Decoding Multifaceted Roles of Sleep-Related Genes as Molecular Bridges in Chronic Disease Pathogenesis
Abstract
1. Introduction
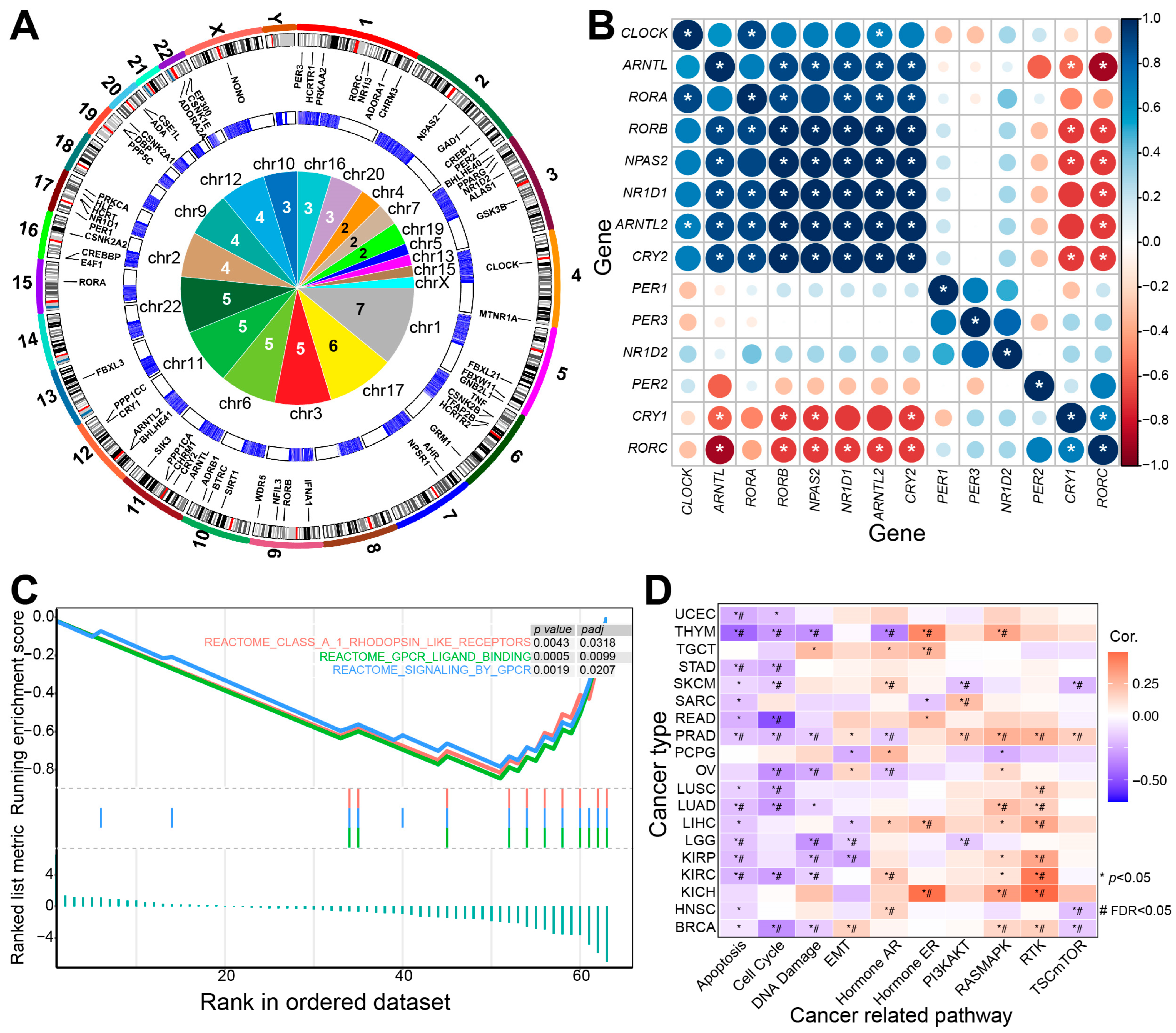
2. Sleep-Related Genes Contribute to Multiple Biological Processes
2.1. Circadian Rhythm-Related Genes Are Associated with Sleep
2.2. Other Sleep-Related Regulatory Genes
2.3. Sleep-Related Genes Are Associated with Multiple Biological Processes
3. Dysregulated Sleep-Related Genes Have a Critical Role in Human Diseases
3.1. Sleep-Related Genes May Be Associated with Metabolism-Related Diseases
3.2. Sleep-Related Genes May Be Associated with Cardiovascular Diseases
3.3. Sleep-Related Genes Are Associated with Cancer
3.4. Sleep and Mental Health
4. Conclusions
Funding
Conflicts of Interest
References
- Siegel, J.M. Clues to the functions of mammalian sleep. Nature 2005, 437, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Sugita, Y.; Koga, E.; Shirakawa, S.; Inoue, K.; Uchida, S.; Kuwahara, H.; Kousaka, M.; Kobayashi, T.; Tsuji, Y.; et al. Proposed supplements and amendments to ’A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects’, the Rechtschaffen & Kales (1968) standard. Psychiatry Clin. Neurosci. 2001, 55, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, L.; Yang, G.; Gan, W.B. REM sleep selectively prunes and maintains new synapses in development and learning. Nat. Neurosci. 2017, 20, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Filho, D.G.; Queiroz, C.M.; Ribeiro, S. Memory corticalization triggered by REM sleep: Mechanisms of cellular and systems consolidation. Cell Mol. Life Sci. 2018, 75, 3715–3740. [Google Scholar] [CrossRef]
- Cade, B.E.; Gottlieb, D.J.; Lauderdale, D.S.; Bennett, D.A.; Buchman, A.S.; Buxbaum, S.G.; De Jager, P.L.; Evans, D.S.; Fülöp, T.; Gharib, S.A.; et al. Common variants in DRD2 are associated with sleep duration: The CARe consortium. Hum. Mol. Genet. 2016, 25, 167–179. [Google Scholar] [CrossRef]
- Chemelli, R.M.; Willie, J.T.; Sinton, C.M.; Elmquist, J.K.; Scammell, T.; Lee, C.; Richardson, J.A.; Williams, S.C.; Xiong, Y.; Kisanuki, Y.; et al. Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell 1999, 98, 437–451. [Google Scholar] [CrossRef]
- Naganuma, F.; Nakamura, T.; Yoshikawa, T.; Iida, T.; Miura, Y.; Kárpáti, A.; Matsuzawa, T.; Yanai, A.; Mogi, A.; Mochizuki, T.; et al. Histamine N-methyltransferase regulates aggression and the sleep-wake cycle. Sci. Rep. 2017, 7, 15899. [Google Scholar] [CrossRef]
- Tatsuki, F.; Sunagawa, G.A.; Shi, S.; Susaki, E.A.; Yukinaga, H.; Perrin, D.; Sumiyama, K.; Ukai-Tadenuma, M.; Fujishima, H.; Ohno, R.; et al. Involvement of Ca2+-Dependent Hyperpolarization in Sleep Duration in Mammals. Neuron 2016, 90, 70–85. [Google Scholar] [CrossRef]
- Schenck, C.H.; Boeve, B.F.; Mahowald, M.W. Delayed emergence of a parkinsonian disorder or dementia in 81% of older men initially diagnosed with idiopathic rapid eye movement sleep behavior disorder: A 16-year update on a previously reported series. Sleep Med. 2013, 14, 744–748. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, J.; Lam, S.P.; Chan, J.W.; Mok, V.; Chan, A.; Li, S.X.; Liu, Y.; Tang, X.; Yung, W.H.; et al. Excessive Daytime Sleepiness Predicts Neurodegeneration in Idiopathic REM Sleep Behavior Disorder. Sleep 2017, 40, zsx041. [Google Scholar] [CrossRef]
- Ju, Y.E.; McLeland, J.S.; Toedebusch, C.D.; Xiong, C.; Fagan, A.M.; Duntley, S.P.; Morris, J.C.; Holtzman, D.M. Sleep quality and preclinical Alzheimer disease. JAMA Neurol. 2013, 70, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.S.; Kowgier, M.; Yu, L.; Buchman, A.S.; Bennett, D.A. Sleep Fragmentation and the Risk of Incident Alzheimer’s Disease and Cognitive Decline in Older Persons. Sleep 2013, 36, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Hahn, E.A.; Wang, H.X.; Andel, R.; Fratiglioni, L. A change in sleep pattern may predict Alzheimer disease. Am. J. Geriatr. Psychiatry 2014, 22, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, P.N.G.; Villaraza, S.G.; Rosa, J.C.D. The association between sleep and Alzheimer’s disease: A systematic review. Dement. Neuropsychol. 2024, 18, e20230049. [Google Scholar] [CrossRef]
- Lacerda, R.A.V.; Desio, J.A.F.; Kammers, C.M.; Henkes, S.; Freitas de Sa, M.; de Souza, E.F.; da Silva, D.M.; Teixeira Pinheiro Gusmao, C.; Santos, J. Sleep disorders and risk of alzheimer’s disease: A two-way road. Ageing Res. Rev. 2024, 101, 102514. [Google Scholar] [CrossRef]
- Kang, J.E.; Lim, M.M.; Bateman, R.J.; Lee, J.J.; Smyth, L.P.; Cirrito, J.R.; Fujiki, N.; Nishino, S.; Holtzman, D.M. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science 2009, 326, 1005–1007. [Google Scholar] [CrossRef]
- Di Meco, A.; Joshi, Y.B.; Pratico, D. Sleep deprivation impairs memory, tau metabolism, and synaptic integrity of a mouse model of Alzheimer’s disease with plaques and tangles. Neurobiol. Aging 2014, 35, 1813–1820. [Google Scholar] [CrossRef]
- Scott-Massey, A.; Boag, M.K.; Magnier, A.; Bispo, D.; Khoo, T.K.; Pountney, D.L. Glymphatic System Dysfunction and Sleep Disturbance May Contribute to the Pathogenesis and Progression of Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 12928. [Google Scholar] [CrossRef]
- Xu, Y.; Toh, K.L.; Jones, C.R.; Shin, J.Y.; Fu, Y.H.; Ptácek, L.J. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 2007, 128, 59–70. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.; Possovre, M.L.; Bandarabadi, M.; Moseholm, K.F.; Justinussen, J.L.; Bozic, I.; Lemcke, R.; Arribat, Y.; Amati, F.; Silahtaroglu, A.; et al. The evolutionarily conserved miRNA-137 targets the neuropeptide hypocretin/orexin and modulates the wake to sleep ratio. Proc. Natl. Acad. Sci. USA 2022, 119, e2112225119. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.; Bang-Berthelsen, C.H.; Knudsen, S.; Kornum, B.R.; Modvig, S.; Jennum, P.; Gammeltoft, S. miRNA profiles in plasma from patients with sleep disorders reveal dysregulation of miRNAs in narcolepsy and other central hypersomnias. Sleep 2014, 37, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, K.; Wiatrek, D.; Khan, A.; Michalik, D.; Sambrani, N.; Sedmík, J.; Vukić, D.; O’Connell, M.A.; Keegan, L.P. ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Saavedra, M.; Antoun, G.; Yanagiya, A.; Oliva-Hernandez, R.; Cornejo-Palma, D.; Perez-Iratxeta, C.; Sonenberg, N.; Cheng, H.Y. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 2011, 20, 731–751. [Google Scholar] [CrossRef]
- Weissová, K.; Škrabalová, J.; Skálová, K.; Červená, K.; Bendová, Z.; Miletínová, E.; Kopřivová, J.; Šonka, K.; Dudysová, D.; Bartoš, A.; et al. Circadian rhythms of melatonin and peripheral clock gene expression in idiopathic REM sleep behavior disorder. Sleep Med. 2018, 52, 1–6. [Google Scholar] [CrossRef]
- Laposky, A.; Easton, A.; Dugovic, C.; Walisser, J.; Bradfield, C.; Turek, F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 2005, 28, 395–409. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Chung, S.; Park, N.; Son, G.H.; An, H.; Jang, J.; Chang, D.J.; Suh, Y.G.; Kim, K. Identification of a novel circadian clock modulator controlling BMAL1 expression through a ROR/REV-ERB-response element-dependent mechanism. Biochem. Biophys. Res. Commun. 2016, 469, 580–586. [Google Scholar] [CrossRef]
- Ehlen, J.C.; Brager, A.J.; Baggs, J.; Pinckney, L.; Gray, C.L.; DeBruyne, J.P.; Esser, K.A.; Takahashi, J.S.; Paul, K.N. Bmal1 function in skeletal muscle regulates sleep. Elife 2017, 6, e26557. [Google Scholar] [CrossRef]
- Park, N.; Kim, H.D.; Cheon, S.; Row, H.; Lee, J.; Han, D.H.; Cho, S.; Kim, K. A Novel Bmal1 Mutant Mouse Reveals Essential Roles of the C-Terminal Domain on Circadian Rhythms. PLoS ONE 2015, 10, e0138661. [Google Scholar] [CrossRef]
- Sochal, M.; Ditmer, M.; Tarasiuk-Zawadzka, A.; Binienda, A.; Turkiewicz, S.; Wysokiński, A.; Karuga, F.F.; Białasiewicz, P.; Fichna, J.; Gabryelska, A. Circadian Rhythm Genes and Their Association with Sleep and Sleep Restriction. Int. J. Mol. Sci. 2024, 25, 10445. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, D.; Shostak, A.; Oster, H. Clock genes and sleep. Pflug. Arch. 2012, 463, 3–14. [Google Scholar] [CrossRef]
- Piggins, H.D. Human clock genes. Ann. Med. 2002, 34, 394–400. [Google Scholar] [CrossRef]
- Hasan, S.; van der Veen, D.R.; Winsky-Sommerer, R.; Hogben, A.; Laing, E.E.; Koentgen, F.; Dijk, D.J.; Archer, S.N. A human sleep homeostasis phenotype in mice expressing a primate-specific PER3 variable-number tandem-repeat coding-region polymorphism. FASEB J. 2014, 28, 2441–2454. [Google Scholar] [CrossRef] [PubMed]
- Okano, S.; Akashi, M.; Hayasaka, K.; Nakajima, O. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neurosci. Lett. 2009, 451, 246–251. [Google Scholar] [CrossRef]
- van der Horst, G.T.; Muijtjens, M.; Kobayashi, K.; Takano, R.; Kanno, S.; Takao, M.; de Wit, J.; Verkerk, A.; Eker, A.P.; van Leenen, D.; et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999, 398, 627–630. [Google Scholar] [CrossRef]
- Jagannath, A.; Taylor, L.; Wakaf, Z.; Vasudevan, S.R.; Foster, R.G. The genetics of circadian rhythms, sleep and health. Hum. Mol. Genet. 2017, 26, R128–R138. [Google Scholar] [CrossRef]
- Chowdhury, S.; Matsubara, T.; Miyazaki, T.; Ono, D.; Fukatsu, N.; Abe, M.; Sakimura, K.; Sudo, Y.; Yamanaka, A. GABA neurons in the ventral tegmental area regulate non-rapid eye movement sleep in mice. Elife 2019, 8, e44928. [Google Scholar] [CrossRef]
- Hu, Y.; Bringmann, H. Tfap2b acts in GABAergic neurons to control sleep in mice. Sci. Rep. 2023, 13, 8026. [Google Scholar] [CrossRef]
- Funato, H.; Miyoshi, C.; Fujiyama, T.; Kanda, T.; Sato, M.; Wang, Z.; Ma, J.; Nakane, S.; Tomita, J.; Ikkyu, A.; et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 2016, 539, 378–383. [Google Scholar] [CrossRef]
- Niwa, Y.; Kanda, G.N.; Yamada, R.G.; Shi, S.; Sunagawa, G.A.; Ukai-Tadenuma, M.; Fujishima, H.; Matsumoto, N.; Masumoto, K.H.; Nagano, M.; et al. Muscarinic Acetylcholine Receptors Chrm1 and Chrm3 Are Essential for REM Sleep. Cell Rep. 2018, 24, 2231–2247.e2237. [Google Scholar] [CrossRef] [PubMed]
- Mieda, M.; Hasegawa, E.; Kisanuki, Y.Y.; Sinton, C.M.; Yanagisawa, M.; Sakurai, T. Differential roles of orexin receptor-1 and -2 in the regulation of non-REM and REM sleep. J. Neurosci. 2011, 31, 6518–6526. [Google Scholar] [CrossRef] [PubMed]
- Willie, J.T.; Chemelli, R.M.; Sinton, C.M.; Tokita, S.; Williams, S.C.; Kisanuki, Y.Y.; Marcus, J.N.; Lee, C.; Elmquist, J.K.; Kohlmeier, K.A.; et al. Distinct narcolepsy syndromes in Orexin receptor-2 and Orexin null mice: Molecular genetic dissection of Non-REM and REM sleep regulatory processes. Neuron 2003, 38, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Dudley, C.A.; Estill, S.J.; Barakat, M.; Thomason, R.; O’Hara, B.F.; McKnight, S.L. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc. Natl. Acad. Sci. USA 2006, 103, 7118–7123. [Google Scholar] [CrossRef]
- Gamble, M.C.; Chuan, B.; Gallego-Martin, T.; Shelton, M.A.; Puig, S.; O’Donnell, C.P.; Logan, R.W. A role for the circadian transcription factor NPAS2 in the progressive loss of non-rapid eye movement sleep and increased arousal during fentanyl withdrawal in male mice. Psychopharmacology 2022, 239, 3185–3200. [Google Scholar] [CrossRef]
- Alam, M.N.; Szymusiak, R.; Gong, H.; King, J.; McGinty, D. Adenosinergic modulation of rat basal forebrain neurons during sleep and waking: Neuronal recording with microdialysis. J. Physiol. 1999, 521 Pt 3, 679–690. [Google Scholar] [CrossRef]
- Oishi, Y.; Huang, Z.L.; Fredholm, B.B.; Urade, Y.; Hayaishi, O. Adenosine in the tuberomammillary nucleus inhibits the histaminergic system via A1 receptors and promotes non-rapid eye movement sleep. Proc. Natl. Acad. Sci. USA 2008, 105, 19992–19997. [Google Scholar] [CrossRef]
- Gallopin, T.; Luppi, P.H.; Cauli, B.; Urade, Y.; Rossier, J.; Hayaishi, O.; Lambolez, B.; Fort, P. The endogenous somnogen adenosine excites a subset of sleep-promoting neurons via A2A receptors in the ventrolateral preoptic nucleus. Neuroscience 2005, 134, 1377–1390. [Google Scholar] [CrossRef]
- Niu, L.; Li, Y.; Zong, P.; Liu, P.; Shui, Y.; Chen, B.; Wang, Z.W. Melatonin promotes sleep by activating the BK channel in C. elegans. Proc. Natl. Acad. Sci. USA 2020, 117, 25128–25137. [Google Scholar] [CrossRef]
- He, Y.; Jones, C.R.; Fujiki, N.; Xu, Y.; Guo, B.; Holder, J.L., Jr.; Rossner, M.J.; Nishino, S.; Fu, Y.H. The transcriptional repressor DEC2 regulates sleep length in mammals. Science 2009, 325, 866–870. [Google Scholar] [CrossRef]
- Shi, G.; Yin, C.; Fan, Z.; Xing, L.; Mostovoy, Y.; Kwok, P.Y.; Ashbrook, L.H.; Krystal, A.D.; Ptáček, L.J.; Fu, Y.H. Mutations in Metabotropic Glutamate Receptor 1 Contribute to Natural Short Sleep Trait. Curr. Biol. 2021, 31, 13–24.e14. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Xing, L.; Wu, D.; Bhattacharyya, B.J.; Jones, C.R.; McMahon, T.; Chong, S.Y.C.; Chen, J.A.; Coppola, G.; Geschwind, D.; et al. A Rare Mutation of β(1)-Adrenergic Receptor Affects Sleep/Wake Behaviors. Neuron 2019, 103, 1044–1055.e1047. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Shi, G.; Mostovoy, Y.; Gentry, N.W.; Fan, Z.; McMahon, T.B.; Kwok, P.Y.; Jones, C.R.; Ptáček, L.J.; Fu, Y.H. Mutant neuropeptide S receptor reduces sleep duration with preserved memory consolidation. Sci. Transl. Med. 2019, 11, eaax2014. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, L.; Sek, K.; Henderson, M.A.; Lai, J.; Chen, A.X.Y.; Meyran, D.; Todd, K.L.; Petley, E.V.; Mardiana, S.; Mølck, C.; et al. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances CAR T cell efficacy. Nat. Commun. 2021, 12, 3236. [Google Scholar] [CrossRef]
- Leone, R.D.; Sun, I.M.; Oh, M.H.; Sun, I.H.; Wen, J.; Englert, J.; Powell, J.D. Inhibition of the adenosine A2a receptor modulates expression of T cell coinhibitory receptors and improves effector function for enhanced checkpoint blockade and ACT in murine cancer models. Cancer Immunol. Immunother. 2018, 67, 1271–1284. [Google Scholar] [CrossRef]
- Globig, A.M.; Zhao, S.; Roginsky, J.; Maltez, V.I.; Guiza, J.; Avina-Ochoa, N.; Heeg, M.; Araujo Hoffmann, F.; Chaudhary, O.; Wang, J.; et al. The β(1)-adrenergic receptor links sympathetic nerves to T cell exhaustion. Nature 2023, 622, 383–392. [Google Scholar] [CrossRef]
- Lewis, D.A. GABAergic local circuit neurons and prefrontal cortical dysfunction in schizophrenia. Brain Res. Brain Res. Rev. 2000, 31, 270–276. [Google Scholar] [CrossRef]
- de las Casas-Engel, M.; Domínguez-Soto, A.; Sierra-Filardi, E.; Bragado, R.; Nieto, C.; Puig-Kroger, A.; Samaniego, R.; Loza, M.; Corcuera, M.T.; Gómez-Aguado, F.; et al. Serotonin skews human macrophage polarization through HTR2B and HTR7. J. Immunol. 2013, 190, 2301–2310. [Google Scholar] [CrossRef]
- Pack, A.I.; Pien, G.W. Update on sleep and its disorders. Annu. Rev. Med. 2011, 62, 447–460. [Google Scholar] [CrossRef]
- Konttinen, H. Emotional eating and obesity in adults: The role of depression, sleep and genes. Proc. Nutr. Soc. 2020, 79, 283–289. [Google Scholar] [CrossRef]
- Reutrakul, S.; Van Cauter, E. Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metabolism 2018, 84, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yeghiazaryan, G.; Hess, S.; Klemm, P.; Sieben, A.; Kleinridders, A.; Morgan, D.A.; Wunderlich, F.T.; Rahmouni, K.; Kong, D.; et al. Orexin receptors 1 and 2 in serotonergic neurons differentially regulate peripheral glucose metabolism in obesity. Nat. Commun. 2021, 12, 5249. [Google Scholar] [CrossRef] [PubMed]
- He, S.K.; Wang, J.H.; Li, T.; Yin, S.; Cui, J.W.; Xiao, Y.F.; Tang, Y.; Wang, J.; Bai, Y.J. Sleep and circadian rhythm disturbance in kidney stone disease: A narrative review. Front. Endocrinol. 2023, 14, 1293685. [Google Scholar] [CrossRef]
- Russell, A.L.; Miller, L.; Yi, H.; Keil, R.; Handa, R.J.; Wu, T.J. Knockout of the circadian gene, Per2, disrupts corticosterone secretion and results in depressive-like behaviors and deficits in startle responses. BMC Neurosci. 2021, 22, 5. [Google Scholar] [CrossRef]
- Gréchez-Cassiau, A.; Feillet, C.; Guérin, S.; Delaunay, F. The hepatic circadian clock regulates the choline kinase α gene through the BMAL1-REV-ERBα axis. Chronobiol. Int. 2015, 32, 774–784. [Google Scholar] [CrossRef]
- Bonney, S.; Kominsky, D.; Brodsky, K.; Eltzschig, H.; Walker, L.; Eckle, T. Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS ONE 2013, 8, e71493. [Google Scholar] [CrossRef]
- Bhaskara, M.; Anjorin, O.; Yoniles, A.; Liu, J.; Wang, M. Importance of Per2 in cardiac mitochondrial protection during stress. Sci. Rep. 2024, 14, 1290. [Google Scholar] [CrossRef]
- Lu, T.; Jiang, B.; Wang, X.L.; Lee, H.C. Coronary arterial BK channel dysfunction exacerbates ischemia/reperfusion-induced myocardial injury in diabetic mice. Appl. Physiol. Nutr. Metab. 2016, 41, 992–1001. [Google Scholar] [CrossRef]
- Viola, A.U.; James, L.M.; Archer, S.N.; Dijk, D.J. PER3 polymorphism and cardiac autonomic control: Effects of sleep debt and circadian phase. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H2156–H2163. [Google Scholar] [CrossRef]
- Mikulska, A.A.; Grzelak, T.; Pelczyńska, M.; Bogdański, P.; Czyżewska, K. Assessment of Selected Clock Proteins (CLOCK and CRY1) and Their Relationship with Biochemical, Anthropometric, and Lifestyle Parameters in Hypertensive Patients. Biomolecules 2021, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Nebigil, C.G.; Hickel, P.; Messaddeq, N.; Vonesch, J.L.; Douchet, M.P.; Monassier, L.; György, K.; Matz, R.; Andriantsitohaina, R.; Manivet, P.; et al. Ablation of serotonin 5-HT(2B) receptors in mice leads to abnormal cardiac structure and function. Circulation 2001, 103, 2973–2979. [Google Scholar] [CrossRef] [PubMed]
- Lairez, O.; Cognet, T.; Schaak, S.; Calise, D.; Guilbeau-Frugier, C.; Parini, A.; Mialet-Perez, J. Role of serotonin 5-HT2A receptors in the development of cardiac hypertrophy in response to aortic constriction in mice. J. Neural Transm. 2013, 120, 927–935. [Google Scholar] [CrossRef]
- Mogavero, M.P.; DelRosso, L.M.; Fanfulla, F.; Bruni, O.; Ferri, R. Sleep disorders and cancer: State of the art and future perspectives. Sleep Med. Rev. 2021, 56, 101409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Meng, X.; Li, Y.; Liu, L.; He, Q.; Jiang, J.; Chen, Y.; Li, X.; Li, Y.; Tang, Y.; et al. Circadian clock gene BMAL1 inhibits the proliferation and tumor-formation ability of nasopharyngeal carcinoma cells and increases the sensitivity of radiotherapy. Chronobiol. Int. 2022, 39, 1340–1351. [Google Scholar] [CrossRef]
- Yang, G.; Yang, Y.; Tang, H.; Yang, K. Loss of the clock gene Per1 promotes oral squamous cell carcinoma progression via the AKT/mTOR pathway. Cancer Sci. 2020, 111, 1542–1554. [Google Scholar] [CrossRef]
- Gong, X.; Tang, H.; Yang, K. PER1 suppresses glycolysis and cell proliferation in oral squamous cell carcinoma via the PER1/RACK1/PI3K signaling complex. Cell Death Dis. 2021, 12, 276. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Q.; Hu, X.; Zhang, S.; Jiang, Y.; Yao, G.; Hu, K.; Xu, X.; Liang, B.; Wu, Q.; et al. Disrupting Circadian Rhythm via the PER1-HK2 Axis Reverses Trastuzumab Resistance in Gastric Cancer. Cancer Res. 2022, 82, 1503–1517. [Google Scholar] [CrossRef]
- Logan, R.W.; Wynne, O.; Levitt, D.; Price, D.; Sarkar, D.K. Altered circadian expression of cytokines and cytolytic factors in splenic natural killer cells of Per1(-/-) mutant mice. J. Interferon Cytokine Res. 2013, 33, 108–114. [Google Scholar] [CrossRef]
- Hernandez-Rosas, F.; Hernandez-Oliveras, A.; Flores-Peredo, L.; Rodriguez, G.; Zarain-Herzberg, A.; Caba, M.; Santiago-Garcia, J. Histone deacetylase inhibitors induce the expression of tumor suppressor genes Per1 and Per2 in human gastric cancer cells. Oncol. Lett. 2018, 16, 1981–1990. [Google Scholar] [CrossRef]
- Gery, S.; Komatsu, N.; Baldjyan, L.; Yu, A.; Koo, D.; Koeffler, H.P. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 2006, 22, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, L.; Liu, X.; Ma, Z.; Lv, L. PER1 Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Ovarian Cancer. Front. Genet. 2021, 12, 697471. [Google Scholar] [CrossRef] [PubMed]
- Bellet, M.M.; Stincardini, C.; Costantini, C.; Gargaro, M.; Pieroni, S.; Castelli, M.; Piobbico, D.; Sassone-Corsi, P.; Della-Fazia, M.A.; Romani, L.; et al. The Circadian Protein PER1 Modulates the Cellular Response to Anticancer Treatments. Int. J. Mol. Sci. 2021, 22, 2974. [Google Scholar] [CrossRef]
- Li, H.X.; Fu, X.J.; Yang, K.; Chen, D.; Tang, H.; Zhao, Q. The clock gene PER1 suppresses expression of tumor-related genes in human oral squamous cell carcinoma. Oncotarget 2016, 7, 20574–20583. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, Q.; Hu, Y.; Wang, F. The Circadian Gene Per1 Plays an Important Role in Radiation-Induced Apoptosis and DNA Damage in Glioma. Asian Pac. J. Cancer Prev. 2019, 20, 2195–2201. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef]
- Xiang, S.; Coffelt, S.B.; Mao, L.; Yuan, L.; Cheng, Q.; Hill, S.M. Period-2: A tumor suppressor gene in breast cancer. J. Circadian Rhythm. 2008, 6, 4. [Google Scholar] [CrossRef]
- Pavithra, S.; Aich, A.; Chanda, A.; Zohra, I.F.; Gawade, P.; Das, R.K. PER2 gene and its association with sleep-related disorders: A review. Physiol. Behav. 2024, 273, 114411. [Google Scholar] [CrossRef]
- Dong, P.; Wang, Y.; Liu, Y.; Zhu, C.; Lin, J.; Qian, R.; Hua, L.; Lu, C. BMAL1 induces colorectal cancer metastasis by stimulating exosome secretion. Mol. Biol. Rep. 2022, 49, 373–384. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, L.; Sun, L.; Jin, H.; Ren, K.; Liu, S.; Qian, Y.; Li, S.; Li, F.; Zhu, C.; et al. BMAL1 collaborates with CLOCK to directly promote DNA double-strand break repair and tumor chemoresistance. Oncogene 2023, 42, 967–979. [Google Scholar] [CrossRef]
- Shan, L.; Zheng, W.; Bai, B.; Hu, J.; Lv, Y.; Chen, K.; Wang, X.; Pan, Y.; Huang, X.; Zhu, H.; et al. BMAL1 promotes colorectal cancer cell migration and invasion through ERK- and JNK-dependent c-Myc expression. Cancer Med. 2023, 12, 4472–4485. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Li, M.; Su, D.; Fan, H.; Xia, H. Bmal1 regulates the stemness and tumorigenesis of gliomas with the Wnt/beta-catenin signaling pathway. Gene 2025, 933, 148940. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, Y.; Quan, P.; Zhang, J.; Han, S.; Wang, G.; Qi, R.; Zhang, X.; Wang, F.; Yuan, J.; et al. NPAS2 promotes aerobic glycolysis and tumor growth in prostate cancer through HIF-1A signaling. BMC Cancer 2023, 23, 280. [Google Scholar] [CrossRef]
- Yuan, P.; Li, J.; Zhou, F.; Huang, Q.; Zhang, J.; Guo, X.; Lyu, Z.; Zhang, H.; Xing, J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017, 8, e2704. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, L.; Kothandan, G.; Manoharan, R. Berberine and Emodin abrogates breast cancer growth and facilitates apoptosis through inactivation of SIK3-induced mTOR and Akt signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165897. [Google Scholar] [CrossRef]
- Wangari-Talbot, J.; Wall, B.A.; Goydos, J.S.; Chen, S. Functional effects of GRM1 suppression in human melanoma cells. Mol. Cancer Res. 2012, 10, 1440–1450. [Google Scholar] [CrossRef]
- Huang, D.; Wang, Y.; Thompson, J.W.; Yin, T.; Alexander, P.B.; Qin, D.; Mudgal, P.; Wu, H.; Liang, Y.; Tan, L.; et al. Cancer-cell-derived GABA promotes β-catenin-mediated tumour growth and immunosuppression. Nat. Cell Biol. 2022, 24, 230–241. [Google Scholar] [CrossRef]
- Miro, C.; Docimo, A.; Barrea, L.; Verde, L.; Cernea, S.; Sojat, A.S.; Marina, L.V.; Docimo, G.; Colao, A.; Dentice, M.; et al. “Time” for obesity-related cancer: The role of the circadian rhythm in cancer pathogenesis and treatment. Semin. Cancer Biol. 2023, 91, 99–109. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Z.; Nice, E.; Huang, C.; Zhang, W.; Tang, Y. Circadian rhythms and cancers: The intrinsic links and therapeutic potentials. J. Hematol. Oncol. 2022, 15, 21. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xie, G.Y.; Miao, Y.R.; Li, X.W.; Zeng, Y.; Guo, A.Y. GSCA: An integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinform. 2023, 24, bbac558. [Google Scholar] [CrossRef] [PubMed]
- Monti, J.M.; Monti, D. Sleep disturbance in schizophrenia. Int. Rev. Psychiatry 2005, 17, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.S.; Owe-Larsson, B.; Hetta, J.; Lundkvist, G.B. Altered circadian clock gene expression in patients with schizophrenia. Schizophr. Res. 2016, 174, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, P.W.; Nadarajah, C.J.; Kanan, M.F.; Patterson, J.N.; Novotny, B.; Lawrence, J.H.; King, M.W.; Brase, L.; Inman, C.E.; Yuede, C.M.; et al. An astrocyte BMAL1-BAG3 axis protects against alpha-synuclein and tau pathology. Neuron 2023, 111, 2383–2398.e2387. [Google Scholar] [CrossRef]
- González-Maeso, J.; Ang, R.L.; Yuen, T.; Chan, P.; Weisstaub, N.V.; López-Giménez, J.F.; Zhou, M.; Okawa, Y.; Callado, L.F.; Milligan, G.; et al. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature 2008, 452, 93–97. [Google Scholar] [CrossRef]
- Wang, M.; Li, P.; Li, Z.; da Silva, B.S.; Zheng, W.; Xiang, Z.; He, Y.; Xu, T.; Cordeiro, C.; Deng, L.; et al. Lateral septum adenosine A(2A) receptors control stress-induced depressive-like behaviors via signaling to the hypothalamus and habenula. Nat. Commun. 2023, 14, 1880. [Google Scholar] [CrossRef]
- Xia, G.; Han, Y.; Meng, F.; He, Y.; Srisai, D.; Farias, M.; Dang, M.; Palmiter, R.D.; Xu, Y.; Wu, Q. Reciprocal control of obesity and anxiety-depressive disorder via a GABA and serotonin neural circuit. Mol. Psychiatry 2021, 26, 2837–2853. [Google Scholar] [CrossRef]
- Trollope, A.F.; Gutièrrez-Mecinas, M.; Mifsud, K.R.; Collins, A.; Saunderson, E.A.; Reul, J.M. Stress, epigenetic control of gene expression and memory formation. Exp. Neurol. 2012, 233, 3–11. [Google Scholar] [CrossRef]
- Liu, D.; Nanclares, C.; Simbriger, K.; Fang, K.; Lorsung, E.; Le, N.; Amorim, I.S.; Chalkiadaki, K.; Pathak, S.S.; Li, J.; et al. Autistic-like behavior and cerebellar dysfunction in Bmal1 mutant mice ameliorated by mTORC1 inhibition. Mol. Psychiatry 2023, 28, 3727–3738. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, G.; He, Y.; Tang, Q.; Yin, Y.; Jie, Y. BMAL1 deficiency provokes dry mouth and eyes by down-regulating ITPR2/3. Ocul. Surf. 2024, 34, 430–440. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Zhao, L.; He, Z.; Zhao, Y.; Jiang, G.; Gong, C.; Zhang, Y.; Yu, J.; Liang, T.; Guo, L. Decoding Multifaceted Roles of Sleep-Related Genes as Molecular Bridges in Chronic Disease Pathogenesis. Int. J. Mol. Sci. 2025, 26, 2872. https://doi.org/10.3390/ijms26072872
Wang W, Zhao L, He Z, Zhao Y, Jiang G, Gong C, Zhang Y, Yu J, Liang T, Guo L. Decoding Multifaceted Roles of Sleep-Related Genes as Molecular Bridges in Chronic Disease Pathogenesis. International Journal of Molecular Sciences. 2025; 26(7):2872. https://doi.org/10.3390/ijms26072872
Chicago/Turabian StyleWang, Wenyuan, Linjie Zhao, Zhiheng He, Yang Zhao, Guijie Jiang, Chengjun Gong, Yan Zhang, Jiafeng Yu, Tingming Liang, and Li Guo. 2025. "Decoding Multifaceted Roles of Sleep-Related Genes as Molecular Bridges in Chronic Disease Pathogenesis" International Journal of Molecular Sciences 26, no. 7: 2872. https://doi.org/10.3390/ijms26072872
APA StyleWang, W., Zhao, L., He, Z., Zhao, Y., Jiang, G., Gong, C., Zhang, Y., Yu, J., Liang, T., & Guo, L. (2025). Decoding Multifaceted Roles of Sleep-Related Genes as Molecular Bridges in Chronic Disease Pathogenesis. International Journal of Molecular Sciences, 26(7), 2872. https://doi.org/10.3390/ijms26072872






