Early Progression Prediction in Korean Crohn’s Disease Using a Korean-Specific PrediXcan Model
Abstract
1. Introduction
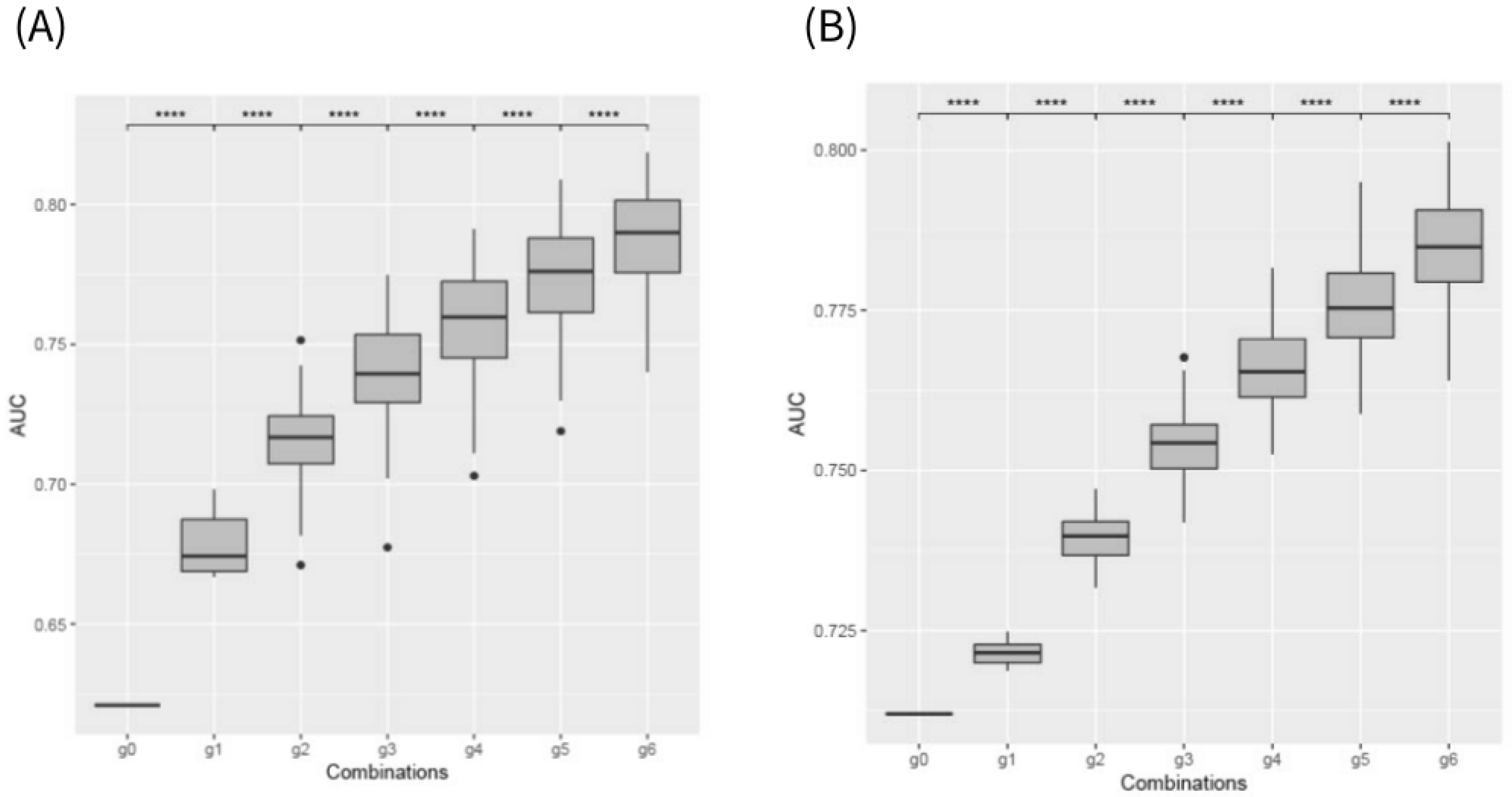
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Genotyping
4.3. RNA-Seq
4.4. Development of the Korean PrediXcan Model
4.5. Development of Early Progression Model Method
4.5.1. Gene Expression
4.5.2. Prediction Model and Performance Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin. Proc. 2019, 94, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Silverberg, M.S.; Satsangi, J.; Ahmad, T.; Arnott, I.D.; Bernstein, C.N.; Brant, S.R.; Caprilli, R.; Colombel, J.F.; Gasche, C.; Geboes, K.; et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. Hepatol. 2005, 19 (Suppl. A), 5A–36A. [Google Scholar] [CrossRef] [PubMed]
- Roda, G.; Chien Ng, S.; Kotze, P.G.; Argollo, M.; Panaccione, R.; Spinelli, A.; Kaser, A.; Peyrin-Biroulet, L.; Danese, S. Crohn’s disease. Nat. Rev. Dis. Primers 2020, 6, 22. [Google Scholar] [CrossRef]
- Louis, E.; Collard, A.; Oger, A.F.; Degroote, E.; El Yafi, F.A.N.; Belaiche, J. Behaviour of Crohn’s disease according to the Vienna classification: Changing pattern over the course of the disease. Gut 2001, 49, 777–782. [Google Scholar] [CrossRef]
- Cosnes, J.; Cattan, S.; Blain, A.; Beaugerie, L.; Carbonnel, F.; Parc, R.; Gendre, J.P. Long-term evolution of disease behavior of Crohn’s disease. Inflamm. Bowel Dis. 2002, 8, 244–250. [Google Scholar] [CrossRef]
- Park, S.K.; Kim, Y.B.; Kim, S.; Lee, C.W.; Choi, C.H.; Kang, S.B.; Kim, T.O.; Bang, K.B.; Chun, J.; Cha, J.M.; et al. Development of a Machine Learning Model to Predict Non-Durable Response to Anti-TNF Therapy in Crohn’s Disease Using Transcriptome Imputed from Genotypes. J. Pers. Med. 2022, 12, 947. [Google Scholar] [CrossRef]
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; GTEx Consortium; Nicolae, D.L.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef]
- Tarrant, K.M.; Barclay, M.L.; Frampton, C.M.; Gearry, R.B. Perianal disease predicts changes in Crohn’s disease phenotype-results of a population-based study of inflammatory bowel disease phenotype. Am. J. Gastroenterol. 2008, 103, 3082–3093. [Google Scholar] [CrossRef]
- Lakatos, P.L.; Czegledi, Z.; Szamosi, T.; Banai, J.; David, G.; Zsigmond, F.; Pandur, T.; Erdelyi, Z.; Gemela, O.; Papp, J.; et al. Perianal disease, small bowel disease, smoking, prior steroid or early azathioprine/biological therapy are predictors of disease behavior change in patients with Crohn’s disease. World J. Gastroenterol. 2009, 15, 3504–3510. [Google Scholar] [CrossRef]
- Tang, L.Y.; Rawsthorne, P.; Bernstein, C.N. Are perineal and luminal fistulas associated in Crohn’s disease? A population-based study. Clin. Gastroenterol. Hepatol. 2006, 4, 1130–1134. [Google Scholar] [CrossRef]
- Cosnes, J.; Seksik, P.; Nion-Larmurier, I.; Beaugerie, L.; Gendre, J.P. Prior appendectomy and the phenotype and course of Crohn’s disease. World J. Gastroenterol. 2006, 12, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ma, J.; Ben, Q.; Lu, L.; Wan, X. Prior Appendectomy and the Onset and Course of Crohn’s Disease in Chinese Patients. Gastroenterol. Res. Pract. 2019, 2019, 8463926. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, S.; Borowski, K.; Espin-Garcia, O.; Milgrom, R.; Kabakchiev, B.; Stempak, J.; Panikkath, D.; Eksteen, B.; Xu, W.; Steinhart, A.H.; et al. The Unsolved Link of Genetic Markers and Crohn’s Disease Progression: A North American Cohort Experience. Inflamm. Bowel Dis. 2019, 25, 1541–1549. [Google Scholar] [CrossRef]
- Pernat Drobez, C.; Repnik, K.; Gorenjak, M.; Ferkolj, I.; Weersma, R.K.; Potocnik, U. DNA polymorphisms predict time to progression from uncomplicated to complicated Crohn’s disease. Eur. J. Gastroenterol. Hepatol. 2018, 30, 447–455. [Google Scholar] [CrossRef]
- Pernat Drobez, C.; Ferkolj, I.; Potocnik, U.; Repnik, K. Crohn’s Disease Candidate Gene Alleles Predict Time to Progression from Inflammatory B1 to Stricturing B2, or Penetrating B3 Phenotype. Genet. Test. Mol. Biomarkers 2018, 22, 143–151. [Google Scholar] [CrossRef]
- Ditrich, F.; Blumel, S.; Biedermann, L.; Fournier, N.; Rossel, J.B.; Ellinghaus, D.; Franke, A.; Stange, E.F.; Rogler, G.; Scharl, M.; et al. Genetic risk factors predict disease progression in Crohn’s disease patients of the Swiss inflammatory bowel disease cohort. Ther. Adv. Gastroenterol. 2020, 13, 1756284820959252. [Google Scholar] [CrossRef]
- Lee, J.-A.; Suh, D.-C.; Kang, J.-E.; Kim, M.-H.; Park, H.; Lee, M.-N.; Kim, J.-M.; Jeon, B.-N.; Roh, H.-E.; Yu, M.-Y.; et al. Transcriptional activity of Sp1 is regulated by molecular interactions between the zinc finger DNA binding domain and the inhibitory domain with corepressors, and this interaction is modulated by MEK. J. Biol. Chem. 2005, 280, 28061–28071. [Google Scholar] [CrossRef]
- Vellingiri, B.; Iyer, M.; Devi Subramaniam, M.; Jayaramayya, K.; Siama, Z.; Giridharan, B.; Narayanasamy, A.; Abdal Dayem, A.; Cho, S.G. Understanding the Role of the Transcription Factor Sp1 in Ovarian Cancer: From Theory to Practice. Int. J. Mol. Sci. 2020, 21, 1153. [Google Scholar] [CrossRef]
- Hou, J.J.; Ma, A.H.; Qin, Y.H. Activation of the aryl hydrocarbon receptor in inflammatory bowel disease: Insights from gut microbiota. Front. Cell Infect. Microbiol. 2023, 13, 1279172. [Google Scholar] [CrossRef]
- Glas, J.; Konrad, A.; Schmechel, S.; Dambacher, J.; Seiderer, J.; Schroff, F.; Wetzke, M.; Roeske, D.; Torok, H.P.; Tonenchi, L.; et al. The ATG16L1 gene variants rs2241879 and rs2241880 (T300A) are strongly associated with susceptibility to Crohn’s disease in the German population. Am. J. Gastroenterol. 2008, 103, 682–691. [Google Scholar] [CrossRef]
- Gammoh, N. The multifaceted functions of ATG16L1 in autophagy and related processes. J. Cell Sci. 2020, 133, jcs249227. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.C.; Xu, B.; Finkelstein, D.; Fan, Y.; Carroll, P.A.; Cheng, P.F.; Eisenman, R.N.; Demontis, F. The glucose-sensing transcription factor MLX promotes myogenesis via myokine signaling. Genes. Dev. 2015, 29, 2475–2489. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Kim, H.N.; Choi, C.H.; Im, J.P.; Cha, J.M.; Eun, C.S.; Kim, T.O.; Kang, S.B.; Bang, K.B.; Kim, H.G.; et al. Differentially Abundant Bacterial Taxa Associated with Prognostic Variables of Crohn’s Disease: Results from the IMPACT Study. J. Clin. Med. 2020, 9, 1748. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Kim, S.; Lee, G.Y.; Kim, S.Y.; Kim, W.; Lee, C.W.; Park, J.L.; Choi, C.H.; Kang, S.B.; Kim, T.O.; et al. Development of a Machine Learning Model to Distinguish between Ulcerative Colitis and Crohn’s Disease Using RNA Sequencing Data. Diagnostics 2021, 11, 2365. [Google Scholar] [CrossRef]
- Kim, H.; Na, J.E.; Kim, S.; Kim, T.O.; Park, S.K.; Lee, C.W.; Kim, K.O.; Seo, G.S.; Kim, M.S.; Cha, J.M.; et al. A Machine Learning-Based Diagnostic Model for Crohn’s Disease and Ulcerative Colitis Utilizing Fecal Microbiome Analysis. Microorganisms 2023, 12, 36. [Google Scholar] [CrossRef]
- Kim, E.S.; Kim, S.K.; Park, D.I.; Kim, H.J.; Lee, Y.J.; Koo, J.S.; Kim, E.S.; Yoon, H.; Lee, J.H.; Kim, J.W.; et al. Comparison of the Pharmacokinetics of CT-P13 Between Crohn’s Disease and Ulcerative Colitis. J. Clin. Gastroenterol. 2023, 57, 601–609. [Google Scholar] [CrossRef]
| Characteristics | CD, n (%) (n = 430) | |
|---|---|---|
| Age at diagnosis, year, median ± SD | 26.9 ± 12.1 | |
| Gender, male | 313 (72.8) | |
| History of smoking | 78 (18.1) | |
| Family history of IBD | 11 (2.6) | |
| Disease location | Terminal ileum | 118 (27.4) |
| Colon | 52 (12.1) | |
| Ileocolon | 244 (56.7) | |
| Terminal ileum + upper GI | 4 (0.9) | |
| Colon + upper GI | 1 (0.2) | |
| Ileocolon + upper GI | 11 (2.6) | |
| Extra GI involvement | Arthritis | 27 (6.3) |
| Iritis/Uveitis | 4 (0.9) | |
| Erythema nodosum/Stomatitis | 4 (0.9) | |
| Perianal disease | 134 (31.2) | |
| Appendectomy history | 36 (8.4) | |
| History of UC diagnosis | 14 (3.3) | |
| Anti-TNF-α treatment within 2 years | 79 (18.4) | |
| Disease behavior changes within 2 years | B1 | 297 (69.1) |
| B2 | 60 (14.0) | |
| B3 | 73 (16.9) | |
| Model | Estimate | Standard Error | Z-Value | p-Value | |
|---|---|---|---|---|---|
| B2 | CCDC154 | −5.446 | 1.710 | −3.184 | 0.001 |
| FAM189A2 | −0.842 | 0.315 | −2.671 | 0.008 | |
| TAS2R19 | −1.012 | 0.358 | −2.827 | 0.005 | |
| FCSK | 0.895 | 0.331 | 2.703 | 0.006 | |
| SP1 | 2.109 | 0.669 | 3.151 | 0.002 | |
| KCNIP1 | −2.205 | 0.935 | −2.359 | 0.018 | |
| B3 | History of appendectomy | 1.882 | 0.508 | 3.705 | <0.001 |
| Anti-TNF use | −3.025 | 0.811 | −3.730 | <0.001 | |
| Ileocolonic disease | 1.412 | 0.608 | 2.323 | 0.020 | |
| PUS7 | −1.531 | 0.573 | −2.671 | 0.008 | |
| CCDC146 | −0.910 | 0.301 | −3.026 | 0.002 | |
| MLXIP | −2.421 | 0.603 | −4.013 | <0.001 | |
| LRGUK | 0.752 | 0.317 | 2.373 | 0.018 | |
| UROS | −1.476 | 0.533 | −2.771 | 0.006 | |
| TAFA1 | −0.820 | 0.357 | −2.298 | 0.022 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.-w.; Park, S.K.; Chun, J.; Kim, S.; Choi, C.H.; Kang, S.-B.; Bang, K.B.; Kim, T.O.; Seo, G.S.; Cha, J.M.; et al. Early Progression Prediction in Korean Crohn’s Disease Using a Korean-Specific PrediXcan Model. Int. J. Mol. Sci. 2025, 26, 2910. https://doi.org/10.3390/ijms26072910
Kim T-w, Park SK, Chun J, Kim S, Choi CH, Kang S-B, Bang KB, Kim TO, Seo GS, Cha JM, et al. Early Progression Prediction in Korean Crohn’s Disease Using a Korean-Specific PrediXcan Model. International Journal of Molecular Sciences. 2025; 26(7):2910. https://doi.org/10.3390/ijms26072910
Chicago/Turabian StyleKim, Tae-woo, Soo Kyung Park, Jaeyoung Chun, Suji Kim, Chang Hwan Choi, Sang-Bum Kang, Ki Bae Bang, Tae Oh Kim, Geom Seog Seo, Jae Myung Cha, and et al. 2025. "Early Progression Prediction in Korean Crohn’s Disease Using a Korean-Specific PrediXcan Model" International Journal of Molecular Sciences 26, no. 7: 2910. https://doi.org/10.3390/ijms26072910
APA StyleKim, T.-w., Park, S. K., Chun, J., Kim, S., Choi, C. H., Kang, S.-B., Bang, K. B., Kim, T. O., Seo, G. S., Cha, J. M., Jung, Y., Kim, H. G., Im, J. P., Ahn, K. S., Lee, C. K., Kim, H. J., Kim, S., & Park, D. I. (2025). Early Progression Prediction in Korean Crohn’s Disease Using a Korean-Specific PrediXcan Model. International Journal of Molecular Sciences, 26(7), 2910. https://doi.org/10.3390/ijms26072910







