R. The origin and composition of cucurbit “phloem” exudate. Plant Physiol. 2012, 158, 1873–1882. [Google Scholar]
- Hannapel, D.J.; Banerjee, A.K. Multiple Mobile mRNA Signals Regulate Tuber Development in Potato. Plants 2017, 6, 8. [Google Scholar] [CrossRef]
- Rodriguez-Villalon, A. Wiring a plant: Genetic networks for phloem formation in Arabidopsis thaliana roots. New Phytol. 2016, 210, 45–50. [Google Scholar]
- Kim, M.; Canio, W.; Kessler, S.; Sinha, N. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 2001, 293, 287–289. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Chatterjee, M.; Yu, Y.; Suh, S.G.; Miller, W.A.; Hannapel, D.J. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 2006, 18, 3443–3457. [Google Scholar] [PubMed]
- Calderwood, A.; Kopriva, S.; Morris, R.J. Transcript Abundance Explains mRNA Mobility Data in Arabidopsis thaliana. Plant Cell 2016, 28, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Paultre, D.S.G.; Gustin, M.P.; Molnar, A.; Oparka, K.J. Lost in transit: Long-distance trafficking and phloem unloading of protein signals in arabidopsis homografts. Plant Cell 2016, 28, 2016–2025. [Google Scholar]
- Cléry, A.; Blatter, M.; Allain, F.H. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Klug, A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 2010, 43, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, K.; Zhang, D.; Dan, Z.; Bao, L.; Mu, W.; Zhang, J. Identification of Long-Distance Mobile mRNAs Responding to Drought Stress in Heterografted Tomato Plants. Int. J. Mol. Sci. 2025, 26, 3168. https://doi.org/10.3390/ijms26073168
Du K, Zhang D, Dan Z, Bao L, Mu W, Zhang J. Identification of Long-Distance Mobile mRNAs Responding to Drought Stress in Heterografted Tomato Plants. International Journal of Molecular Sciences. 2025; 26(7):3168. https://doi.org/10.3390/ijms26073168
Chicago/Turabian StyleDu, Kanghua, Da Zhang, Zhong Dan, Lingfeng Bao, Wanfu Mu, and Jie Zhang. 2025. "Identification of Long-Distance Mobile mRNAs Responding to Drought Stress in Heterografted Tomato Plants" International Journal of Molecular Sciences 26, no. 7: 3168. https://doi.org/10.3390/ijms26073168
APA StyleDu, K., Zhang, D., Dan, Z., Bao, L., Mu, W., & Zhang, J. (2025). Identification of Long-Distance Mobile mRNAs Responding to Drought Stress in Heterografted Tomato Plants. International Journal of Molecular Sciences, 26(7), 3168. https://doi.org/10.3390/ijms26073168





