Integrated Multi-Omics Analysis Unveils Distinct Molecular Subtypes and a Robust Immune–Metabolic Prognostic Model in Clear Cell Renal Cell Carcinoma
Abstract
1. Introduction
2. Results
2.1. Molecular Subtypes and Prognostic Implications in ccRCC
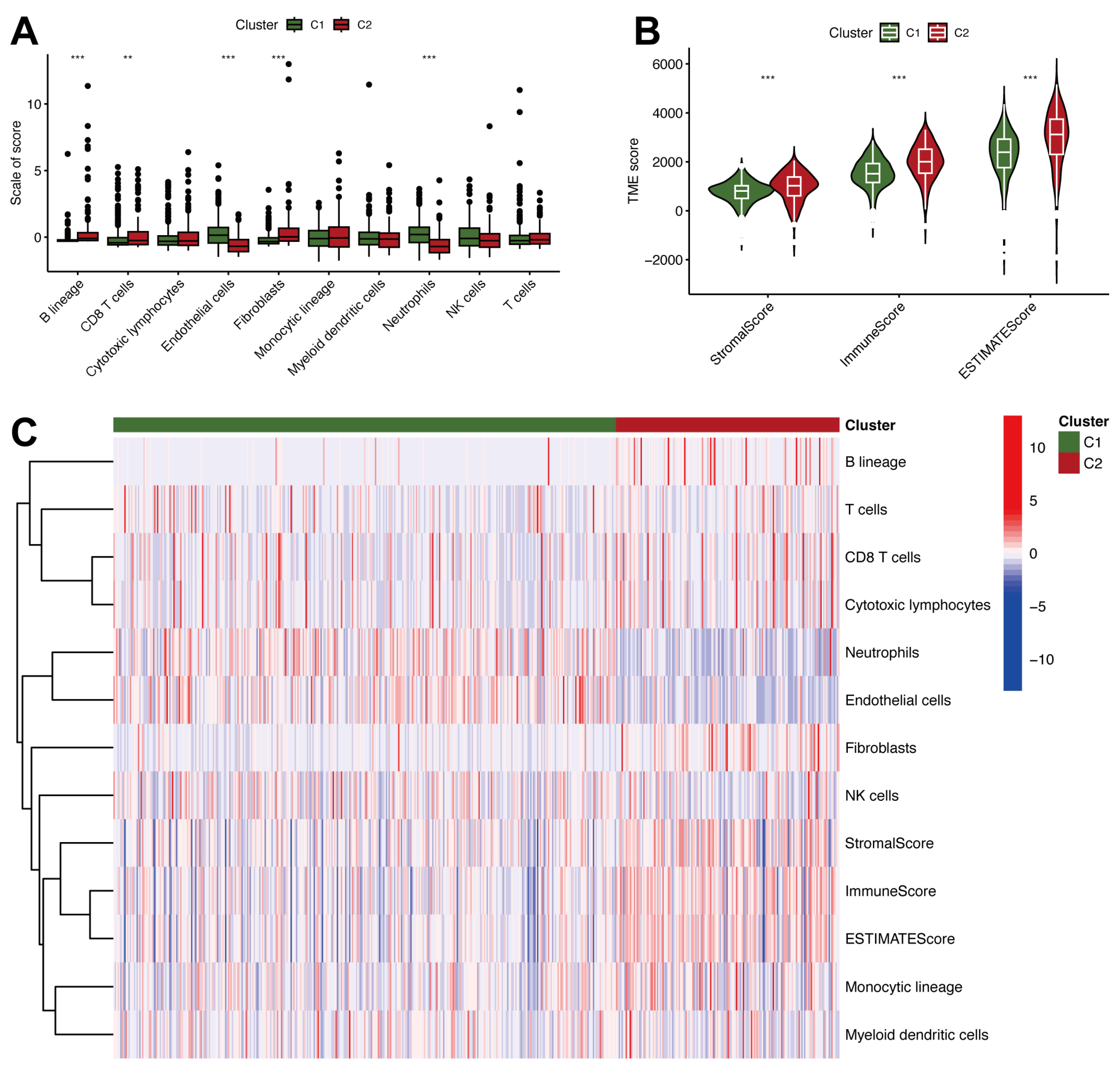
2.2. Tumor Microenvironment Heterogeneity and Immune Characteristics in ccRCC
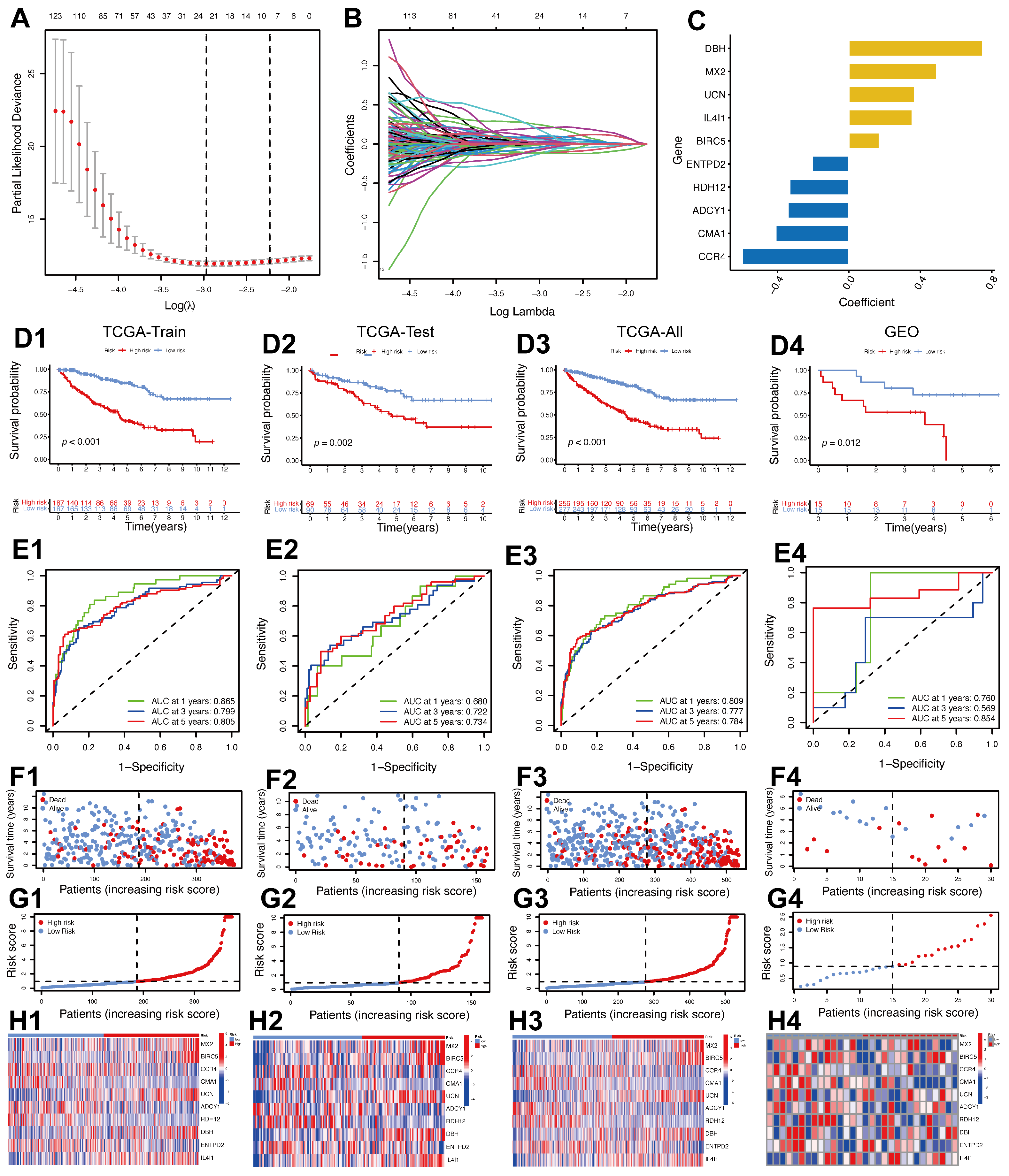
2.3. Creation of a Prognostic Model Utilizing IMRGs in ccRCC
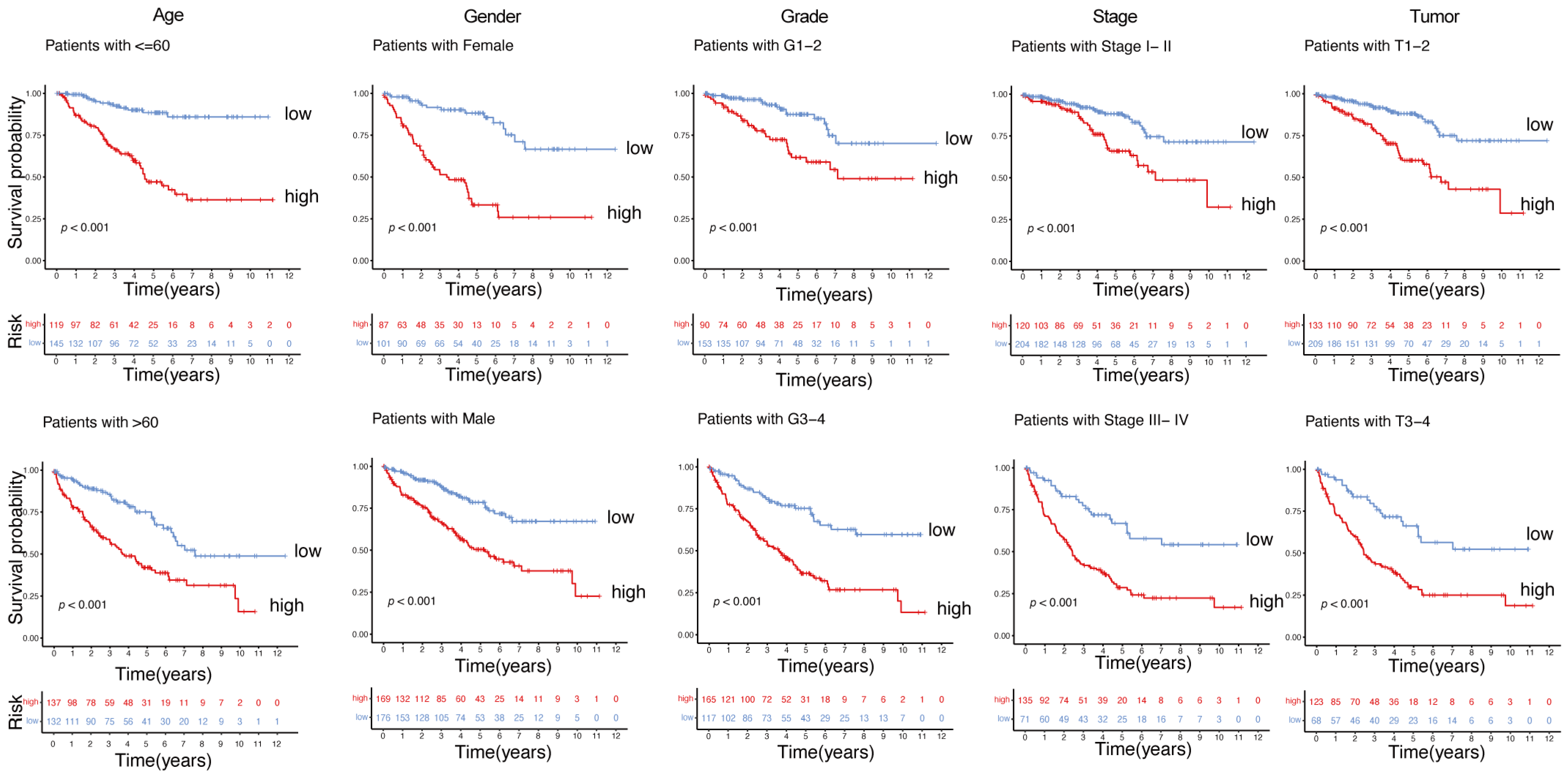
2.4. Robust Prognostic Value of the Immunity- and Metabolism-Related Gene Signature Across Clinical Subgroups
2.5. Comprehensive Prognostic Evaluation of an Immunity- and Metabolism-Related Gene Model Using Nomogram Analysis
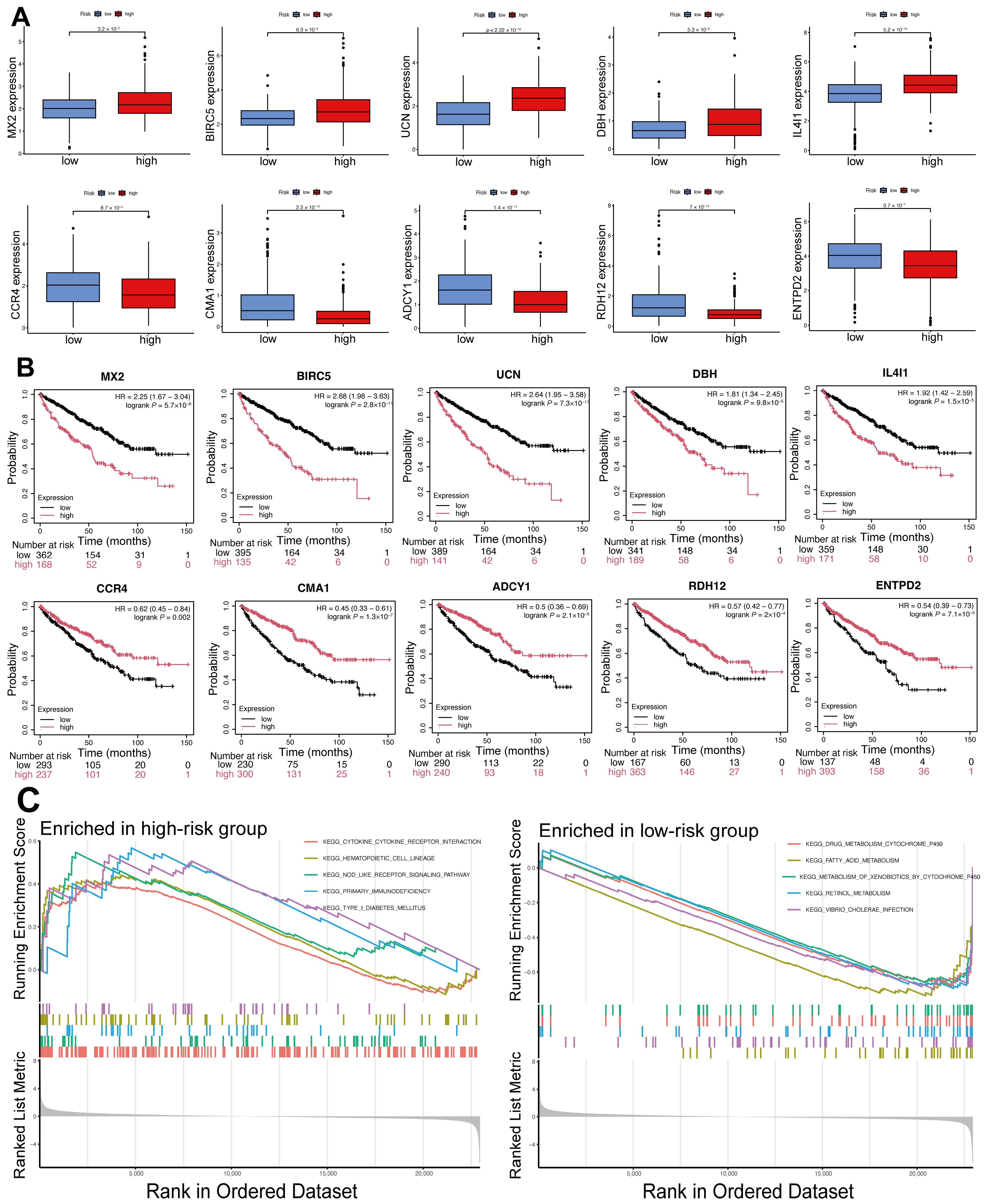
2.6. Differential Expression and Prognostic Significance of Model Genes, and Functional Enrichment Analysis of DEGs in High- and Low-Risk Groups
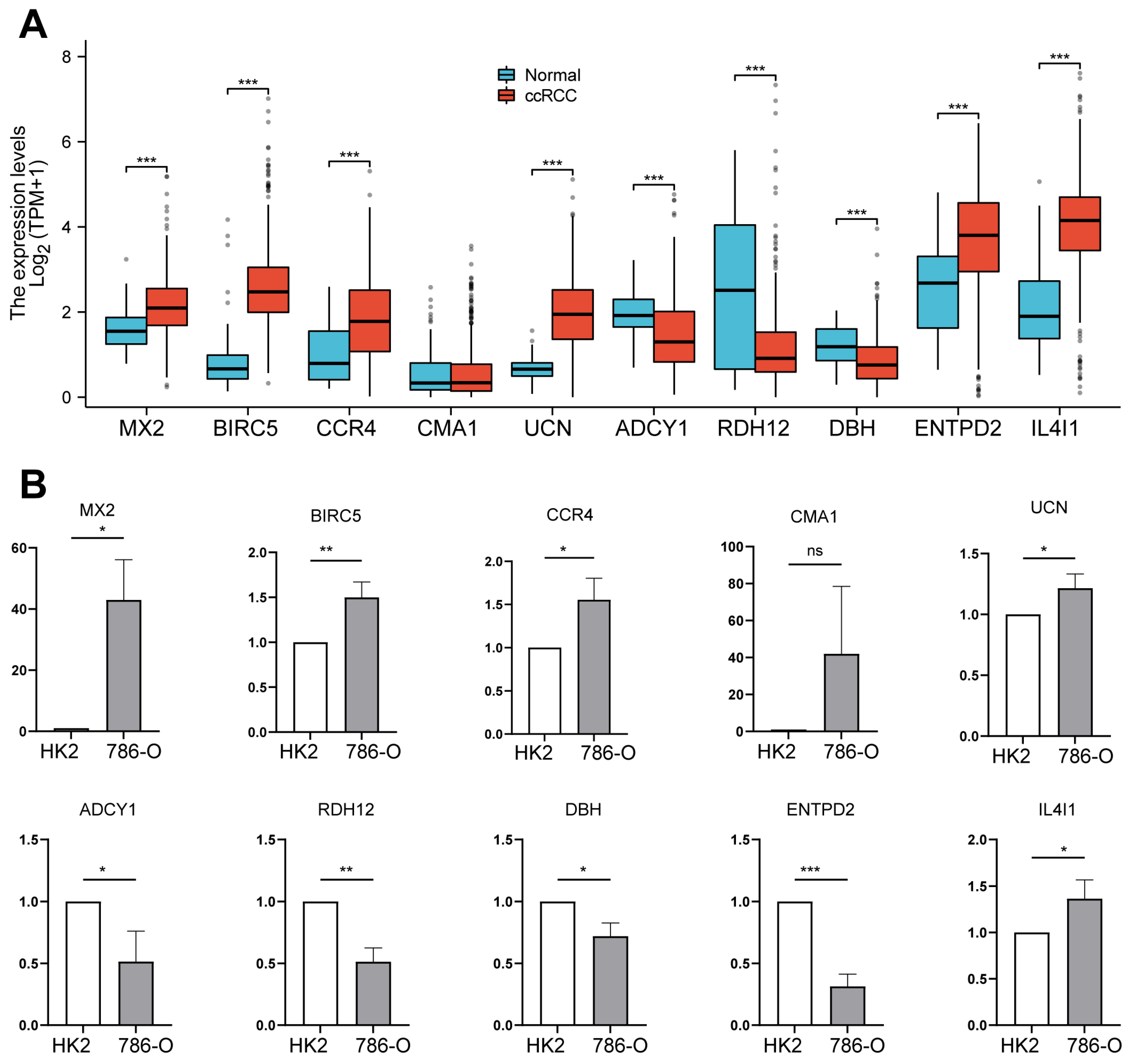
2.7. Analysis of Prognostic Gene Expression Differences Between Tumor and Normal Tissues, with Validation in ccRCC Cell Lines
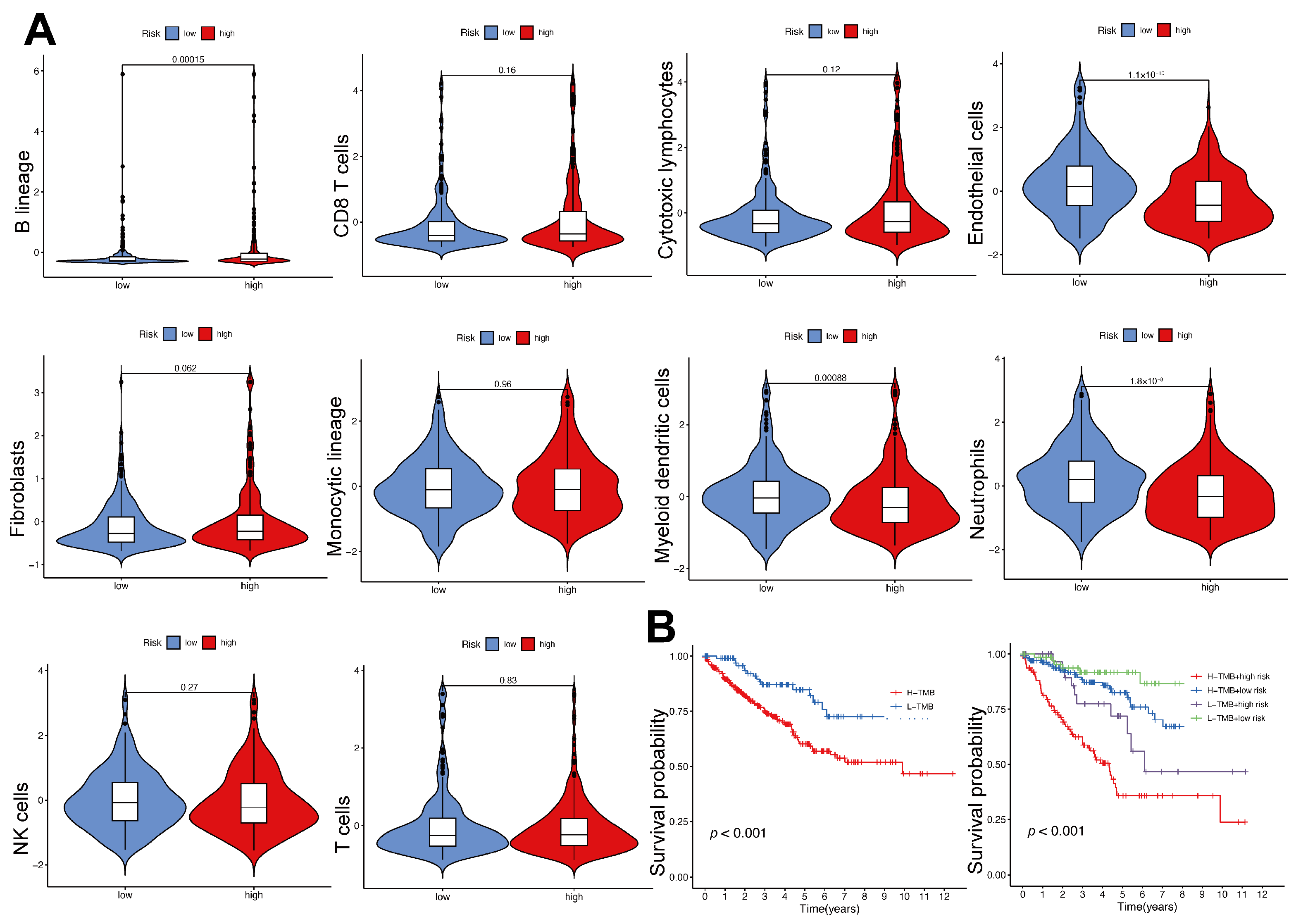
2.8. Immune Cell Infiltration and Tumor Mutational Burden in High- and Low-Risk Groups
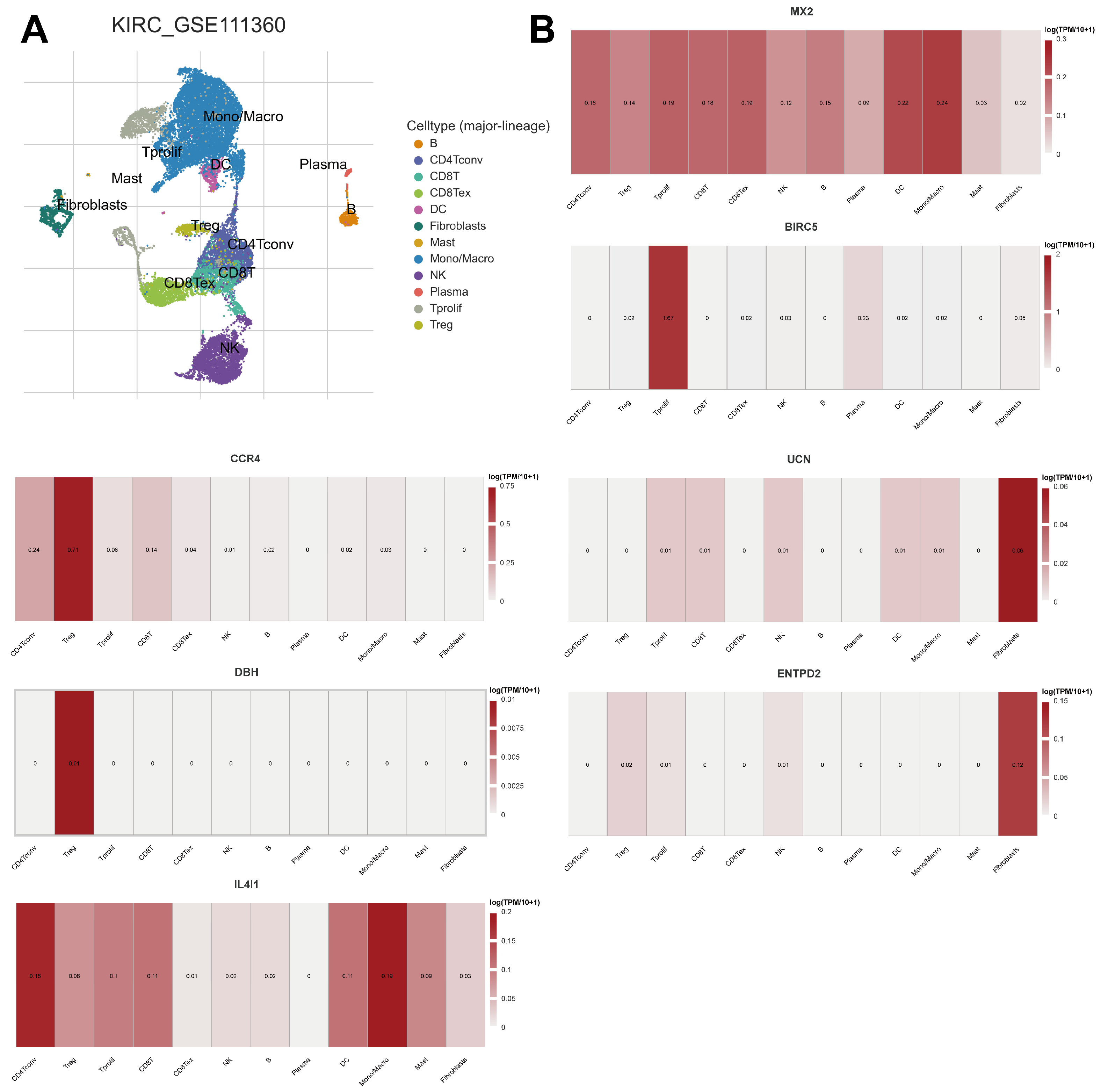
2.9. Expression of Prognostic Model Genes in Immune Cell Subsets in ccRCC
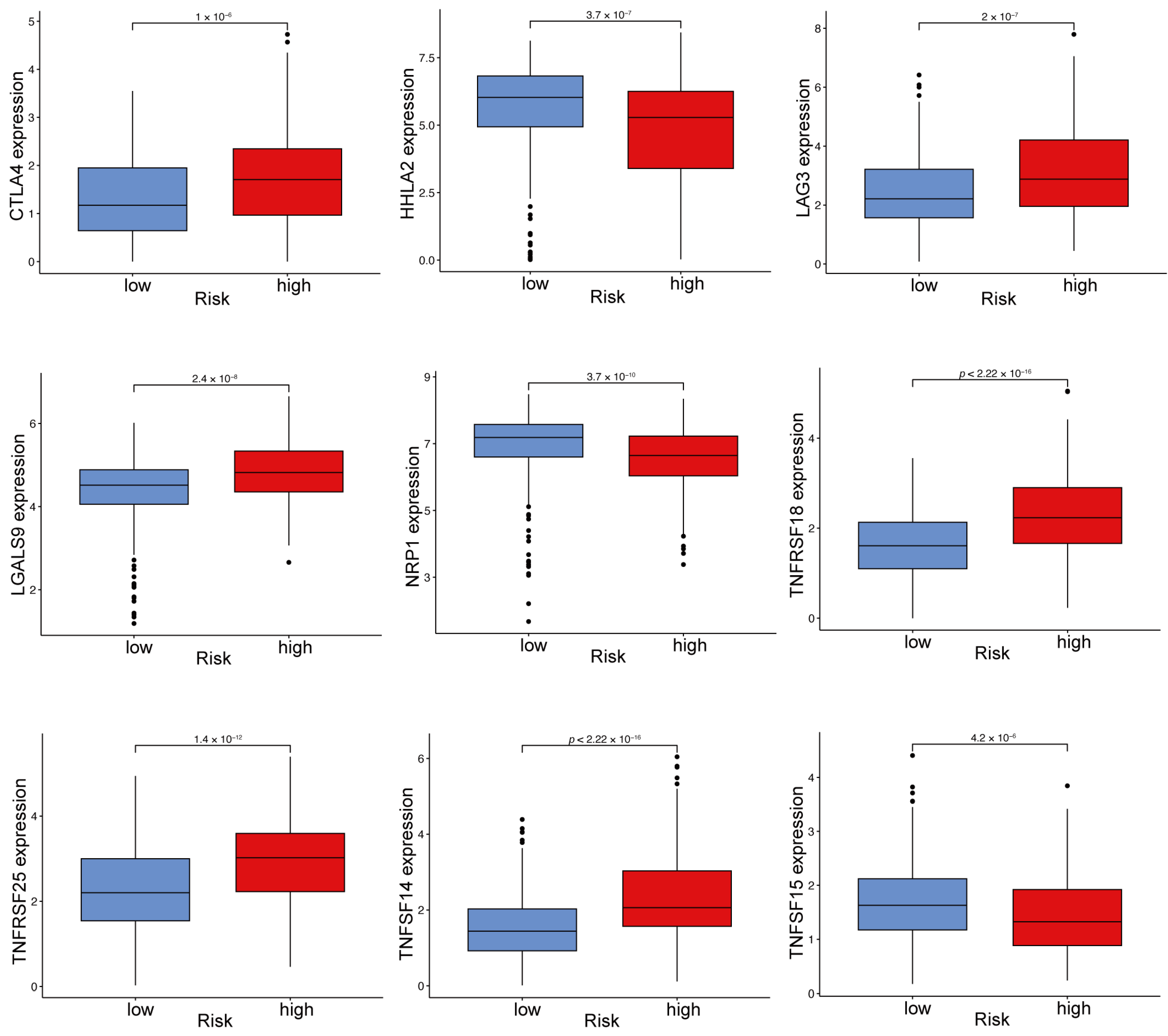
2.10. Immune Checkpoint Gene Expression Varies Between High- and Low-Risk ccRCC Groups
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Preprocessing
4.2. Differential Gene Expression Analysis
4.3. Clustering Using Non-Negative Matrix Factorization (NMF)
4.4. Kaplan–Meier Survival Analysis
4.5. Tumor Microenvironment and Immune Analysis
4.6. Construction of the Prognostic Gene Signature
4.7. Model Validation and Performance Evaluation
4.8. Functional Enrichment Analysis
4.9. Analysis of Immune Checkpoints and TMB
4.10. Nomogram Development and Validation
4.11. Validation of Prognostic Genes in Cell Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ccRCC | Clear cell renal cell carcinoma |
| TME | Tumor microenvironment |
| VHL | von Hippel–Lindau |
| HIF | Hypoxia-inducible factor |
| TAMs | Tumor-associated macrophages |
| Tregs | Regulatory T cells |
| NMF | Non-negative matrix factorization |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| DEGs | Differentially expressed genes |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| GSEA | Gene Set Enrichment Analysis |
| ssGSEA | Single-sample gene set enrichment analysis |
| OS | Overall survival |
| PFS | Progression-free survival |
| TMB | Tumor mutational burden |
| AUC | Area Under the Curve |
| ROC | Receiver Operating Characteristic |
| HR | Hazard ratio |
| CI | Confidence interval |
| C-index | Concordance index |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| KM | Kaplan–Meier |
| TISCH | Tumor Immune Single Cell Hub |
| MSigDB | Molecular Signatures Database |
| IMRGs | Immunity- and metabolism-related genes |
| DCA | Decision Curve Analysis |
| UMAP | Uniform Manifold Approximation and Projection |
References
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bedke, J.; Capitanio, U.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur. Urol. 2022, 82, 399–410. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar]
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal cell carcinoma. BMJ 2014, 349, g4797. [Google Scholar]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar]
- Gossage, L.; Eisen, T.; Maher, E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [PubMed]
- Motzer, R.J.; Jonasch, E.; Agarwal, N.; Alva, A.; Baine, M.; Beckermann, K.; Carlo, M.I.; Choueiri, T.K.; Costello, B.A.; Derweesh, I.H.; et al. Kidney Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 71–90. [Google Scholar]
- Rosellini, M.; Marchetti, A.; Mollica, V.; Rizzo, A.; Santoni, M.; Massari, F. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat. Rev. Urol. 2023, 20, 133–157. [Google Scholar]
- Zhang, S.; Zhang, E.; Long, J.; Hu, Z.; Peng, J.; Liu, L.; Tang, F.; Li, L.; Ouyang, Y.; Zeng, Z. Immune infiltration in renal cell carcinoma. Cancer Sci. 2019, 110, 1564–1572. [Google Scholar]
- Zhang, Z.; Ji, M.; Li, J.; Wu, Q.; Huang, Y.; He, G.; Xu, J. Molecular Classification Based on Prognostic and Cell Cycle-Associated Genes in Patients with Colon Cancer. Front. Oncol. 2021, 11, 636591. [Google Scholar]
- Huo, X.; Yang, M.; Zhang, X.; Wang, S.; Sun, H. Identification of Tumor Microenvironment Scoring Scheme Based on Bioinformatics Analysis of Immune Cell Infiltration Pattern of Ovarian Cancer. J. Oncol. 2022, 2022, 7745675. [Google Scholar] [PubMed]
- Chakiryan, N.H.; Hajiran, A.; Kim, Y.; Aydin, A.M.; Zemp, L.; Katende, E.; Nguyen, J.; Fan, W.; Cheng, C.H.; Lopez-Blanco, N.; et al. Correlating Immune Cell Infiltration Patterns with Recurrent Somatic Mutations in Advanced Clear Cell Renal Cell Carcinoma. Eur. Urol. Focus 2022, 8, 784–793. [Google Scholar]
- Hua, X.; Ge, S.; Zhang, J.; Xiao, H.; Tai, S.; Yang, C.; Zhang, L.; Liang, C. A costimulatory molecule-related signature in regard to evaluation of prognosis and immune features for clear cell renal cell carcinoma. Cell Death Discov. 2021, 7, 252. [Google Scholar] [CrossRef]
- Braun, D.A.; Hou, Y.; Bakouny, Z.; Ficial, M.; Sant’Angelo, M.; Forman, J.; Ross-Macdonald, P.; Berger, A.C.; Jegede, O.A.; Elagina, L.; et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 2020, 26, 909–918. [Google Scholar]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar]
- Motzer, R.J.; Rini, B.I.; McDermott, D.F.; Arén Frontera, O.; Hammers, H.J.; Carducci, M.A.; Salman, P.; Escudier, B.; Beuselinck, B.; Amin, A.; et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: Extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol. 2019, 20, 1370–1385. [Google Scholar] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar]
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N., Jr.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [PubMed]
- Choueiri, T.K.; Motzer, R.J. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar]
- Yang, R.; Hung, M.C. The role of T-cell immunoglobulin mucin-3 and its ligand galectin-9 in antitumor immunity and cancer immunotherapy. Sci. China Life Sci. 2017, 60, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Li, L.; Wang, Y.; Fu, R.; Wang, H.; Shao, Z. Increased TIM3+CD8+ T cells in Myelodysplastic Syndrome patients displayed less perforin and granzyme B secretion and higher CD95 expression. Leuk. Res. 2016, 51, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yu, N.; Li, X.; Wang, L.; Pan, Y.; Li, X.; Tao, J.; Chen, Z.; Wang, G. Aberrant GITR expression on different T cell subsets and the regulation by glucocorticoid in systemic lupus erythematosus. Int. J. Rheum. Dis. 2016, 19, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Slebioda, T.J.; Rowley, T.F.; Ferdinand, J.R.; Willoughby, J.E.; Buchan, S.L.; Taraban, V.Y.; Al-Shamkhani, A. Triggering of TNFRSF25 promotes CD8+ T-cell responses and anti-tumor immunity. Eur. J. Immunol. 2011, 41, 2606–2611. [Google Scholar] [CrossRef]
- Benwell, C.J.; Taylor, J.; Robinson, S.D. Endothelial neuropilin-2 influences angiogenesis by regulating actin pattern development and α5-integrin-p-FAK complex recruitment to assembling adhesion sites. FASEB J. 2021, 35, e21679. [Google Scholar] [CrossRef]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.B.K.; Nicol, D.; et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 2018, 173, 595–610.e11. [Google Scholar]
- Fridman, W.H.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Altieri, D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 2008, 8, 61–70. [Google Scholar] [CrossRef]
- Carbonnelle-Puscian, A.; Copie-Bergman, C.; Baia, M.; Martin-Garcia, N.; Allory, Y.; Haioun, C.; Crémades, A.; Abd-Alsamad, I.; Farcet, J.P.; Gaulard, P.; et al. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia 2009, 23, 952–960. [Google Scholar] [CrossRef]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar]
- Chen, Y.; Zhou, X.; Xie, Y.; Wu, J.; Li, T.; Yu, T.; Pang, Y.; Du, W. Establishment of a Seven-Gene Signature Associated with CD8+ T Cells through the Utilization of Both Single-Cell and Bulk RNA-Sequencing Techniques in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 13729. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [PubMed]
- Sharma, G.; Colantuoni, C.; Goff, L.A.; Fertig, E.J.; Stein-O’Brien, G. projectR: An R/Bioconductor package for transfer learning via PCA, NMF, correlation and clustering. Bioinformatics 2020, 36, 3592–3593. [Google Scholar] [PubMed]
- Hong, K.; Cen, K.; Chen, Q.; Dai, Y.; Mai, Y.; Guo, Y. Identification and validation of a novel senescence-related biomarker for thyroid cancer to predict the prognosis and immunotherapy. Front. Immunol. 2023, 14, 1128390. [Google Scholar]
- Shi, Y.; Wang, Y.; Dong, H.; Niu, K.; Zhang, W.; Feng, K.; Yang, R.; Zhang, Y. Crosstalk of ferroptosis regulators and tumor immunity in pancreatic adenocarcinoma: Novel perspective to mRNA vaccines and personalized immunotherapy. Apoptosis 2023, 28, 1423–1435. [Google Scholar]
- Zhao, M.; Zhang, Q.; Song, Z.; Lei, H.; Li, J.; Peng, F.; Lin, S. ATP2C2 as a novel immune-related marker that defines the tumor microenvironment in triple-negative breast cancer. Transl. Cancer Res. 2023, 12, 1802–1815. [Google Scholar]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [PubMed]
- Wang, T.; Dai, L.; Shen, S.; Yang, Y.; Yang, M.; Yang, X.; Qiu, Y.; Wang, W. Comprehensive Molecular Analyses of a Macrophage-Related Gene Signature with Regard to Prognosis, Immune Features, and Biomarkers for Immunotherapy in Hepatocellular Carcinoma Based on WGCNA and the LASSO Algorithm. Front. Immunol. 2022, 13, 843408. [Google Scholar]
- Li, X.; Lei, J.; Shi, Y.; Peng, Z.; Gong, M.; Shu, X. Developing a RiskScore Model based on Angiogenesis-related lncRNAs for Colon Adenocarcinoma Prognostic Prediction. Curr. Med. Chem. 2024, 31, 2449–2466. [Google Scholar]
- Tastsoglou, S.; Skoufos, G.; Miliotis, M.; Karagkouni, D.; Koutsoukos, I.; Karavangeli, A.; Kardaras, F.S.; Hatzigeorgiou, A.G. DIANA-miRPath v4.0: Expanding target-based miRNA functional analysis in cell-type and tissue contexts. Nucleic Acids Res. 2023, 51, W154–W159. [Google Scholar] [PubMed]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, D.; Miao, C.; Tao, J.; Ge, C.; Chen, T.; Li, H.; Hou, H. The stage-dependent prognostic role of ARID1A in hepatocellular carcinoma. Transl. Cancer Res. 2023, 12, 3088–3104. [Google Scholar]
- Zhang, W.; Ji, L.; Wang, X.; Zhu, S.; Luo, J.; Zhang, Y.; Tong, Y.; Feng, F.; Kang, Y.; Bi, Q. Nomogram Predicts Risk and Prognostic Factors for Bone Metastasis of Pancreatic Cancer: A Population-Based Analysis. Front. Endocrinol. 2021, 12, 752176. [Google Scholar]
- He, Z.; Gu, Y.; Yang, H.; Fu, Q.; Zhao, M.; Xie, Y.; Liu, Y.; Du, W. Identification and verification of a novel anoikis-related gene signature with prognostic significance in clear cell renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 11661–11678. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Yu, S.; Yang, D.; Yu, T.; Liu, Y.; Du, W. Integrated Multi-Omics Analysis Unveils Distinct Molecular Subtypes and a Robust Immune–Metabolic Prognostic Model in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2025, 26, 3125. https://doi.org/10.3390/ijms26073125
Zhu Y, Yu S, Yang D, Yu T, Liu Y, Du W. Integrated Multi-Omics Analysis Unveils Distinct Molecular Subtypes and a Robust Immune–Metabolic Prognostic Model in Clear Cell Renal Cell Carcinoma. International Journal of Molecular Sciences. 2025; 26(7):3125. https://doi.org/10.3390/ijms26073125
Chicago/Turabian StyleZhu, Yilin, Shihui Yu, Dan Yang, Tian Yu, Yi Liu, and Wenlong Du. 2025. "Integrated Multi-Omics Analysis Unveils Distinct Molecular Subtypes and a Robust Immune–Metabolic Prognostic Model in Clear Cell Renal Cell Carcinoma" International Journal of Molecular Sciences 26, no. 7: 3125. https://doi.org/10.3390/ijms26073125
APA StyleZhu, Y., Yu, S., Yang, D., Yu, T., Liu, Y., & Du, W. (2025). Integrated Multi-Omics Analysis Unveils Distinct Molecular Subtypes and a Robust Immune–Metabolic Prognostic Model in Clear Cell Renal Cell Carcinoma. International Journal of Molecular Sciences, 26(7), 3125. https://doi.org/10.3390/ijms26073125




