From Gene to Plate: Molecular Insights into and Health Implications of Rice (Oryza sativa L.) Grain Protein
Abstract
1. Introduction
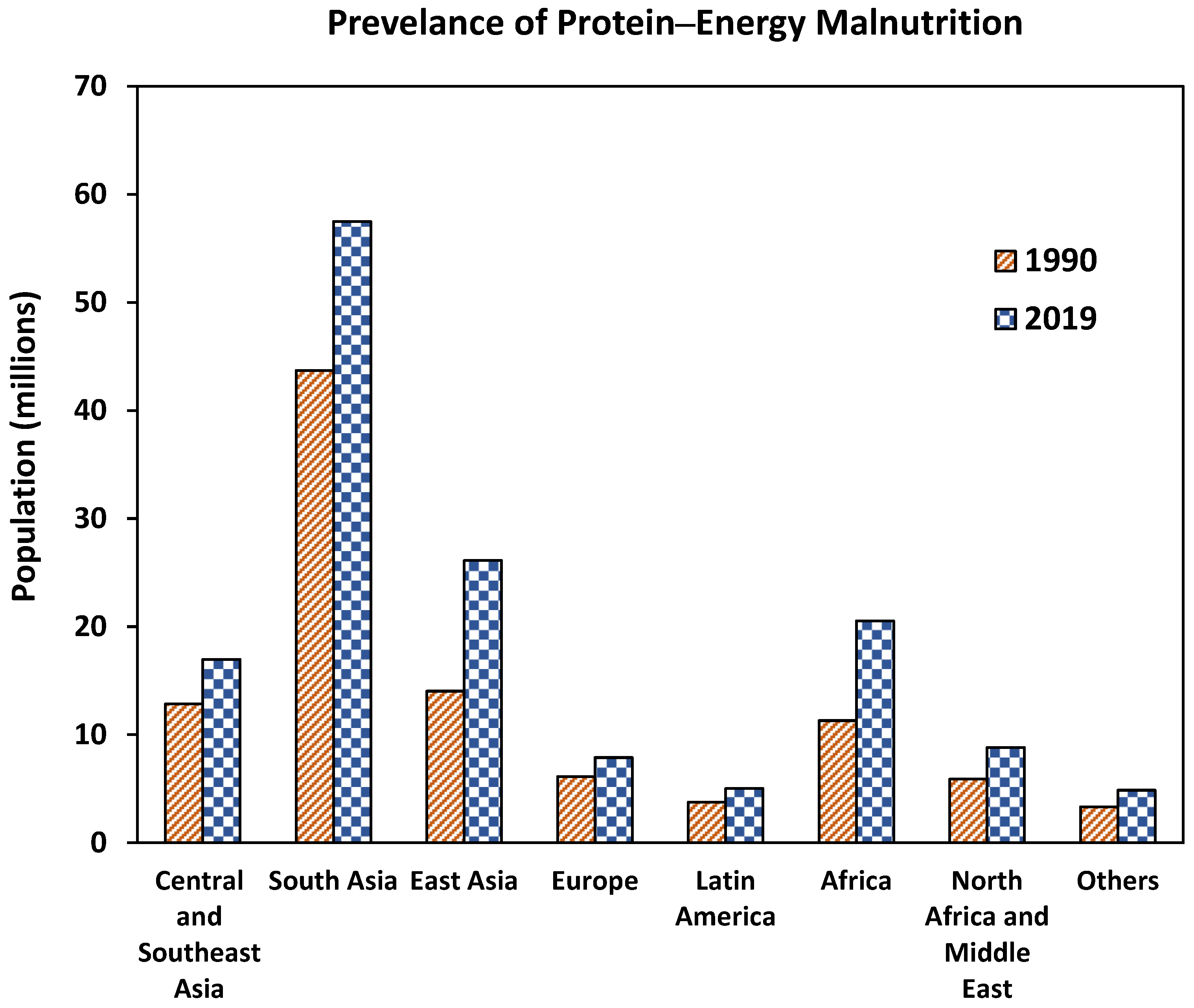
2. Protein–Energy Malnutrition (PEM)
3. Protein Sources, Constitution, and Chemistry
3.1. Available Protein Sources
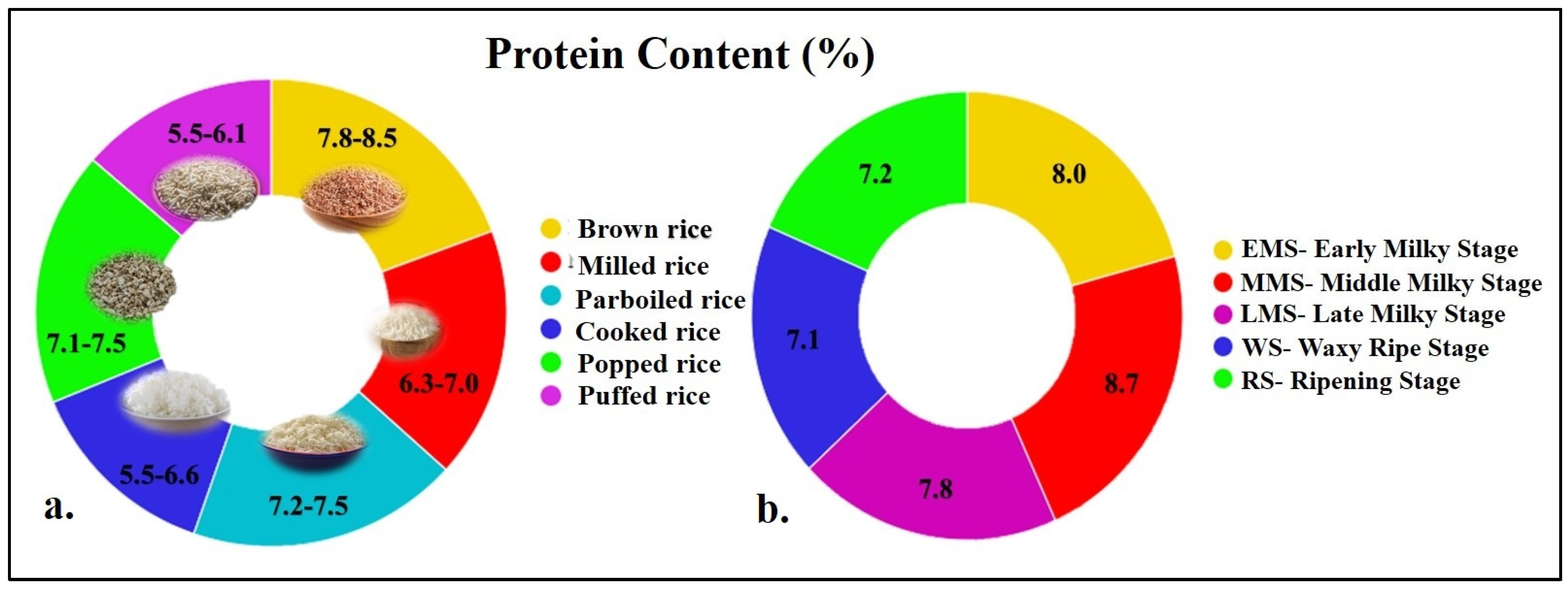
3.2. Protein Content and Quality in Rice
3.3. Composition and Distribution of Grain Protein in Rice
3.3.1. Albumins
3.3.2. Globulins
3.3.3. Prolamins
3.3.4. Glutelins
3.4. Protein Structure
3.5. Amino Acid Composition
3.6. Surface Hydrophobicity
3.7. Impact of Changes in Protein Structure
3.7.1. Solubility
3.7.2. Foaming Capacity (FC) and Foaming Stability (FS)
3.7.3. Emulsifying Capacity and Emulsion Stability
3.7.4. Protein Digestibility
4. Factors Influencing Grain Protein Content in Rice
4.1. Impact of Temperature
4.2. Impact of Carbon Dioxide (CO2)
4.3. Impact of Management Practices
4.4. Impact of Stress Conditions
4.5. Impact of Post-Harvest Processing
5. Genomic Regions Affecting Grain Protein Content, Protein Fractions, Amino Acids, and Protein Index
5.1. QTLs Identified for GPC and Protein Components in Rice
5.2. Candidate Genes Controlling GPC and Protein Components in Rice
6. Health Benefits of Rice Grain Protein and Its Derivatives
6.1. Antioxidant Activity
6.2. Hypocholesterolemic Response
6.3. Anti-Cancer Activity
7. Applications of Rice Grain Protein
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alam, M.; Lou, G.; Abbas, W.; Osti, R.; Ahmad, A.; Bista, S.; Ahiakpa, J.K.; Yuqing He, Y. Improving rice grain quality through ecotype breeding for enhancing food and nutritional security in Asia–Pacific region. Rice 2024, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. 2024. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 17 December 2024).
- Bagchi, T.B.; Chattopadhyay, K.; Sarkar, S.; Sanghamitra, P.; Kumar, A.; Basak, N.; Sivashankari, M.; Priyadarsini, S.; Pathak, H. Rice Products and Their Nutritional Status. Res. Bull. 2020, 21, 1–20. [Google Scholar]
- Ning, H.F.; Liu, Z.G.; Wang, Q.S.; Lin, Z.M.; Chen, S.J.; Li, G.H.; Wang, S.H.; Ding, Y.F. Effect of nitrogen fertilizer application on grain phytic acid and protein concentrations in japonica rice and its variations with genotypes. J. Cereal Sci. 2009, 50, 49–55. [Google Scholar] [CrossRef]
- Stevens, G.A.; Beal, T.; Mbuya, M.N.N.; Luo, H.; Neufeld, L.M. Micronutrient deficiencies among preschool-aged children and women of reproductive age worldwide: A pooled analysis of individual-level data from population-representative surveys. Lancet Glob. Health 2022, 10, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Lonnie, M.; Laurie, I.; Myers, M.; Horgan, G.; Russell, W.R.; Johnstone, A.M. Exploring health-promoting attributes of plant proteins as a functional ingredient for the food sector: A systematic review of human interventional studies. Nutrients 2020, 12, 2291. [Google Scholar] [CrossRef]
- Gardner, C.D.; Hartle, J.C.; Garrett, R.D.; Oringa, L.C.; Wasserman, A.S. Maximizing the intersection of human health and the health of the environment with regard to the amount and type of protein produced and consumed in the United States. Nutr. Rev. 2019, 77, 197–215. [Google Scholar] [CrossRef]
- Rosato, V.; Edefonti, V.; Bravi, F.; Bosetti, C.; Bertuccio, P.; Talamini, R.; Dal Maso, L.; Montella, M.; Ferraroni, M.; La Vecchia, C.; et al. Nutrient-based dietary patterns and prostate cancer risk: A case-control study from Italy. Cancer Causes Control 2014, 25, 525–532. [Google Scholar] [CrossRef]
- Larsson, S.C.; Orsini, N. Red meat and processed meat consumption and all-cause mortality: A meta-analysis. Am. J. Epidemiol. 2014, 17, 282–289. [Google Scholar] [CrossRef]
- Jannasch, F.; Kröger, J.; Schulze, M.B. Dietary patterns and type 2 diabetes: A systematic literature review and meta-analysis of prospective studies. J. Nutr. 2017, 147, 1174–1182. [Google Scholar] [CrossRef]
- Cui, K.; Liu, Y.; Zhu, L.; Mei, X.; Jin, P.; Luo, Y. Association between intake of red and processed meat and the risk of heart failure: A meta-analysis. BMC Public Health 2019, 19, 354. [Google Scholar] [CrossRef]
- Zhong, V.W.; Van Horn, L.; Greenland, P.; Carnethon, M.R.; Wilkins, J.T.; Lloyd-Jones, M.D.; Allen, N.B. Associations of processed meat, unprocessed red meat, poultry, or fish intake with incident cardiovascular disease and all-cause mortality. JAMA Intern. Med. 2020, 180, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ahmedna, M. Functional components of grape pomace: Their composition, biological properties and potential applications. Int. J. Food Sci. Technol. 2013, 48, 221–237. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/FBS (accessed on 19 August 2024).
- Scully, A.; Neacsu, M.; Ruddel, W.; Vaughan, N.; Fyfe, C.; Hudson, K.; Taylor, K.; Johnstone, A.M. Plant protein influence on appetite and food intake in healthy subjects. Proc. Nutr. Soc. 2017, 76, E44. [Google Scholar] [CrossRef]
- Gorissen, S.H.M.; Witard, O.C. Characterising the muscle anabolic potential of dairy, meat and plant-based protein sources in older adults. Proc. Nutr. Soc. 2018, 77, 20–31. [Google Scholar] [CrossRef]
- FAO; IFAD; UNICEF; WFP; WHO. The State of Food Security and Nutrition in the World 2024—Financing to End Hunger, Food Insecurity and Malnutrition in All Its Forms; FAO: Rome, Italy, 2024. [Google Scholar] [CrossRef]
- Crichton, M.; Craven, D.; Mackay, H.; Marx, W.; de van der Schueren, M.; Marshall, S. A systematic review, meta-analysis and meta-regression of the prevalence of protein-energy malnutrition: Associations with geographical region and sex. Age Ageing 2019, 48, 38–48. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Pu, Y.; Sun, M.; Zhao, Y.; Zhang, D.; Wang, X.; Li, Y.; Guo, D.; He, S. Global, regional, and national burden of protein–energy malnutrition: A systematic analysis for the global burden of disease study. Nutrients 2022, 14, 2592. [Google Scholar] [CrossRef]
- WHO. Levels and Trends in Child Malnutrition: UNICEF/WHO/The World Bank Group Joint Child Malnutrition Estimates: Key Findings of the 2021 Edition. Available online: https://www.who.int/publications/i/item/9789240025257 (accessed on 19 August 2024).
- Raynaud-Simon, A.; Revel-Delhom, C.; Hébuterne, X. Clinical practice guidelines from the French health high authority: Nutritional support strategy in protein-energy malnutrition in the elderly. Clin. Nutr. 2011, 30, 312–319. [Google Scholar] [CrossRef]
- Amagliani, L.; O’Regan, J.; Kelly, A.L.; O’Mahony, J.A. The composition, extraction, functionality and applications of rice proteins: A review. Trends Food Sci. Technol. 2017, 64, 1–12. [Google Scholar] [CrossRef]
- Champagne, E.T.; Wood, D.F.; Juliano, B.O.; Bechtel, D.B. The rice grain and its gross composition. In Rice: Chemistry and Technology, 3rd ed.; Champagne, E.T., Ed.; American Association of Cereal Chemists: St. Paul, MN, USA, 2004; pp. 77–107. [Google Scholar]
- Juliano, B.O. Polysaccharides, proteins, and lipids of rice. In Rice: Chemistry and Technology, 2nd ed.; Juliano, B.O., Ed.; American Association of Cereal Chemists: St. Paul, MN, USA, 1985; pp. 59–174. [Google Scholar]
- Paraman, I.; Wagner, M.E.; Rizvi, S.S. Micronutrient and protein-fortified whole grain puffed rice made by supercritical fluid extrusion. J. Agric. Food Chem. 2012, 60, 11188–11194. [Google Scholar] [CrossRef]
- Khan, H.; Devi, S.S.; Aparna, K.; Kameswari, S.L. Nutritional properties of three selected varieties of puffed rice: WGL44, WGL283 and RNR2458. Pharma. Innov. 2017, 6, 458–460. [Google Scholar]
- Adi, A.; Haryana, N.; Adhika, D.; Suwandi, A.; Rachmawati, H. Chemical and physical characterizations of cooked rice using different cooking methods. J. Food Nutri. Res. 2020, 8, 638–645. [Google Scholar]
- Itagi, H.; Sartagoda, K.J.D.; Pratap, V.; Roy, P.; Tiozon, R.N.; Regina, A.; Sreenivasulu, N. Popped rice with distinct nutraceutical properties. LWT 2023, 173, 114346. [Google Scholar] [CrossRef]
- Li, D.; Yao, M.; Yang, Y.; Wang, B.; Zhang, D.; Zhang, N. Changes of structure and functional properties of rice protein in the fresh edible rice during the seed development. Food Sci. Hum. Wellness 2023, 12, 1850–1860. [Google Scholar] [CrossRef]
- Mohanty, A.; Marndi, B.C.; Sharma, S.; Das, A. Biochemical characterization of two high protein rice cultivars from Assam rice collections. ORYZA-Int. J. Rice 2011, 48, 171–174. [Google Scholar]
- Banerjee, S.; Chandel, G.; Mandal, N.; Meena, B.M.; Saluja, T. Assessment of nutritive value in milled rice grain of some Indian rice landraces and their molecular characterization. Bangladesh J. Agril. Res. 2011, 36, 369–380. [Google Scholar] [CrossRef]
- Islam, M.Z.; Arifuzzaman, M.; Banik, S.; Hossain, M.A.; Ferdous, J.; Khalequzzaman, M.; Pittendrigh, B.R.; Tomita, M.; Ali, M.P. Mapping QTLs underpin nutrition components in aromatic rice germplasm. PLoS ONE 2020, 15, e0234395. [Google Scholar] [CrossRef]
- Gorinstein, S.; Pawelzik, E.; Delgado-Licon, E.; Haruenkit, R.; Weisz, M.; Trakhtenberg, S. Characterisation of pseudocereal and cereal proteins by protein and amino acid analyses. J. Sci. Food Agric. 2002, 82, 886–891. [Google Scholar] [CrossRef]
- Zilić, S.; Barać, M.; Pešić, M.; Dodig, D.; Ignjatović-Micić, D. Characterization of proteins from grain of different bread and durum wheat genotypes. Int. J. Mol. Sci. 2011, 12, 5878–5894. [Google Scholar] [CrossRef]
- Siddiqi, R.A.; Singh, T.P.; Rani, M.; Sogi, D.S.; Bhat, M.A. Diversity in grain, flour, amino acid composition, protein profiling, and proportion of total flour proteins of different wheat cultivars of North India. Front. Nutr. 2020, 7, 141. [Google Scholar] [CrossRef]
- Langyan, S.; Bhardwaj, R.; Kumari, J.; Jacob, S.R.; Bisht, I.S.; Pandravada, S.R.; Singh, A.; Singh, P.B.; Dar, Z.A.; Kumar, A.; et al. Nutritional diversity in native germplasm of maize collected from three different fragile ecosystems of India. Front. Nutr. 2022, 9, 812599. [Google Scholar] [CrossRef]
- Pantoa, T.; Baricevic-Jones, I.; Suwannaporn, P.; Kadowaki, M.; Kubota, M.; Roytrakul, S.; Mills, E.N.C. Young rice protein as a new source of low allergenic plant-base protein. J. Cereal Sci. 2020, 93, 102970. [Google Scholar] [CrossRef]
- Sharma, N.; Bhatia, S.; Chunduri, V.; Kaur, S.; Sharma, S.; Kapoor, P.; Kumari, A.; Garg, M. Pathogenesis of celiac disease and other gluten related disorders in wheat and strategies for mitigating them. Front. Nutr. 2020, 7, 6. [Google Scholar]
- Scibilia, J.; Pastorello, E.A.; Zisa, G.; Ottolenghi, A.; Ballmer-Weber, B.; Pravettoni, V.; Scovena, E.; Robino, A.; Ortolani, C. Maize food allergy: A double-blind placebo-controlled study. Clin. Exp. Allergy 2008, 38, 1943–1949. [Google Scholar] [CrossRef]
- Lou, G.; Bhat, M.A.; Tan, X.; Wang, Y.; He, Y. Research progress on the relationship between rice protein content and cooking and eating quality and its influencing factors. Seed Biol. 2023, 2, 16. [Google Scholar]
- Mandal, S.; Mandal, R.K. Seed storage proteins and approaches for improvement of their nutritional quality by genetic engineering. Curr. Sci. 2000, 79, 576–589. [Google Scholar]
- Zhao, L.X.; Pan, T.; Cai, C.H.; Wang, J.; Wei, C.X. Application of whole sections of mature cereal seeds to visualize the morphology of endosperm cell and starch and the distribution of storage protein. J. Cereal Sci. 2016, 71, 19–27. [Google Scholar]
- Long, X.; Guan, C.; Wang, L.; Jia, L.; Fu, X.; Lin, Q.; Huang, Z.; Liu, C. Rice storage proteins: Focus on composition, distribution, genetic improvement and effects on rice quality. Rice Sci. 2023, 30, 207–221. [Google Scholar]
- Tanaka, K.; Sugimoto, T.; Ogawa, M.; Kasai, Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric. Biol. Chem. 1980, 44, 1633–1639. [Google Scholar]
- Kubota, M.; Saito, Y.; Masumura, T.; Kumagai, T.; Watanabe, R.; Fujimura, S.; Kadowaki, M. Improvement in the in vivo digestibility of rice protein by alkali extraction is due to structural changes in prolamin/protein body-I particle. Biosci. Biotechnol. Biochem. 2010, 74, 614–619. [Google Scholar] [CrossRef]
- Iwasaki, T.; Shibuya, N.; Suzuki, T.; Chikubu, S. Gel filtration and electrophoresis of soluble rice proteins extracted from long, medium, and short grain varieties. Cereal Chem. 1982, 59, 192–195. [Google Scholar]
- Kawakatsu, T.; Takaiwa, F. Rice proteins and essential amino acids. In Rice, 4th ed.; Bao, J., Ed.; AACC International Press: Washington, DC, USA, 2019; pp. 109–130. [Google Scholar]
- Zhou, L.; Zhang, Y.; Zhao, C.; Lin, H.; Wang, Z.; Wu, F. Structural and functional properties of rice bran protein oxidized by peroxyl radicals. Int. J. Food Prop. 2017, 20, 1456–1467. [Google Scholar] [CrossRef]
- Adachi, T.; Izumi, H.; Yamada, T.; Tanaka, K.; Takeuchi, S.; Nakamura, R.; Matsuda, T. Gene structure and expression of rice seed allergenic proteins belonging to the α-amylase/trypsin inhibitor family. Plant Mol. Biol. 1993, 21, 239–248. [Google Scholar]
- Swamy, B.; Rahman, M.A.; Inabangan-Asilo, M.A.; Amparado, A.; Manito, C.; Chadha-Mohanty, P.; Reinke, R.; Slamet-Loedin, I.H. Advances in breeding for high grain zinc in rice. Rice 2016, 9, 49. [Google Scholar]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [PubMed]
- Jayaprakash, G.; Bains, A.; Chawla, P.; Fogarasi, M.; Fogarasi, S. A narrative review on rice proteins: Current scenario and food industrial application. Polymers 2022, 14, 3003. [Google Scholar] [CrossRef]
- Nakase, M.; Yamada, T.; Kira, T.; Yamaguchi, J.; Aoki, N.; Nakamura, R.; Matsuda, T.; Adachi, T. The same nuclear proteins bind to the 5′-flanking regions of genes for the rice seed storage protein: 16 kDa albumin, 13 kDa prolamin and type II glutelin. Plant Mol. Biol. 1996, 32, 621–630. [Google Scholar]
- Sun, J.L.; Nakagawa, H.; Karita, S.; Ohmiya, K.; Hattori, T. Rice embryo globulins: Amino-terminal amino acid sequences, cDNA cloning and expression. Plant Cell Physiol. 1996, 37, 612–620. [Google Scholar] [PubMed]
- Padhye, V.W.; Salunkhe, D.K. Extraction and characterization of rice proteins. Cereal Chem. 1979, 56, 389. [Google Scholar]
- He, W.; Wang, L.; Lin, Q.L.; Yu, F. Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J. Integr. Plant Biol. 2021, 63, 1999–2019. [Google Scholar]
- Hibino, T.; Kidzu, K.; Masumura, T.; Ohtsuki, K.; Tanaka, K.; Kawabat, M.; Fujii, S. Amino acid composition of rice prolamin polypeptides. Agric. Biol. Chem. 1989, 53, 513–518. [Google Scholar]
- Xu, J.H.; Messing, J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 2009, 119, 1397–1412. [Google Scholar]
- Yamagata, H.; Tanaka, K.; Kasai, Z. Evidence for a precursor form of rice glutelin subunits. Agric. Biol. Chem. 1982, 46, 321–322. [Google Scholar]
- Sugimoto, T.; Tanaka, K.; Kasai, Z. Molecular species in the protein body II (PB-II) of developing rice endosperm. Agric. Biol. Chem. 1986, 50, 3031–3035. [Google Scholar]
- Ghanghas, N.; MT, M.; Sharma, S.; Prabhakar, P.K. Classification, composition, extraction, functional modification, and application of rice (Oryza sativa) seed protein: A comprehensive review. Food Rev. Int. 2022, 38, 354–383. [Google Scholar]
- Kawakatsu, T.; Yamamoto, M.P.; Hirose, S.; Yano, M.; Takaiwa, F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J. Exp. Bot. 2008, 59, 4233–4245. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, M.; Ouwerkerk, P.B.F. Molecular and environmental factors determining grain quality in rice. Food Energy Sec. 2012, 1, 111–132. [Google Scholar]
- Malecki, J.; Muszynski, S.; Solowiej, B.G. Proteins in food systems—Bionanomaterials, conventional and unconventional sources, functional properties, and development opportunities. Polymers 2021, 13, 2506. [Google Scholar] [CrossRef]
- Larkins, B.A.; Wu, Y.; Song, R.; Messing, J. Maize seed storage proteins. In Maize Kernel Development; Larkins, B.A., Ed.; CABI International: Wallingford, UK, 2017; p. 175. [Google Scholar]
- Kimball, S.R.; Jefferson, L.S. New functions for amino acids: Effects on gene transcription and translation. Am. J. Clin. Nutr. 2006, 83, 500S–507S. [Google Scholar] [CrossRef]
- Ray, R.M.; Viar, M.J.; Johnson, L.R. Amino acids regulate expression of antizyme-1 to modulate ornithine decarboxylase activity. J. Biol. Chem. 2012, 287, 3674–3690. [Google Scholar] [CrossRef]
- Singh, T.P.; Sogi, D.S. Comparative study of structural and functional characterization of bran protein concentrates from superfine, fine and coarse rice cultivars. Int. J. Biol. Macromol. 2018, 111, 281–288. [Google Scholar] [CrossRef]
- Shewry, P.R. Improving the protein content and composition of cereal grain. J. Cereal Sci. 2007, 46, 239–250. [Google Scholar] [CrossRef]
- Yoo, S.-Y. Quantitative trait loci controlling the amino acid content in rice (Oryza sativa L.). J. Plant Biotechnol. 2017, 44, 349–355. [Google Scholar] [CrossRef]
- Kalman, D.S. Amino acid composition of an organic brown rice protein concentrate and isolate compared to soy and whey concentrates and isolates. Foods 2014, 3, 394–402. [Google Scholar] [CrossRef]
- Sekhar, B.P.S.; Reddy, G.M. Amino acid profiles in some scented rice varieties. Theor. Appl. Genet. 1982, 62, 35–37. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, J.; Chen, F. Heat-induced changes in the physicochemical properties and in vitro digestibility of rice protein fractions. J. Food Sci. Technol. 2021, 58, 1368–1377. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Zhong, J.; Luo, L.; Liu, C.; Luo, S.; Chen, L. Binding interaction between rice glutelin and amylose: Hydrophobic interaction and conformational changes. Int. J. Biol. Macromol. 2015, 81, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Meng, D.; Wu, Z.; Chen, J.; Xue, L. Modification and solubility enhancement of rice protein and its application in food processing: A review. Molecules 2023, 28, 4078. [Google Scholar] [CrossRef]
- Al-Doury, M.K.W.; Hettiarachchy, N.S.; Horax, R. Rice-endosperm and rice-bran proteins: A review. J. Am. Oil Chem. Soc. 2018, 95, 95943–95956. [Google Scholar] [CrossRef]
- Park, J.; Sung, J.M.; Choi, Y.S.; Park, J.D. pH-dependent pasting and texture properties of rice flour subjected to limited protein hydrolysis. Food Hydrocoll. 2021, 117, 106754. [Google Scholar] [CrossRef]
- Jia, X.; Zhao, M.; Xia, N.; Teng, J.; Jia, C.; Wei, B.; Chen, D. Interaction between plant phenolics and rice protein improved oxidative stabilities of emulsion. J. Cereal Sci. 2019, 89, 102818. [Google Scholar] [CrossRef]
- Jiménez-Munoz, L.M.; Tavares, G.M.; Corredig, M. Design future foods using plant protein blends for best nutritional and technological functionality. Trends Food Sci. Technol. 2021, 113, 139–150. [Google Scholar]
- Thongkong, S.; Klangpetch, W.; Unban, K.; Tangjaidee, P.; Phimolsiripol, Y.; Rachtanapun, P.; Jantanasakulwong, K.; Schönlechner, R.; Thipchai, P.; Phongthai, S. Impacts of electroextraction using the pulsed electric field on properties of rice bran protein. Foods 2023, 12, 835. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.; Ouyang, S.; Wang, S. Effect of radio frequency treatment on functional, structural and thermal behaviors of protein isolates in rice bran. Food Chem. 2019, 289, 537–544. [Google Scholar] [PubMed]
- Zhao, Q.; Lin, J.; Wang, C.; Yousaf, L.; Xue, Y.; Shen, Q. Protein structural properties and proteomic analysis of rice during storage at different temperatures. Food Chem. 2021, 361, 130028. [Google Scholar]
- Liu, K.; Zheng, J.; Chen, F. Effect of domestic cooking on rice protein digestibility. Food Sci. Nutr. 2019, 24, 608–616. [Google Scholar] [CrossRef]
- Sanadya, A.; Yadu, A.; Raj, J.; Chandrakar, H.; Singh, R. Effect of temperature on growth, quality, yield attributing characters and yield of rice—A Review. Int. J. Environ. Clim. Chang. 2023, 13, 804–814. [Google Scholar]
- Shimoyanagi, R.; Abo, M.; Shiotsu, F. Higher temperatures during grain filling affect grain chalkiness and rice nutrient contents. Agronomy 2021, 11, 1360. [Google Scholar] [CrossRef]
- Liang, C.G.; Liu, J.; Wang, Y.; Xiong, D.; Ding, C.B.; Li, T. Low light during grain filling stage Deteriorates rice cooking quality, but not nutritional value. Rice Sci. 2015, 22, 197–206. [Google Scholar]
- Goufo, P.; Falco, V.; Brites, C.; Wessel, D.F.; Kratz, S.; Rosa, E.A.S.; Carranca, C.; Trindade, H. Effect of elevated carbon dioxide concentration on rice quality: Nutritive value, color, milling, cooking, and eating qualities. Cereal Chem. 2014, 91, 513–521. [Google Scholar] [CrossRef]
- Wang, J.; Hasegawa, T.; Li, L.; Lam, S.K.; Zhang, X.; Liu, X.; Pan, G. Changes in grain protein and amino acids composition of wheat and rice under short-term increased [CO2] and temperature of canopy air in a paddy from East China. New Phytol. 2019, 222, 726–734. [Google Scholar] [CrossRef]
- Taub, D.R.; Miller, B.; Allen, H. Effects of elevated CO2 on the protein concentration of food crops: A meta-analysis. Glob. Chang. Biol. 2008, 14, 565–575. [Google Scholar] [CrossRef]
- Uprety, D.C.; Sen, S.; Dwivedi, N. Rising atmospheric carbon dioxide on grain quality in crop plants. Physiol. Mol. Biol. Plants 2010, 16, 215–227. [Google Scholar] [CrossRef]
- Jing, L.; Wang, J.; Shen, S.; Wang, Y.; Zhu, J.; Wang, Y.; Yang, L. The impact of elevated CO2 and temperature on grain quality of rice grown under open-air field conditions. J. Sci. Food Agric. 2016, 96, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Bahuguna, R.N.; Pal, M.; Shah, D.; Maurya, S.; Jagadish, K.S. Elevated CO2 and heat stress interactions affect grain yield, quality and mineral nutrient composition in rice under field conditions. Field Crop Res. 2017, 206, 149–157. [Google Scholar] [CrossRef]
- Cheng, W.; Zhang, G.; Zhao, G.; Yao, H.; Xu, H. Variation in rice quality of different cultivars and grain positions as affected by water management. Field Crops Res. 2003, 80, 245–252. [Google Scholar] [CrossRef]
- Gomez, K.A.; De Datta, S.K. Influence of environment on protein content of rice. Agronomy 1975, 67, 565–568. [Google Scholar] [CrossRef]
- Upadhyay, R.; Banjara, M.; Thombare, D.; Yankanchi, S.; Chandel, G. Deciphering the effect of different nitrogen doses on grainprotein content, quality attributes and yield related traits of rice. ORYZA-Int. J. Rice 2021, 58, 530–539. [Google Scholar]
- De Datta, S.K.; Kerim, M.S.A.A.A. Water and nitrogen economy of rainfed rice as affected by soil puddling. Soil Sci. Soc. Am. Proc. 1974, 38, 515–518. [Google Scholar]
- Siscar-Lee, J.J.H.; Juliano, B.O.; Qureshi, R.H.; Akbar, M. Effect of saline soil on grain quality of rices differing in salinity tolerance. Plant Foods Hum. Nutr. 1990, 40, 31–36. [Google Scholar]
- Zhao, S.Y.; Shi, J.; Cai, S.; Xiong, T.; Cai, F.; Li, S.; Chen, X.; Fan, C.; Mei, X.; Sui, Y. Effects of milling degree on nutritional, sensory, gelatinization, and taste quality of different rice varieties. LWT-Food Sci. Technol. 2023, 186, 115244. [Google Scholar] [CrossRef]
- Sandhu, R.S.; Singh, N.; Kaler, R.S.S.; Kaur, A.; Shevkani, K. Effect of degree of milling on physicochemical, structural, pasting and cooking properties of short and long grain Indica rice cultivars. Food Chem. 2018, 260, 231–238. [Google Scholar] [CrossRef]
- Shenoy, V.V.; Seshu, D.V.; Sachan, J.K.S. Inheritance of protein per grain in rice. Indian J. Genet. 1991, 51, 214–220. [Google Scholar]
- Mizobuchi, R.; Fukuoka, S.; Tsushima, S.; Yano, M.; Sato, H. QTLs for resistance to major rice diseases exacerbated by global warming: Brown spot, bacterial seedling rot, and bacterial grain rot. Rice 2016, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Han, J.-H.; Lee, Y.K.; Shin, N.-H.; Kang, Y.J.; Kim, C.-K.; Chin, J.H. Mapping and validation of QTLs for the amino acid and total protein content in brown rice. Front. Genet. 2020, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.-H.; Zhu, Y.-J.; Tang, S.-Q.; Wei, X.-J.; Sheng, Z.-H.; Jiao, G.-A.; Hu, P.-S.; Zhuang, J.-Y. Pleiotropic effects of rice florigen gene RFT1 on the amino acid content of unmilled rice. Front. Genet. 2020, 11, 13. [Google Scholar] [CrossRef]
- Bruno, E.; Choi, Y.-S.; Chung, I.K.; Kim, K.-M. QTLs and analysis of the candidate gene for amylose, protein, and moisture content in rice (Oryza sativa L.). 3 Biotech 2017, 7, 40. [Google Scholar] [CrossRef]
- Liu, X.; Wan, X.; Ma, X.; Wan, J. Dissecting the genetic basis for the effect of rice chalkiness, amylose content, protein content, and rapid viscosity analyzer profile characteristics on the eating quality of cooked rice using the chromosome segment substitution line population across eight environments. Genome 2011, 54, 64–80. [Google Scholar]
- Yu, Y.H.; Li, G.; Fan, Y.Y.; Zhang, K.Q.; Min, J.; Zhu, Z.W.; Zhuang, J.Y. Genetic relationship between grain yield and the contents of protein and fat in a recombinant inbred population of rice. J. Cereal Sci. 2009, 50, 121–125. [Google Scholar] [CrossRef]
- Aluko, G.; Martinez, C.; Tohme, J.; Castano, C.; Bergman, C.; Oard, J.H. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor. Appl. Genet. 2004, 109, 630–639. [Google Scholar] [CrossRef]
- Hu, Z.L.; Li, P.; Zhou, M.Q.; Zhang, Z.H.; Wang, L.X.; Zhu, L.H.; Zhu, Y.G. Mapping of quantitative trait loci (QTLs) for rice protein and fat content using doubled haploid lines. Euphytica 2004, 135, 47–54. [Google Scholar] [CrossRef]
- Tan, Y.F.; Sun, M.; Xing, Y.Z.; Hua, J.P.; Sun, X.L.; Zhang, Q.F.; Corke, H. Mapping quantitative trait loci for milling quality, protein content and color characteristics of rice using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 2001, 103, 1037–1045. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, C.F.; Zhou, L.H.; Yao, S.; Zhao, Q.Y.; Chen, T.; Wang, C.L. Mapping QTLs for rice (Oryza sativa L.) grain protein content via chromosome segment substitution lines. Cereal Res. Commun. 2022, 50, 699–708. [Google Scholar] [CrossRef]
- Fiaz, S.; Sheng, Z.; Zeb, A.; Barman, H.N.; Shar, T.; Ali, U.; Tang, S. Analysis of genomic regions for crude protein and fractions of protein using a recombinant inbred population in Rice (Oryza sativa L.). J. Taibah Univ. Sci. 2021, 15, 579–588. [Google Scholar] [CrossRef]
- Yoshida, S.; Ikegami, M.; Kuze, J.; Sawada, K.; Hashimoto, Z.; Ishii, T.; Kamijima, O. QTL analysis for plant and grain characters of sake-brewing rice using a doubled haploid population. Breed. Sci. 2002, 52, 309–317. [Google Scholar] [CrossRef][Green Version]
- Zhang, W.; Bi, J.; Chen, L.; Zheng, L.; Ji, S.; Xia, Y.; Wan, J. QTL mapping for crude protein and protein fraction contents in rice (Oryza sativa L.). J. Cereal Sci. 2008, 48, 539–547. [Google Scholar] [CrossRef]
- Badoni, S.; Pasion-Uy, E.A.; Kora, S.; Kima, S.-R.; Tiozon Jr, R.N.; Misra, G.; Buenafea, R.J.Q.; Labarga, L.M.; Ramos-Castrosanto, A.R.; Pratap, V.; et al. Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content. Proc. Natl. Acad. Sci. USA 2024, 121, e2410598121. [Google Scholar] [CrossRef]
- He, L.; Sui, Y.; Che, Y.; Liu, L.; Liu, S.; Wang, X.; Cao, G. New insights into the genetic basis of lysine accumulation in rice revealed by multi-model GWAS. Int. J. Mol. Sci. 2024, 25, 4667. [Google Scholar] [CrossRef]
- Ming, Z.; Wang, L.Q.; Yuan, D.J.; Luo, L.J.; Xu, C.G.; He, Y.Q. Identification of QTL affecting protein and amino acid contents in rice. Rice Sci. 2011, 18, 187–195. [Google Scholar]
- Wang, L.; Zhong, M.; Li, X.; Yuan, D.; Xu, Y.; Liu, H.; Zhang, Q. The QTL controlling amino acid content in grains of rice (Oryza sativa) are co-localized with the regions involved in the amino acid metabolism pathway. Mol. Breeding 2008, 21, 127–137. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, W.; Liu, S.; Chen, L.; Liu, X.; Chen, X.; Ma, J.; Chen, W.; Zhao, Z.; Jiang, L.; et al. Genetic relationship between grain chalkiness, protein content, and paste viscosity properties in a backcross inbred population of rice. J. Cereal Sci. 2012, 56, 153–160. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Lee, J.; Shin, D.; Marmagne, A.; Lim, P.O.; Masclaux-Daubresse, C.; An, G.; Nam, H.G. OsASN1 overexpression in rice increases grain protein content and yield under nitrogen-limiting conditions. Plant Cell Physiol. 2020, 61, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Kumazawa, K. Assimilation and transport of nitrogen in rice I. 15N-labelled ammonium nitrogen. Plant Cell Physiol. 1974, 15, 747–758. [Google Scholar]
- Santiago, J.P.; Tegeder, M. Connecting source with sink: The role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef]
- Tegeder, M.; Hammes, U.Z. The way out and in: Phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 2018, 43, 16–21. [Google Scholar] [CrossRef]
- Guo, N.; Hu, J.; Yan, M.; Luo, L.; Qu, H.; Tegeder, M.; Xu, G. Oryza sativa lysine-histidine-type transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 2020, 103, 395–411. [Google Scholar] [CrossRef]
- Guo, N.; Gu, M.; Hu, J.; Qu, H.; Xu, G. Rice OsLHT1 functions in leaf-to-panicle nitrogen allocation for grain yield and quality. Front. Plant Sci. 2020, 11, 1150. [Google Scholar] [CrossRef]
- Peng, B.; Kong, H.; Li, Y.; Wang, L.; Zhong, M.; Sun, L.; Gao, G.; Zhang, Q.; Luo, L.; Wang, G.; et al. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 2014, 5, 4847. [Google Scholar] [CrossRef]
- Chattopadhyay, K.; Sharma, S.; Bagchi, T.B.; Mohanty, B.; Sardar, S.S.; Sarkar, S.; Singh, O.N. High-protein rice in high-yielding background, cv. Naveen. Curr. Sci. 2019, 117, 1722–1727. [Google Scholar] [CrossRef]
- Ruthsatz, M.; Candeias, V. Non-communicable disease prevention, nutrition and aging. Acta Biomed. 2020, 91, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Bigna, J.J.; Noubiap, J.J. The rising burden of non-communicable diseases in sub-Saharan Africa. Lancet Glob. Health 2019, 7, 1295–1296. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Hira, T.; Inoue, D.; Harada, Y.; Hashimoto, H.; Fujii, M.; Kadowaki, M.; Hara, H. Rice protein hydrolysates stimulate GLP-1 secretion, reduce GLP-1 degradation, and lower the glycemic response in rats. Food Funct. 2015, 6, 2525–2534. [Google Scholar]
- Hosojima, M.; Kaseda, R.; Kondo, H.; Fujii, M.; Kubota, M.; Watanabe, R.; Tanabe, N.; Kadowaki, M.; Suzuki, Y.; Saito, A. Beneficial effects of rice endosperm protein intake in Japanese men with risk factors for metabolic syndrome: A randomized, crossover clinical trial. BMC Nutr. 2016, 2, 25. [Google Scholar] [CrossRef][Green Version]
- Higuchi, Y.; Hosojima, M.; Kabasawa, H.; Kuwahara, S.; Goto, S.; Toba, K.; Kaseda, R.; Tanaka, T.; Kitamura, N.; Takihara, H.; et al. Rice endosperm protein administration to juvenile mice regulates gut microbiota and suppresses the development of high-fat diet-induced obesity and related disorders in adulthood. Nutrients 2019, 11, 2919. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, R.; Nishi, D.; Sato, M.; Ito, A.; Uchiyama, K.; Higuchi, Y.; Takahashi, H.; Ohinata, K. The effect of the rice endosperm protein hydrolysate on the subjective negative mood status in healthy humans: A randomized, double-blind, and placebo-controlled clinical trial. Nutrients 2023, 15, 3491. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.; Fassett, R.G.; Geraghty, D.P.; Kunde, D.A.; Ball, M.J.; Robertson, I.K.; Coombes, J.S. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 2012, 501, 89–103. [Google Scholar] [CrossRef]
- Schülke, S.; Dreidax, D.; Malik, A.; Burmester, T.; Nevo, E.; Band, M.; Avivi, A.; Hankeln, T. Living with stress: Regulation of antioxidant defense genes in the subterranean, hypoxia-tolerant mole rat, Spalax. Gene 2012, 500, 199–206. [Google Scholar] [CrossRef]
- Malik, A.I.; Storey, K.B. Activation of antioxidant defense during dehydration stress in the African clawed frog. Gene 2009, 442, 99–107. [Google Scholar]
- Fang, Y.Z.; Yang, S.; Wu, G. Free radicals, antioxidants, and nutrition. Nutrition 2002, 18, 872–879. [Google Scholar] [CrossRef]
- Yang, L.; Chen, J.-H.; Xu, T.; Zhou, A.-S.; Yang, H.-K. Rice protein improves oxidative stress by regulating glutathione metabolism and attenuating oxidative damage to lipids and proteins in rats. Life Sci. 2012, 91, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, L.; He, H.J.; Xu, T.; Liu, H.B.; Wu, Q.; Ma, Y.; Liu, Q.H.; Nie, M.H. Antioxidant capacity responsible for a hypocholesterolemia is independent of dietary cholesterol in adult rats fed rice protein. Gene 2014, 533, 57–66. [Google Scholar] [CrossRef]
- Li, H.; He, H.; Wang, Z.; Cai, J.; Sun, B.; Wu, Q.; Zhang, Y.; Zhou, G.; Yang, L. Rice protein suppresses ROS generation and stimulates antioxidant gene expression via Nrf2 activation in adult rats. Gene 2016, 585, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Li, H.; Yang, L. Rice proteins, extracted by alkali and α-amylase, differently affect in vitro antioxidant activity. Food Chem. 2016, 206, 137–145. [Google Scholar] [CrossRef]
- Wang, Z.; Li, H.; Liang, M.; Yang, L. Glutelin and prolamin, different components of rice protein, exert differently in vitro antioxidant activities. J. Cereal Sci. 2016, 72, 108–116. [Google Scholar] [CrossRef]
- Saisavoey, T.; Sangtanoo, P.; Reamtong, O.; Karnchanatat, A. Antioxidant and anti-inflammatory effects of defatted rice bran (Oryza sativa L.) protein hydrolysates on raw 264.7 macrophage cells. J. Food Biochem. 2016, 40, 731–740. [Google Scholar] [CrossRef]
- Thamnarathip, P.; Jangchud, K.; Nitisinprasert, S.; Vardhanabhuti, B. Identification of peptide molecular weight from rice bran protein hydrolysate with high antioxidant activity. J. Cereal Sci. 2016, 69, 329–335. [Google Scholar] [CrossRef]
- Wattanasiritham, L.; Theerakulkait, C.; Wickramasekara, S.; Maier, C.S.; Stevens, J.F. Isolation and identification of antioxidant peptides from enzymatically hydrolyzed rice bran protein. Food Chem. 2016, 192, 156–162. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Liang, M.; Cai, L.; Yang, L. Methionine augments antioxidant activity of rice protein during gastrointestinal digestion. Int. J. Mol. Sci. 2019, 20, 868. [Google Scholar] [CrossRef]
- Yang, L.; Chen, J.-H.; Xu, T.; Nie, M.-H.; Yang, H.-K. Hypocholesterolemic effect of rice protein is due to regulating hepatic cholesterol metabolism in adult rats. Gene 2013, 512, 470–476. [Google Scholar] [CrossRef]
- Yang, L.; Han, G.; Liu, Q.H.; Wu, Q.; He, H.J.; Cheng, C.Z.; Duan, Y.J. Rice protein exerts a hypocholesterolemic effect through regulating cholesterol metabolism-related gene expression and enzyme activity in adult rats fed a cholesterol-enriched diet. Int. J. Food Sci. Nutr. 2013, 64, 836–842. [Google Scholar] [CrossRef]
- Li, H.; Yang, L.; Yang, H.-K.; Sun, S.-H.; Liu, H.-B.; Wu, Q.; Chen, J.-H.; Zhuang, T.-C. Rice protein regulates HDL metabolism-related gene expression and enzyme activity in adult rats. Food Biosci. 2014, 8, 1–7. [Google Scholar]
- Yang, L.; Chen, J.; Xu, T.; Qiu, W.; Zhang, Y.; Zhang, L.; Xu, F.; Liu, H. Rice protein extracted by different methods affects cholesterol metabolism in rats due to its lower digestibility. Int. J. Mol. Sci. 2011, 12, 7594–7608. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Chen, Y.-Y.; Wu, C.-T.; Yu, C.-C.; Liao, H.-F. Prolamin, a rice protein, augments anti-leukaemia immune response. Food Chem. Toxicol. 2010, 51, 189–197. [Google Scholar] [CrossRef]
- Kannan, A.; Hettiarachchy, N.S.; Lay, J.O.; Liyanage, R. Human cancer cell proliferation inhibition by a pentapeptide isolated and characterized from rice bran. Peptides 2010, 31, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hettiarachchy, N.; Rayaprolu, S.; Mahadevan, M. Rice bran derived pentapeptide-induced apoptosis in human breast cancer cell models (MCF-7 and MDA-MB-231). Int. J. Biomed. Res. 2014, 5, 599–605. [Google Scholar] [CrossRef]
- Park, H.-Y.; Yoon, T.J.; Lee, W.; Kim, Y.; Choi, H.-D. Antimetastatic effect of glycoprotein isolated from rice bran on colon 26-M3. 1 cell line. J. Funct. Foods. 2017, 32, 278–284. [Google Scholar] [CrossRef]
- Park, H.Y.; Yu, A.R.; Hong, H.D.; Kim, H.H.; Lee, K.W.; Choi, H.D. Immunomodulatory effects of nontoxic glycoprotein fraction isolated from rice bran. Planta Med. 2016, 82, 606–611. [Google Scholar] [CrossRef]
- Langers, I.; Renoux, V.M.; Thiry, M.; Delvenne, P.; Jacobs, N. Natural killer cells: Role in local tumor growth and metastasis. Biologics 2012, 6, 73–82. [Google Scholar] [CrossRef]
- Wattayagorn, V.; Kongsema, M.; Tadakittisarn, S.; Chumnanpuen, P. Riceberry rice bran protein hydrolyzed fractions induced apoptosis, senescence and G1/S cell cycle arrest in human colon cancer cell lines. Appl. Sci. 2022, 12, 6917. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, J.; Wang, J.; Sun, B. The anti-cancer activity and potential clinical application of rice bran extracts and fermentation products. RSC Adv. 2019, 9, 8060. [Google Scholar] [CrossRef] [PubMed]
- Boonloh, K.; Kukongviriyapan, V.; Kongyingyoes, B.; Kukongviriyapan, U.; Thawornchinsombut, S.; Pannangpetch, P. Rice bran protein hydrolysates improve insulin resistance and decrease pro-inflammatory cytokine gene expression in rats fed a high carbohydrate-high fat diet. Nutrients 2015, 7, 6313–6329. [Google Scholar] [CrossRef] [PubMed]
- Marconi, O.; Sileoni, V.; Ceccaroni, D.; Perretti, G. The use of rice in brewing. In Advances in International Rice Research; Li, J.Q., Ed.; InTech: Janeza, Croatia, 2017; pp. 49–66. [Google Scholar]
- Dupont, C.; Alain Bocquet, A.; Tomé, D.; Marie Bernard, M.; Campeotto, F.; Dumond, P.; Essex, A.; Frelut, M.-L.; Guénard-Bilbault, L.; Lack, G.; et al. Hydrolyzed rice protein-based formulas, a vegetal alternative in cow’s milk allergy. Nutrients 2020, 12, 2654. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, K.; Rakkapao, N.; Lekjing, S. Physicochemical and antimicrobial characterization of chitosan and native glutinous rice starch-based composite edible films: Influence of different essential oils incorporation. Membranes 2023, 13, 161. [Google Scholar] [CrossRef]
- Pintado, T.; Delgado-Pando, G. Towards more sustainable meat products: Extenders as a way of reducing meat content. Foods 2020, 9, 1044. [Google Scholar] [CrossRef]
- Alam, M.; Wang, Y.Y.; Chen, J.; Lou, G.; Yang, H.; Zhou, Y.; Luitel, S.; Jiang, G.; He, Y. QTL detection for rice grain storage protein content and genetic effect verifications. Mol. Breeding 2023, 43, 89. [Google Scholar]
- Chen, P.; Lou, G.; Wang, Y.; Chen, J.; Chen, W.; Fan, Z.; Liu, Q.; Sun, B.; Mao, X.; Yu, H.; et al. The genetic basis of grain protein content in rice by genome-wide association analysis. Mol. Breeding 2023, 43, 1. [Google Scholar]
- Peng, B.; Sun, X.; Tian, X.; Kong, D.; He, L.; Peng, J.; Liu, Y.; Guo, G.; Sun, Y.; Pang, R.; et al. OsNAC74 affects grain protein content and various biological traits by regulating OsAAP6 expression in rice. Mol. Breeding 2023, 43, 87. [Google Scholar]
- Nayak, D.K.; Sahoo, S.; Barik, S.R.; Sanghamitra, P.; Sangeeta, S.; Pandit, E.; Raj, K.R.R.; Basak, N.; Pradhan, S.K. Association mapping for protein, total soluble sugars, starch, amylose and chlorophyll content in rice. BMC Plant Biol. 2022, 22, 620. [Google Scholar]
- Wu, Y.B.; Li, G.; Zhu, Y.J.; Cheng, Y.C.; Yang, J.Y.; Chen, H.Z.; Ying, J.Z. Genome-wide identification of QTLs for grain protein content based on genotyping-by-resequencing and verification of qGPC1-1 in rice. Int. J. Mol. Sci. 2020, 21, 408. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Kato, M.; Koyasaki, K.; Kawashima, T.; Nishimura, T.; Hirayama, Y.; Takamure, I.; Sato, T.; Kato, K. Identification of quantitative trait loci for rice grain quality and yield-related traits in two closely related Oryza sativa L. subsp. japonica cultivars grown near the northernmost limit for rice paddy cultivation. Breed. Sci. 2017, 67, 191–206. [Google Scholar] [CrossRef]
- Lee, G.H.; Yun, B.W.; Kim, K.M. Analysis of QTLs associated with the rice quality related gene by double haploid populations. Int. J. Genomics 2014, 2014, 781832. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.-W.; Kim, M.-G.; Handoyo, T.; Kim, K.-M. Analysis of rice grain quality-associated quantitative trait loci by using genetic mapping. Am. J. Plant Sci. 2014, 5, 1125–1132. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, W.; Chen, X.; Ma, J.; Chen, W.; Zhao, Z.; Zhai, H.; Wan, J. Dynamic QTL analysis of rice protein content and protein index using recombinant inbred lines. J. Plant Biol. 2011, 54, 321–328. [Google Scholar] [CrossRef]
- Qin, Y.; Min, S.-M.; Sohn, J.-K. QTL analysis of protein content in double-haploid lines of rice. Korean J. Crop Sci. 2009, 54, 165–171. [Google Scholar]
- Lou, J.; Chen, L.; Yue, G.; Lou, Q.; Mei, H.; Xiong, L.; Luo, L. QTL mapping of grain quality traits in rice. J. Cereal Sci. 2009, 50, 145–151. [Google Scholar] [CrossRef]
- Chen, P.; Shen, Z.; Ming, L.; Li, Y.; Dan, W.; Lou, G.; Peng, B.; Wu, B.; Li, Y.; Zhao, D.; et al. Genetic basis of variation in rice seed storage protein (albumin, globulin, prolamin, and glutelin) content revealed by genome-wide association analysis. Front. Plant Sci. 2018, 9, 612. [Google Scholar] [CrossRef]
| Region | Rice * | Wheat | Maize | Millet | Sorghum |
|---|---|---|---|---|---|
| Africa | 42.76 | 26.34 | 95.00 | 13.13 | 25.95 |
| Asia | 716.77 | 352.34 | 403.73 | 17.31 | 7.67 |
| Europe | 3.33 | 269.26 | 119.03 | 0.66 | 1.03 |
| Central America | 1.37 | 3.47 | 31.75 | - | 5.05 |
| North America | 9.90 | 81.26 | 404.77 | 0.44 | 8.07 |
| South America | 24.19 | 24.68 | 186.27 | 0.01 | 7.18 |
| Oceania | 0.50 | 41.59 | 0.57 | 0.04 | 2.33 |
| Region | Rice | Wheat | Maize | Millet | Sorghum |
|---|---|---|---|---|---|
| Africa | 37.07 | 45.67 | 40.56 | 6.92 | 14.37 |
| Asia | 117.12 | 68.67 | 7.76 | 2.64 | 2.36 |
| Europe | 8.95 | 113.28 | 5.96 | 0.20 | - |
| Central America | 16.92 | 37.98 | 106.50 | - | 0.59 |
| North America | 12.20 | 90.95 | 12.01 | - | 0.23 |
| South America | 37.98 | 62.59 | 29.56 | - | - |
| Oceania | 28.24 | 74.70 | 3.07 | - | 0.51 |
| Region | Plant | Meat | Eggs | Dairy | Fish and Seafood |
|---|---|---|---|---|---|
| Africa | 50.61 | 7.83 | 0.70 | 3.53 | 2.57 |
| Asia | 58.83 | 15.30 | 3.65 | 7.15 | 6.78 |
| Europe | 44.22 | 32.54 | 4.25 | 23.02 | 6.10 |
| Central America | 42.89 | 29.65 | 5.57 | 10.25 | 3.51 |
| North America | 40.18 | 49.28 | 4.96 | 22.42 | 4.77 |
| South America | 38.71 | 36.21 | 3.74 | 11.64 | 2.69 |
| Oceania | 38.58 | 39.29 | 1.81 | 12.46 | 5.62 |
| Region | Rice | Wheat | Maize | Millet | Sorghum |
|---|---|---|---|---|---|
| Africa | 4.81 | 10.86 | 7.69 | 1.51 | 3.34 |
| Asia | 15.42 | 18.66 | 1.16 | 0.62 | 0.54 |
| Europe | 1.04 | 25.14 | 0.94 | 0.04 | - |
| Central America | 2.14 | 7.89 | 18.51 | - | 0.15 |
| North America | 1.69 | 18.62 | 1.57 | - | 0.06 |
| South America | 5.00 | 13.42 | 4.99 | - | - |
| Oceania | 3.33 | 15.48 | 0.48 | - | 0.11 |
| Essential Amino Acid | Milled Rice [24] | Milled Rice [69] | Milled Rice [22] | Milled Rice [70] | Milled Rice [52] | Brown Rice [24] | Brown Rice Isolate [71] | Scented Rice (Milled) [72] |
|---|---|---|---|---|---|---|---|---|
| Histidine | 2.3–2.7 | 2.4 | 2.5 | 1.18–2.53 | 1.19–3.49 | 2.4–2.6 | 1.6–1.8 | 2.2 |
| Isoleucine | 3.7–4.8 | 3.8 | 3.8 | 3.75–8.51 | 2.69–5.18 | 3.6–4.6 | 3.2–3.4 | 3.0 |
| Leucine | 8.4–8.6 | 8.2 | 8.2 | 1.68–4.19 | 5.30–9.51 | 8.3–8.9 | 6.2–6.4 | 7.5 |
| Lysine | 3.4–4.2 | 3.7 | 3.3 | 1.28–3.49 | 2.2–6.24 | 3.9–4.3 | 2.1–2.4 | 4.2 |
| Cysteine | 1.8–2.6 | 1.6 | 3.9 | - | 0.13–3.42 | 2.2–2.4 | 1.6–1.8 | - |
| Methionine | 2.3–3.0 | 2.1 | 0.16–2.25 | 0.65–3.49 | 2.3–2.5 | 2.2–2.8 | 2.7 | |
| Phenylalanine | 5.3–5.5 | 4.8 | 10.1 | 2.31–5.71 | 3.5–6.30 | 5.0–5.3 | 4.1–4.4 | 5.2 |
| Tyrosine | 4.4–5.5 | 2.5 | 0.81–3.21 | 1.33–6.0 | 3.8–4.6 | 4.3–6.2 | 3.1 | |
| Threonine | 3.7–3.9 | 3.4 | 3.5 | 1.98–4.92 | 2.09–5.06 | 3.9–4.0 | 2.8–3.0 | 4.0 |
| Tryptophan | 1.3–1.8 | 1.3 | 0.8 | - | - | 1.3–1.5 | 1.1–1.2 | - |
| Valine | 4.9–6.8 | 5.8 | 5.1 | 2.53–6.02 | 3.78–6.80 | 5.0–6.6 | 4.3–4.6 | 4.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jukanti, A.K.; Karapati, D.; Bharali, V.; Gudla, M.; Thati, S.; Yadla, S.; Kumar, M.; Sundaram, R.M. From Gene to Plate: Molecular Insights into and Health Implications of Rice (Oryza sativa L.) Grain Protein. Int. J. Mol. Sci. 2025, 26, 3163. https://doi.org/10.3390/ijms26073163
Jukanti AK, Karapati D, Bharali V, Gudla M, Thati S, Yadla S, Kumar M, Sundaram RM. From Gene to Plate: Molecular Insights into and Health Implications of Rice (Oryza sativa L.) Grain Protein. International Journal of Molecular Sciences. 2025; 26(7):3163. https://doi.org/10.3390/ijms26073163
Chicago/Turabian StyleJukanti, Aravind Kumar, Divya Karapati, Violina Bharali, Mahesh Gudla, Srinivas Thati, Suneetha Yadla, Manoj Kumar, and Raman Meenakshi Sundaram. 2025. "From Gene to Plate: Molecular Insights into and Health Implications of Rice (Oryza sativa L.) Grain Protein" International Journal of Molecular Sciences 26, no. 7: 3163. https://doi.org/10.3390/ijms26073163
APA StyleJukanti, A. K., Karapati, D., Bharali, V., Gudla, M., Thati, S., Yadla, S., Kumar, M., & Sundaram, R. M. (2025). From Gene to Plate: Molecular Insights into and Health Implications of Rice (Oryza sativa L.) Grain Protein. International Journal of Molecular Sciences, 26(7), 3163. https://doi.org/10.3390/ijms26073163






