Unveiling Genetic Markers for Milk Yield in Xinjiang Donkeys: A Genome-Wide Association Study and Kompetitive Allele-Specific PCR-Based Approach
Abstract
1. Introduction
2. Results
2.1. Descriptive Statistics
2.2. Resequencing of Xinjiang Donkey
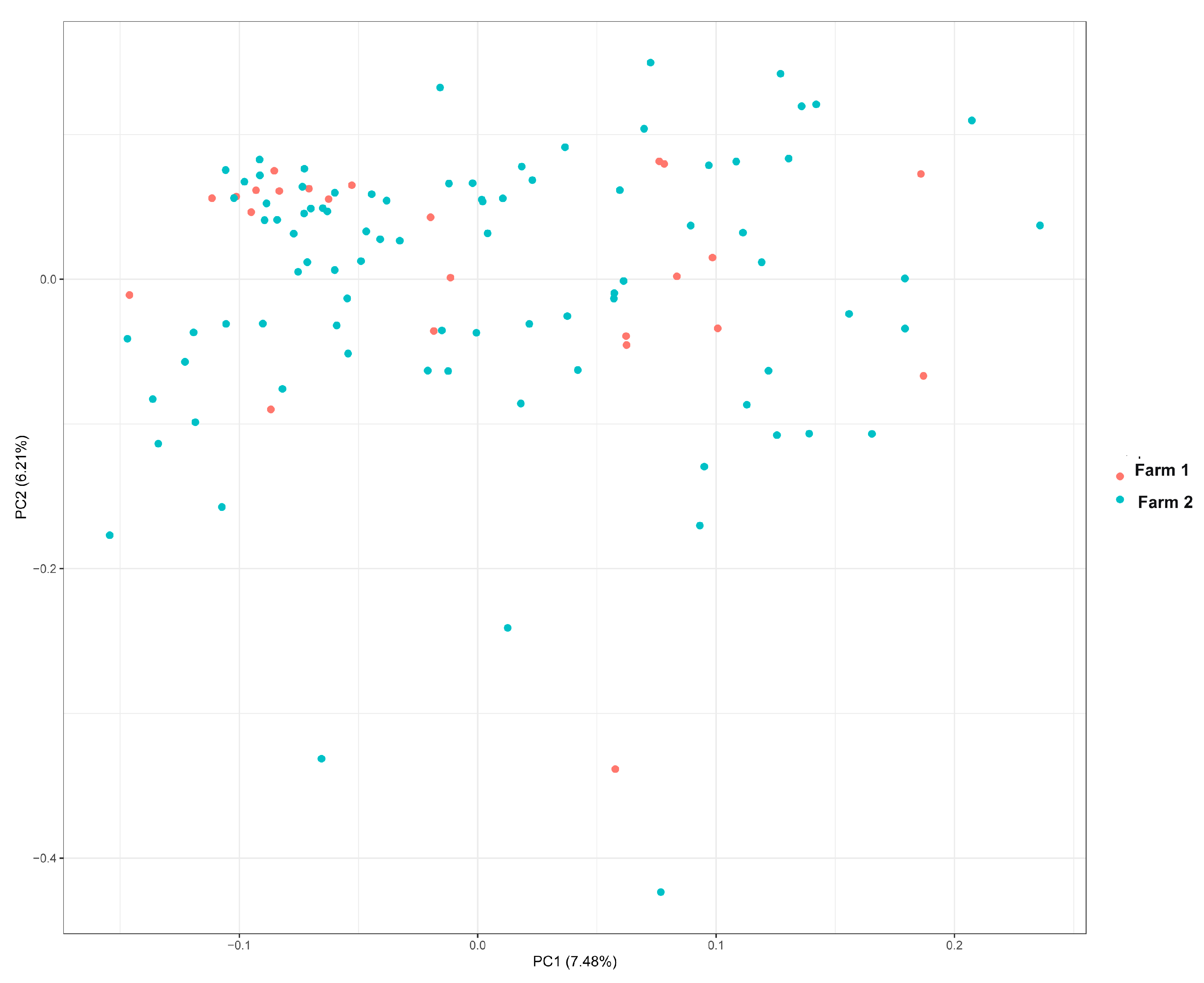
2.3. Principal Component Analysis
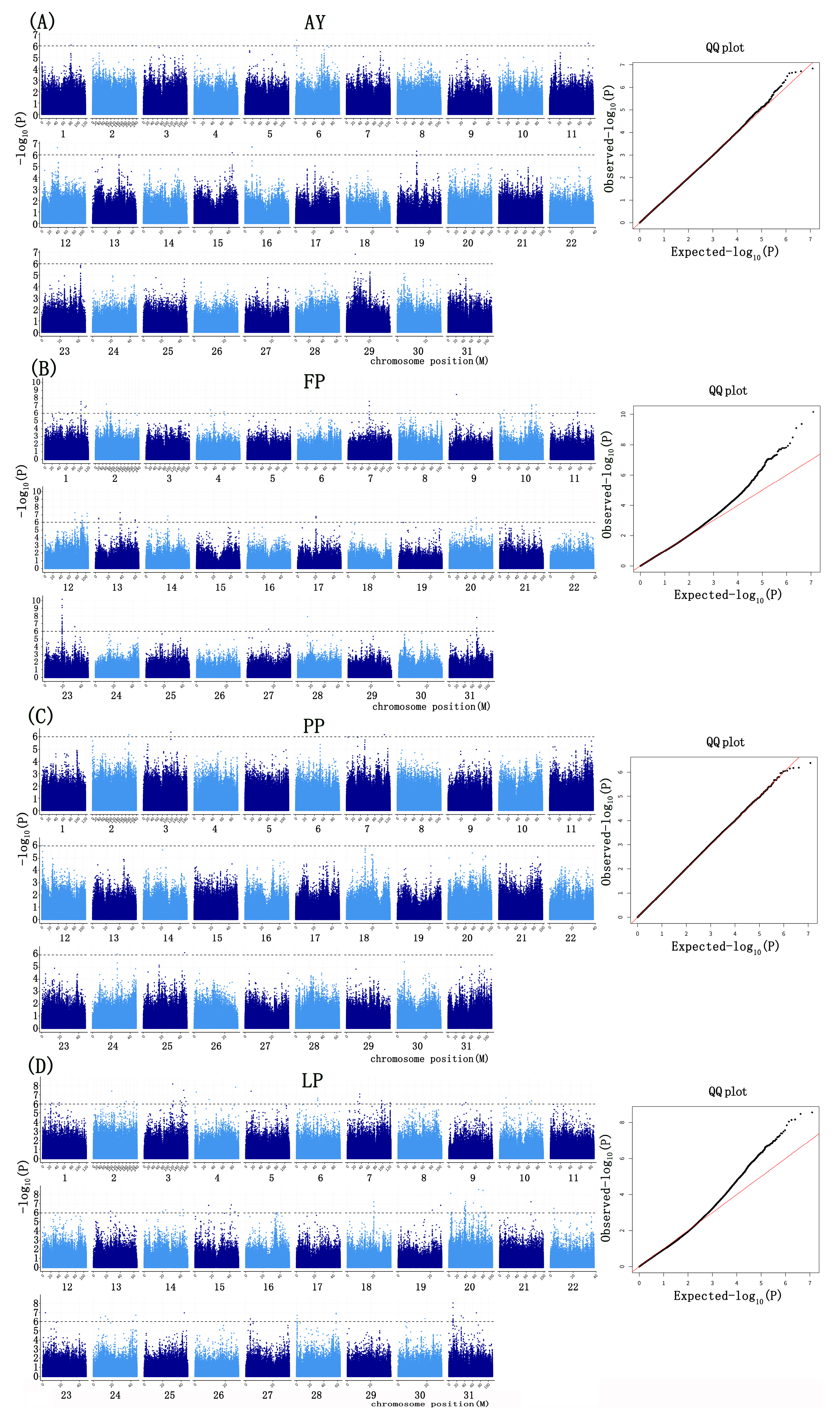
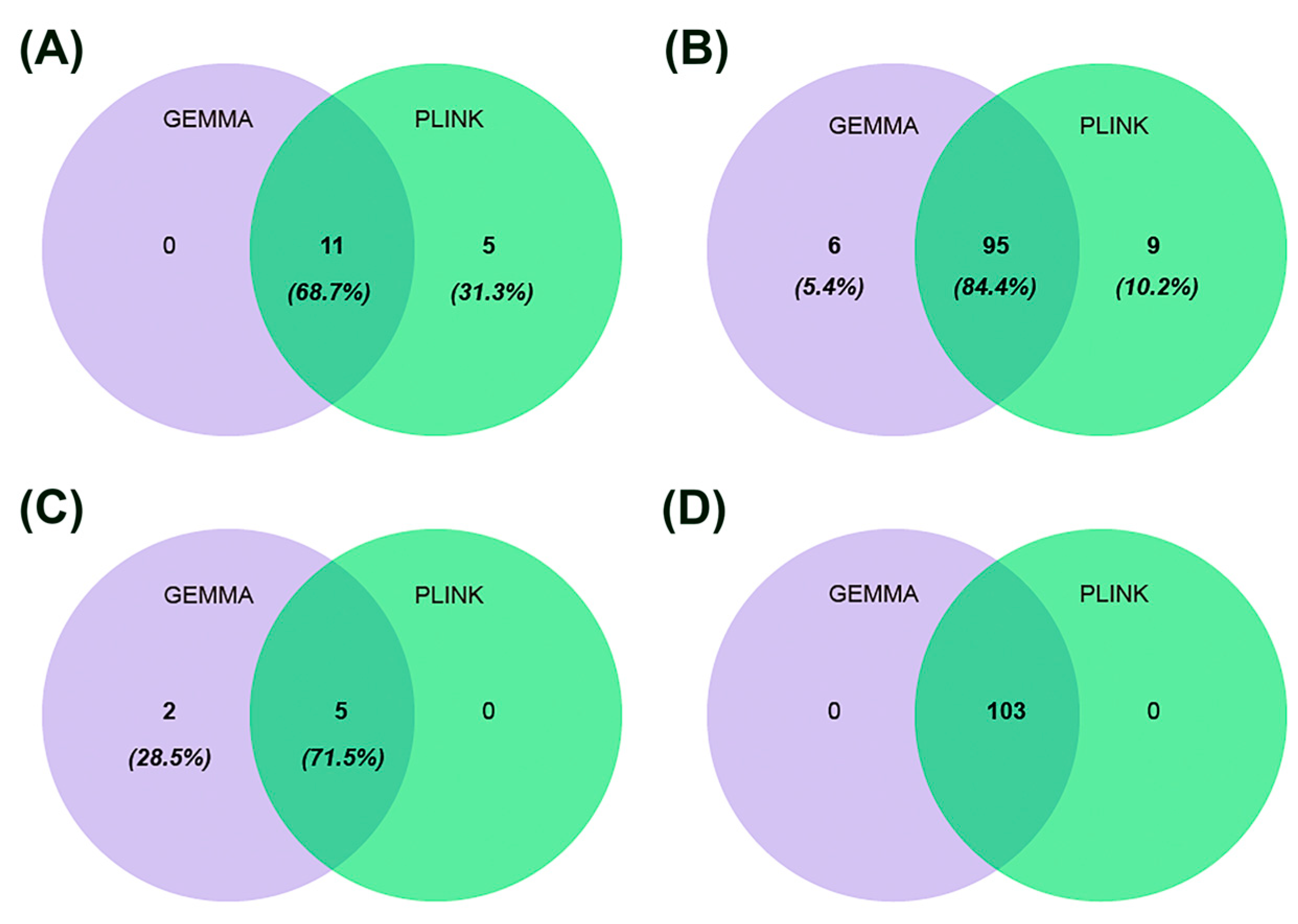
2.4. GWAS Based on GEMMA and PLINK
2.5. Candidate Genes
2.6. Validation of Candidate Genes Using KASP Technology
2.6.1. Locus Information of Candidate Genes
2.6.2. Genotyping Results by KASP
2.6.3. Polymorphism Analysis
2.6.4. Correlation Analysis of Genotype and Milk Yield in Xinjiang Donkey
3. Discussion
3.1. Feasibility of GWAS Analysis Software
3.2. Candidate Genes Related to Milk Yield (AY) in Xinjiang Donkeys
3.3. Candidate Genes Related to Milk Composition (FP, PP, and LP) in Xinjiang Donkeys
4. Materials and Methods
4.1. Animals and Phenotyping
4.2. Blood Sample Collection and DNA Extraction
4.3. Monitoring of Genomic DNA
4.4. Library Construction
4.5. Variant Sites Detection
4.6. Variant Sites Filtering
4.7. Genome-Wide Association Study
4.8. KASP
4.9. Primer Design and PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blasi, F.; Montesano, D.; De Angelis, M.; Maurizi, A.; Ventura, F.; Cossignani, L.; Simonetti, M.; Damiani, P. Results of stereospecific analysis of triacylglycerol fraction from donkey, cow, ewe, goat and buffalo milk. J. Food Compos. Anal. 2008, 21, 1–7. [Google Scholar] [CrossRef]
- Fantuz, F.; Ferraro, S.; Todini, L.; Cimarelli, L.; Fatica, A.; Marcantoni, F.; Salimei, E. Distribution of calcium, phosphorus, sulfur, magnesium, potassium, and sodium in major fractions of donkey milk. J. Dairy Sci. 2020, 103, 8741–8749. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Su, D.Q.; Ji, C.F.; Ding, Y.S.; Zhang, L.; Yu, D. Study on health protection efficacy of fresh donkey’s milk. Food Sci. 2008, 29, 423–426. [Google Scholar]
- Cimmino, F.; Catapano, A.; Villano, I.; Di Maio, G.; Petrella, L.; Traina, G.; Pizzella, A.; Tudisco, R.; Cavaliere, G. Invited review: Human, cow, and donkey milk comparison: Focus on metabolic effects. J. Dairy Sci. 2023, 106, 3072–3085. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, X.; Guo, H. The nutritional ingredients and antioxidant activity of donkey milk and donkey milk powder. Food Sci. Biotechnol. 2017, 27, 393–400. [Google Scholar] [CrossRef]
- Dalajiruga, D.; Zhang, Y.C.; Narenhua, N.; Cha, G.; Wu, H.Q.; Liu, Y.B. Research status on milk replacer for donkey foal. Anim. Husb. Feed. Sci. 2017, 38, 27–30. [Google Scholar]
- Selionova, M.; Trukhachev, V.; Aibazov, M.; Sermyagin, A.; Belous, A.; Gladkikh, M.; Zinovieva, N. Genome-wide association study of milk composition in Karachai goats. Animals 2024, 14, 327. [Google Scholar] [CrossRef]
- Yao, H.; Dou, Z.; Zhao, Z.; Liang, X.; Yue, H.; Ma, W.; Su, Z.; Wang, Y.; Hao, Z.; Yan, H.; et al. Transcriptome analysis of the Bactrian camel (Camelus bactrianus) reveals candidate genes affecting milk production traits. BMC Genom. 2023, 24, 660. [Google Scholar] [CrossRef]
- Tara, A.; Singh, P.; Gautam, D.; Tripathi, G.; Uppal, C.; Malhotra, S.; De, S.; Singh, M.K.; Telugu, B.P.; Selokar, N.L. CRISPR-mediated editing of β-lactoglobulin (BLG) gene in buffalo. Sci. Rep. 2024, 14, 14822. [Google Scholar] [CrossRef]
- Du, A.; Guo, Z.; Chen, A.; Xu, L.; Sun, D.; Han, B. PC gene affects milk production traits in dairy Cattle. Genes 2024, 15, 708. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Li, J.; Li, H.; Yang, L. Genome-wide identification of Diacylglycerol Acyltransferases (DGAT) family genes influencing milk production in Buffalo. BMC Genet. 2020, 21, 26. [Google Scholar] [CrossRef]
- Tian, H.; Luo, J.; Guo, P.; Li, C.; Zhang, X. C/EBPα promotes triacylglycerol synthesis via regulating PPARG promoter activity in goat mammary epithelial cells. J. Anim. Sci. 2023, 101, skac412. [Google Scholar] [CrossRef]
- Balia, F.; Pazzola, M.; Dettori, M.L.; Mura, M.C.; Luridiana, S.; Carcangiu, V.; Piras, G.; Vacca, G.M. Effect of CSN1S1 gene polymorphism and stage of lactation on milk yield and composition of extensively reared goats. J. Dairy Res. 2013, 80, 129–137. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, S.; Liu, S.; Dong, J.; Cao, Y.; Sun, Y. Single nucleotide polymorphisms (SNPs) and indels identified from whole-genome re-sequencing of four Chinese donkey breeds. Anim. Biotechnol. 2023, 34, 1828–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.L.; Zhang, S.E.; Sun, Y.J.; Wang, J.J.; Shen, W. Comparative transcriptomics uncover the uniqueness of oocyte development in the Donkey. Front. Genet. 2022, 13, 839207–839218. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, T.; Liang, H.; Wang, L.; Akhtar, F.; Shi, X.; Ren, W.; Huang, B.; Kou, X.; Chen, Y.; et al. A novel SNP in NKX1-2 gene is associated with carcass traits in Dezhou donkey. BMC Genom. Data 2023, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Shi, X.; Liu, Z.; Ren, W.; Wang, X.; Huang, B.; Kou, X.; Liang, H.; Wang, C.; Chai, W. A novel A > G polymorphism in the intron 1 of LCORL gene is significantly associated with hide weight and body size in Dezhou Donkey. Animals 2022, 12, 2581. [Google Scholar] [CrossRef]
- Chang, T.; Li, M.; An, X.; Bai, F.; Wang, F.; Yu, J.; Lei, C.; Dang, R. Association analysis of IGF2 gene polymorphisms with growth traits of Dezhou donkey. Anim. Biotechnol. 2023, 34, 1143–1153. [Google Scholar] [CrossRef]
- Yu, J.; Yang, G.; Li, S.; Li, M.; Ji, C.; Liu, G.; Wang, Y.; Chen, N.; Lei, C.; Dang, R. Identification of Dezhou donkey muscle development-related genes and long non-coding RNA based on differential expression analysis. Anim. Biotechnol. 2023, 34, 2313–2323. [Google Scholar] [CrossRef]
- He, H.Y.; Liu, L.L.; Chen, B.; Xiao, H.X.; Liu, W.J. Study on lactation performance and development of KASP marker for milk traits in Xinjiang donkey (Equus asinus). Anim. Biotechnol. 2023, 34, 2724–2735. [Google Scholar] [CrossRef]
- Zhou, X.L. Research progress of donkey lactation physiology and milk nutritional components. Chin. Dairy Cow 2010, 6, 44–47. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Vogt, F.; Shirsekar, G.; Weigel, D. vcf2gwas-python API for comprehensive GWAS analysis using GEMMA. Bioinformatics 2021, 38, 839–840. [Google Scholar] [CrossRef]
- Hisey, E.A.; Hermans, H.; Lounsberry, Z.T.; Avila, F.; Grahn, R.A.; Knickelbein, K.E.; Duward-Akhurst, S.A.; McCue, M.E.; Kalbfleisch, T.; Lassaline, M.E.; et al. Whole genome sequencing identified a 16 kilobase deletion on ECA13 associated with distichiasis in Friesian horses. BMC Genom. 2020, 21, 848–861. [Google Scholar] [CrossRef]
- Morton, J.M.; Auldist, M.J.; Douglas, M.L.; Macmillan, K.L. Associations between milk protein concentration, milk yield, and reproductive performance in dairy cows. J. Dairy Sci. 2016, 99, 10033–10043. [Google Scholar] [CrossRef]
- D’alessandro, A.G.; Martemucci, G. Lactation curve and effects of milking regimen on milk yield and quality, and udder health in martina franca jennies (Equus asinus). J. Anim. Sci. 2012, 90, 669–681. [Google Scholar] [CrossRef]
- Yu, X.; Fang, C.; Liu, L.; Zhao, X.; Liu, W.; Cao, H.; Lv, S. Transcriptome study underling difference of milk yield during peak lactation of Kazakh horse. J. Equine Vet. Sci. 2021, 102, 103424. [Google Scholar] [CrossRef]
- Liu, T. MicroRNA-26a Inhibits Cell Proliferation, Invasion and Enhances Docetaxel Sensitivity by Targeting FAM98A in Breast Cancer. Master’s Thesis, Anhui Medical University, Hefei, China, 2021. [Google Scholar]
- Liu, Y.; Du, H.; Wang, S.; Lv, Y.; Deng, H.; Chang, K.; Zhou, P.; Hu, C. Grass carp (Ctenopharyngodon idella) TNK1 modulates JAK-STAT signaling through phosphorylating STAT1. Dev. Comp. Immunol. 2021, 116, 103951–103963. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform. Biol. Insights 2011, 5, 83–98. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, H.R.; Wang, M.Z.; Wang, C.; Liu, S.M. Lactogenic hormones regulate mammary proteinsynthesis in bovine mammary epithelial cells via the mTOR and JAK-STAT signal pathways. Anim. Prod. Sci. 2016, 56, 1803–1809. [Google Scholar]
- Fong, L.G.; Young, S.G.; Beigneux, A.P.; Bensadoun, A.; Oberer, M.; Jiang, H.; Ploug, M. GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol. Metab. 2016, 27, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef]
- Dong, W.; Yang, J.; Zhang, Q.; Jiang, L. Role of GPIHBP1 in regulating milk protein traits in dairy cattle. Anim. Biotechnol. 2020, 31, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.Y.; Pang, K.; Zhang, X.Y.; Zhao, L.; Chen, S.W.; Dong, M.L.; Ren, F.Z. Composition, physiochemical properties, nitrogen fraction distribution, and amino acid profile of donkey milk. J. Dairy Sci. 2007, 90, 1635–1643. [Google Scholar] [CrossRef]
- Chiavari, C.; Coloretti, F.; Nanni, M.; Sorrentino, E.; Grazia, L. Use of donkey’s milk for a fermented beverage with lactobacilli. Le Lait 2005, 85, 481–490. [Google Scholar] [CrossRef]
- Salimei, E.; Fantuz, F.; Coppola, R.; Chiofalo, B.; Polidori, P.; Varisco, G. Composition and characteristics of ass’s milk. Anim. Res. 2004, 53, 67–78. [Google Scholar] [CrossRef]
- Haiman, C.A.; Han, Y.; Feng, Y.; Xia, L.; Hsu, C.; Sheng, X.; Pooler, L.C.; Patel, Y.; Kolonel, L.N.; Carter, E.; et al. Genome-wide testing of putative functional exonic variants in relationship with breast and prostate cancer risk in a multiethnic population. PLoS Genet. 2013, 9, e1003419. [Google Scholar] [CrossRef]
- Montaudon, E.; Nikitorowicz-Buniak, J.; Sourd, L.; Morisset, L.; EL Botty, R.; Huguet, L.; Dahmani, A.; Painsec, P.; Nemati, F.; Vacher, S.; et al. PLK1 inhibition exhibits strong anti-tumoral activity in CCND1-driven breast cancer metastases with acquired palbociclib resistance. Nat. Commun. 2020, 11, 4053–4069. [Google Scholar] [CrossRef]
- Zhao, X.; Xie, T.; Dai, T.; Zhao, W.; Li, J.; Xu, R.; Jiang, C.; Li, P.; Deng, J.; Su, X.; et al. CHP2 promotes cell proliferation in breast cancer via suppression of FOXO3a. Mol. Cancer Res. 2018, 16, 1512–1522. [Google Scholar] [CrossRef]
- Li, T.-F.; Zeng, H.-J.; Shan, Z.; Ye, R.-Y.; Cheang, T.-Y.; Zhang, Y.-J.; Lu, S.-H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020, 20, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Li, X.; Li, W.; Wang, Y.; Jiang, C.; Zhou, L.; Gao, J.; Yu, Y.; Shen, Y.; Xu, Q. Intracellular CYTL1, a novel tumor suppressor, stabilizes NDUFV1 to inhibit metabolic reprogramming in breast cancer. Signal Transduct. Target. Ther. 2022, 7, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Vishnubalaji, R.; Alajez, N.M. Epigenetic regulation of triple negative breast cancer (TNBC) by TGF-β signaling. Sci. Rep. 2021, 11, 15410–15423. [Google Scholar] [CrossRef]
- Zeng, Z.; Xu, S.; Wang, F.; Peng, X.; Zhang, W.; Zhan, Y.; Ding, Y.; Liu, Z.; Liang, L. HAO1-mediated oxalate metabolism promotes lung pre-metastatic niche formation by inducing neutrophil extracellular traps. Oncogene 2022, 41, 3719–3731. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, L.; Li, J.; Hu, K. ALKBH family members as novel biomarkers and prognostic factors in human breast cancer. Aging 2022, 14, 6579–6593. [Google Scholar] [CrossRef]
- Mimoto, R.; Taira, N.; Takahashi, H.; Yamaguchi, T.; Okabe, M.; Uchida, K.; Miki, Y.; Yoshida, K. DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. Cancer Lett. 2013, 339, 214–225. [Google Scholar] [CrossRef]
- Xie, Q.; Xiao, Y.-S.; Jia, S.-C.; Zheng, J.-X.; Du, Z.-C.; Chen, Y.-C.; Chen, M.-T.; Liang, Y.-K.; Lin, H.-Y.; Zeng, D. FABP7 is a potential biomarker to predict response to neoadjuvant chemotherapy for breast cancer. Cancer Cell Int. 2020, 20, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Schenkel, F.S.; Miglior, F.; Zhao, X.; Ibeagha-Awemu, E.M. Genome wide association study identifies novel potential candidate genes for bovine milk cholesterol content. Sci. Rep. 2018, 8, 13239–13255. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Zhu, J.; Zhao, Y.; Lin, Y. Molecular characterization of GTP binding protein overexpressed in skeletal muscle (GEM) and its role in promoting adipogenesis in goat intramuscular preadipocytes. Anim. Biotechnol. 2020, 31, 17–24. [Google Scholar] [CrossRef]
- Yu, J.; Ka, S.O.; Kwon, K.B.; Lee, S.M.; Park, J.W.; Park, B.H. Overexpression of the small GTPase Arl4D suppresses adipogenesis. Int. J. Mol. Med. 2011, 28, 793–798. [Google Scholar] [CrossRef]
- Park, B.S.; Im, H.L.; Yoon, N.A.; Tu, T.H.; Park, J.W.; Kim, J.G.; Lee, B.J. Developmentally regulated GTP-binding protein-2 regulates adipocyte differentiation. Biochem. Biophys. Res. Commun. 2021, 578, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Liu, T.; Jin, S.-B.; Ning, C.; Lendahl, U.; Nistér, M.; Zhao, J. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci. Rep. 2017, 7, 880–896. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Cheng, L.; Shi, Y.; Li, J.; Yun, Q.; Yang, H. MIEF2 reprograms lipid metabolism to drive progression of ovarian cancer through ROS/AKT/mTOR signaling pathway. Cell Death Dis. 2021, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef]
- Mazzarotto, F.; Tayal, U.; Buchan, R.J.; Midwinter, W.; Wilk, A.; Whiffin, N.; Govind, R.; Mazaika, E.; de Marvao, A.; Dawes, T.J.; et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation 2020, 141, 387–398. [Google Scholar] [CrossRef]
- Cigliola, V.; Populaire, C.; Pierri, C.L.; Deutsch, S.; Haefliger, J.-A.; Fadista, J.; Lyssenko, V.; Groop, L.; Rueedi, R.; Thorel, F.; et al. A variant of GJD2, encoding for connexin 36, alters the function of insulin producing β-cells. PLoS ONE 2016, 11, e0150880. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Liu, S.; Campanile, G.; Salzano, A.; Gasparrini, B.; Plastow, G.; Zhang, C.; Wang, Z.; Liang, A.; et al. Genome-wide association study for buffalo mammary gland morphology. J. Dairy Res. 2020, 87, 27–31. [Google Scholar] [CrossRef]
- Mcgrath, J.A.; Ohyama, M.; Simpson, M.A. PADI3, hair disorders and genomic investigation. Br. J. Dermatol. 2019, 181, 1115–1116. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, K. Multiple functions of DYRK2 in cancer and tissue development. FEBS Lett. 2019, 593, 2953–2965. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, L.; Chen, C.J.; Zhang, M.; Lu, X.; Zhang, Z.; Huang, X.; Shi, Y. Genome-wide association study of milk and reproductive traits in dual-purpose Xinjiang Brown cattle. BMC Genom. 2019, 20, 827–838. [Google Scholar] [CrossRef]
- Mao, C.; Ju, X.; Cheng, H.; Huang, X.; Jiang, F.; Yao, Y.; Lan, X.; Song, E. Determination of genetic variation within the DYRK2 gene and its associations with milk traits in cattle. Arch. Anim. Breed. 2020, 63, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kwong, S.C.; Jamil, A.H.A.; Rhodes, A.; Taib, N.A.; Chung, I. Metabolic role of fatty acid binding protein 7 in mediating triple-negative breast cancer cell death via PPAR-α signaling. J. Lipid Res. 2019, 60, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, H.; Guo, Y.; Huang, J.; Sun, Y.; Min, J.; Wang, J.; Fang, X.; Zhao, Z.; Wang, S.; et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 2020, 11, 6014–6029. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- VanRanden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar]



| Traits | n | Mean | SD | Minimum | Maximum |
|---|---|---|---|---|---|
| AY (kg) | 112 | 38.51 | 14.87 | 13.60 | 72.75 |
| FP (%) | 112 | 1.02 | 0.93 | 0.12 | 4.52 |
| PP (%) | 112 | 1.44 | 0.19 | 0.99 | 1.94 |
| LP (%) | 112 | 6.46 | 0.39 | 4.47 | 6.96 |
| Official Full Name of Gene | Gene | Gene ID | Allele Substitution | Position | Chromosome |
|---|---|---|---|---|---|
| Glutamine-fructose-6-phosphate transaminase 2 | GFPT2 | 106843727 | C/A | 25060895 | 9 |
| GFPT2 | 106843727 | T/G | 25062599 | 9 | |
| Notch receptor 1 | NOTCH1 | 106845559 | T/C | 9133371 | 10 |
| DNA topoisomerase I mitochondrial | TOP1MT | 106841874 | G/A | 38323301 | 12 |
| Glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 | GPIHBP1 | 106841943 | C/T | 38365122 | 12 |
| Developmentally regulated GTP binding protein 2 | DRG2 | 106837927 | C/A | 4912631 | 13 |
| DRG2 | 106837927 | C/T | 4939345 | 13 | |
| FLII actin remodeling protein | FLII | 106837933 | G/C | 5044716 | 13 |
| FLII | 106837933 | C/T | 5046888 | 13 | |
| Mitochondrial elongation factor 2 | MIEF2 | 106837926 | G/T | 5060681 | 13 |
| Tyrosine kinase non receptor 1 | TNK1 | 106844191 | A/G | 14413548 | 13 |
| TNK1 | 106844191 | A/C | 14416183 | 13 | |
| Polo like kinase 1 | PLK1 | 106845827 | T/C | 23585377 | 14 |
| PLK1 | 106845827 | G/C | 23587802 | 14 | |
| Dual specificity tyrosine phosphorylation regulated kinase 2 | DYRK2 | 106843596 | C/G | 3170005 | 22 |
| DYRK2 | 106843596 | T/C | 3171414 | 22 |
| Gene | Position | Allele Substitution | Genotype | Genotype Frequencies | χ2 | p |
|---|---|---|---|---|---|---|
| GFPT2 | 25060895 | C>A | CC | 0.013 | 0.295 | 0.587 |
| CA | 0.150 | |||||
| AA | 0.837 | |||||
| GFPT2 | 25062599 | T>G | TT | 0.063 | 1.902 | 0.348 |
| TG | 0.225 | |||||
| GG | 0.712 | |||||
| NOTCH1 | 9133371 | T>C | TT | 0.288 | 1.618 | 0.472 |
| TC | 0.400 | |||||
| CC | 0.312 | |||||
| TOP1MT | 38323301 | G>A | GG | 0.500 | 3.005 | 0.232 |
| GA | 0.325 | |||||
| AA | 0.175 | |||||
| GPIHBP1 | 38365122 | C>T | CC | 0.513 | 2.023 | 0.389 |
| CT | 0.337 | |||||
| TT | 0.150 | |||||
| DRG2 | 4912631 | C>A | CC | 0.473 | 14.006 | 0.001 |
| CA | 0.203 | |||||
| AA | 0.324 | |||||
| DRG2 | 4939345 | C>T | CC | 0.400 | 3.453 | 0.188 |
| CT | 0.350 | |||||
| TT | 0.250 | |||||
| FLII | 5044716 | G>C | GG | 0.950 | 1.450 | 0.719 |
| GC | 0.038 | |||||
| CC | 0.012 | |||||
| FLII | 5046888 | C>T | CC | 0.357 | 4.644 | 0.105 |
| CT | 0.314 | |||||
| TT | 0.329 | |||||
| MIEF2 | 5060681 | G>T | GG | 0.738 | 2.236 | 0.338 |
| GT | 0.200 | |||||
| TT | 0.062 | |||||
| TNK1 | 14413548 | A>G | AA | 0.014 | 0.290 | 1.000 |
| AG | 0.043 | |||||
| GG | 0.943 | |||||
| TNK1 | 14416183 | A>C | AA | 0.792 | 35.321 | <0.001 |
| AC | 0.013 | |||||
| CC | 0.195 | |||||
| PLK1 | 23585377 | T>C | TT | 0.571 | 2.311 | 0.330 |
| TC | 0.300 | |||||
| CC | 0.129 | |||||
| PLK1 | 23587802 | G>C | GG | 0.164 | 3.528 | 0.177 |
| GC | 0.288 | |||||
| CC | 0.548 | |||||
| DYRK2 | 3170005 | C>G | CC | 0.914 | 1.293 | 0.764 |
| CG | 0.071 | |||||
| GG | 0.015 | |||||
| DYRK2 | 3171414 | T>C | TT | 0.914 | <0.001 | 1.000 |
| TC | 0.086 | |||||
| CC | 0 |
| Gene | Position | Genotype | Average Daily Milk Yield (kg) | Total Milk Yield (kg) |
|---|---|---|---|---|
| NOTCH1 | 9133371 | TT (23) | 2.67 ± 0.15 Aa | 482.33 ± 28.02 Aa |
| TC (32) | 2.42 ± 0.10 Aa | 435.89 ± 18.51 Aa | ||
| CC (25) | 1.57 ± 0.19 Bb | 254.48 ± 35.16 Bb | ||
| TOP1MT | 38323301 | GG (40) | 1.86 ± 0.12 Bb | 316.47 ± 24.11 Bb |
| GA (26) | 2.60 ± 0.16 Aa | 467.93 ± 28.16 Aa | ||
| AA (14) | 2.61 ± 0.23 Aa | 469.92 ± 41.61 Aa | ||
| GPIHBP1 | 38365122 | CC (41) | 1.88 ± 0.14 Bb | 320.12 ± 26.65 Bb |
| CT (27) | 2.75 ± 0.13 Aa | 495.25 ± 22.99 Aa | ||
| TT (12) | 2.27 ± 0.19 ABab | 408.95 ± 35.05 ABab | ||
| DRG2 | 4912631 | CC (35) | 2.57 ± 0.12 Aa | 462.09 ± 21.32 Aa |
| CA (15) | 1.98 ± 0.21 ABa | 345.35 ± 41.44 ABa | ||
| AA (24) | 1.76 ± 0.18 Bb | 293.94 ± 33.67 Bb | ||
| DRG2 | 4939345 | CC (32) | 1.74 ± 0.17 Bb | 291.96 ± 31.66 Bb |
| CT (28) | 2.42 ± 0.10 Aa | 435.73 ± 18.53 Aa | ||
| TT (20) | 2.74 ± 0.17 Aa | 493.05 ± 31.19 Aa | ||
| FLII | 5046888 | CC (25) | 1.66 ± 0.18 Bb | 270.49 ± 34.51 Bb |
| CT (22) | 2.67 ± 0.13 Aa | 480.44 ± 24.28 Aa | ||
| TT (23) | 2.51 ± 0.17 Aa | 451.18 ± 31.33 Aa | ||
| PLK1 | 23585377 | TT (40) | 1.93 ± 0.15 Ab | 330.86 ± 29.08 Bb |
| TC (21) | 2.65 ± 0.14 Aa | 476.60 ± 25.39 ABa | ||
| CC (9) | 2.74 ± 0.23 Aa | 493.45 ± 41.05 Aa | ||
| PLK1 | 23587802 | GG (12) | 2.62 ± 0.18 Aa | 471.73 ± 32.51 Aa |
| GC (21) | 2.65 ± 0.14 Aa | 476.60 ± 25.39 Aa | ||
| CC (40) | 1.93 ± 0.15 Ab | 330.86 ± 29.08 Bb |
| Gene | Position | Primer Sequence |
|---|---|---|
| GFPT2 | 25060895 | F1: GAAGGTCGGAGTCAACGGATTTCACATGGTCTCTCCTCCCAC |
| F2: GAAGGTGACCAAGTTCATGCTTCACATGGTCTCTCCTCCCAA | ||
| R: CCTTCATGGGGATTCACTGC | ||
| GFPT2 | 25062599 | F1: GAAGGTCGGAGTCAACGGATTGGCTCGGCCTCCTGCTA |
| F2: GAAGGTGACCAAGTTCATGCTGGCTCGGCCTCCTGCTC | ||
| R: GCTGAGGCTCCGGGCTAT | ||
| NOTCH1 | 9133371 | F1: GAAGGTCGGAGTCAACGGATTGATTGTCCTGCTGTTCAAACAC |
| F2: GAAGGTGACCAAGTTCATGCTGATTGTCCTGCTGTTCAAACAT | ||
| R: GCACTGCCCCCTCGC | ||
| TOP1MT | 38323301 | F1: GAAGGTCGGAGTCAACGGATTGGCGAAGACTTTGAGTTCTAAACA |
| F2: GAAGGTGACCAAGTTCATGCTGCGAAGACTTTGAGTTCTAAACG | ||
| R: ATAAACTCCAGTGAGGCACGAG | ||
| GPIHBP1 | 38365122 | F1: GAAGGTCGGAGTCAACGGATTGCCGCAGGACAGGACAT |
| F2: GAAGGTGACCAAGTTCATGCTGCCGCAGGACAGGACAC | ||
| R: TGCCTCCCGCATTCTTC | ||
| DRG2 | 4912631 | F1: GAAGGTCGGAGTCAACGGATTGCCTCTCTACTCGGTCACCG |
| F2: GAAGGTGACCAAGTTCATGCTGGCCTCTCTACTCGGTCACCT | ||
| R: GCCCCTGAGGGATCTGG | ||
| DRG2 | 4939345 | F1: GAAGGTCGGAGTCAACGGATTGGCCTGGGGAAGGCA |
| F2: GAAGGTGACCAAGTTCATGCTGGCCTGGGGAAGGCG | ||
| R: CTCTCCTCCTGCAGCTCTCA | ||
| FLII | 5044716 | F1: GAAGGTCGGAGTCAACGGATTCGTGCAGGTACCCCCAG |
| F2: GAAGGTGACCAAGTTCATGCTCGTGCAGGTACCCCCAC | ||
| R: GCTGGGCATGACATGAGG | ||
| FLII | 5046888 | F1: GAAGGTCGGAGTCAACGGATTGCGCTGCCCTAGGCC |
| F2: GAAGGTGACCAAGTTCATGCTGGCGCTGCCCTAGGCT | ||
| R: CTGCTTCCATGCCTGGG | ||
| MIEF2 | 5060681 | F1: GAAGGTCGGAGTCAACGGATTCACCCGTTCCGAGGCA |
| F2: GAAGGTGACCAAGTTCATGCTCACCCGTTCCGAGGCC | ||
| R: CACTGGGTCACACCATTCACA | ||
| TNK1 | 14413548 | F1: GAAGGTCGGAGTCAACGGATTACAGGGGAAGGGAGGTTTT |
| F2: GAAGGTGACCAAGTTCATGCTACAGGGGAAGGGAGGTTTC | ||
| R: CCAGGCCGGACCCTG | ||
| TNK1 | 14416183 | F1: GAAGGTCGGAGTCAACGGATTTCTGCTTCTTCACCTGGGG |
| F2: GAAGGTGACCAAGTTCATGCTGTCTGCTTCTTCACCTGGGT | ||
| R: ACTGTGCCAGGTTCCGG | ||
| PLK1 | 23585377 | F1: GAAGGTCGGAGTCAACGGATTGAGAGTTCCCAGGAGGCAAT |
| F2: GAAGGTGACCAAGTTCATGCTGAGAGTTCCCAGGAGGCAAC | ||
| R: AGTCCCTGTCCACAGGTGG | ||
| PLK1 | 23587802 | F1: GAAGGTCGGAGTCAACGGATTCAAACTCATCCTGTGCCCG |
| F2: GAAGGTGACCAAGTTCATGCTCAAACTCATCCTGTGCCCC | ||
| R: CGATGTAGGTCACGGCTGC | ||
| DYRK2 | 3170005 | F1: GAAGGTCGGAGTCAACGGATTTGGGAAGGCAAGGTAATTATATG |
| F2: GAAGGTGACCAAGTTCATGCTTGGGAAGGCAAGGTAATTATATC | ||
| R: GGATTACATTTGCAATTATGTATCTG | ||
| DYRK2 | 3171414 | F1: GAAGGTCGGAGTCAACGGATTTGCATCTCCAGGAAGGTCAG |
| F2: GAAGGTGACCAAGTTCATGCTTGCATCTCCAGGAAGGTCAA | ||
| R: GGAGGAGTAAATATTAATTACTTGGTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, C.; Farnir, F.; Liu, L.; Xiao, H. Unveiling Genetic Markers for Milk Yield in Xinjiang Donkeys: A Genome-Wide Association Study and Kompetitive Allele-Specific PCR-Based Approach. Int. J. Mol. Sci. 2025, 26, 2961. https://doi.org/10.3390/ijms26072961
Fang C, Farnir F, Liu L, Xiao H. Unveiling Genetic Markers for Milk Yield in Xinjiang Donkeys: A Genome-Wide Association Study and Kompetitive Allele-Specific PCR-Based Approach. International Journal of Molecular Sciences. 2025; 26(7):2961. https://doi.org/10.3390/ijms26072961
Chicago/Turabian StyleFang, Chao, Frederic Farnir, Lingling Liu, and Haixia Xiao. 2025. "Unveiling Genetic Markers for Milk Yield in Xinjiang Donkeys: A Genome-Wide Association Study and Kompetitive Allele-Specific PCR-Based Approach" International Journal of Molecular Sciences 26, no. 7: 2961. https://doi.org/10.3390/ijms26072961
APA StyleFang, C., Farnir, F., Liu, L., & Xiao, H. (2025). Unveiling Genetic Markers for Milk Yield in Xinjiang Donkeys: A Genome-Wide Association Study and Kompetitive Allele-Specific PCR-Based Approach. International Journal of Molecular Sciences, 26(7), 2961. https://doi.org/10.3390/ijms26072961







