Integrated Transcriptome and Metabolome Analyses Reveal Candidate Genes Associate with Phenolic Compound Biosynthesis in Different Varieties of Perilla frutescens
Abstract
1. Introduction
2. Results
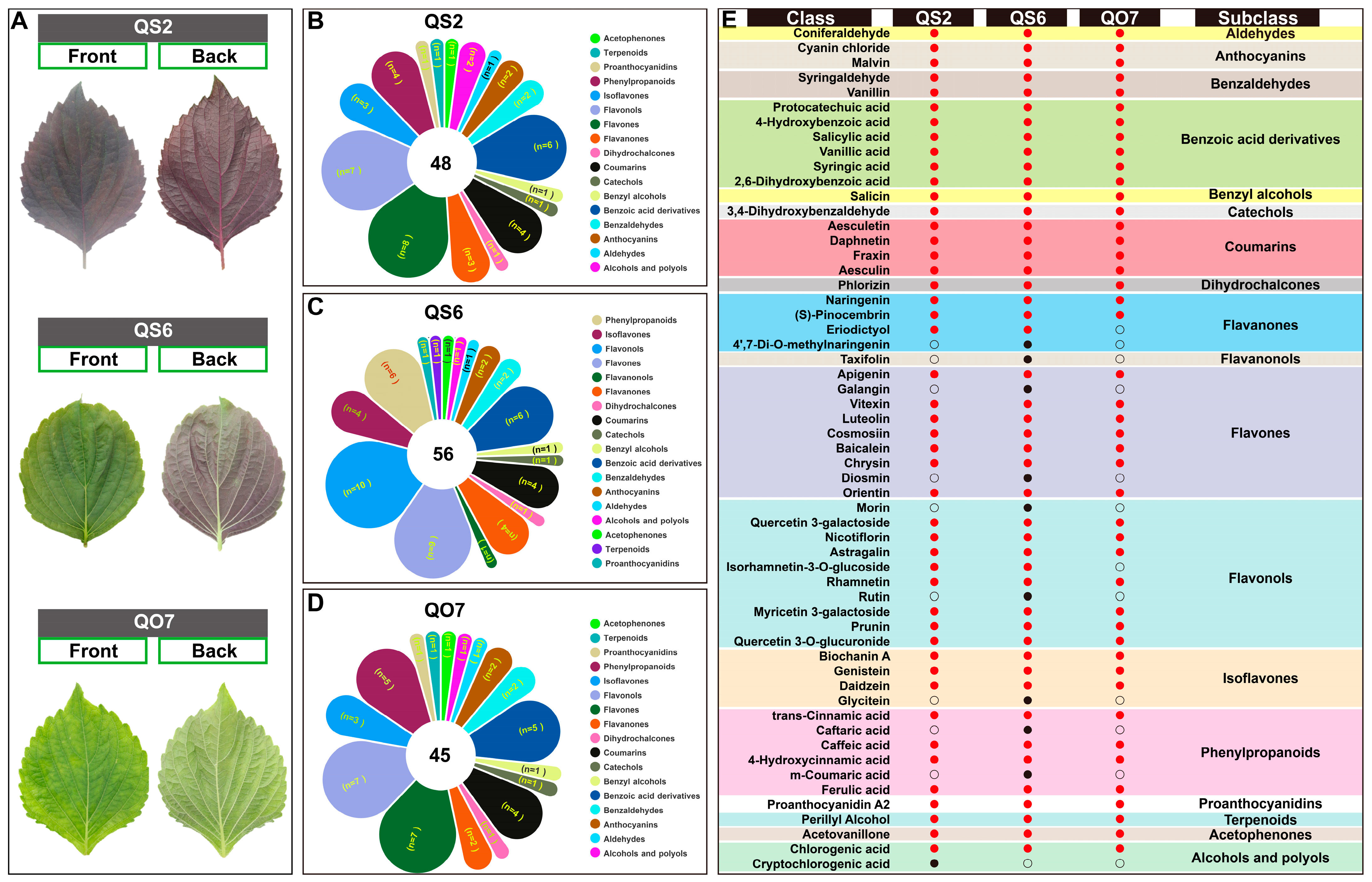
2.1. Differences in Phenolic Compounds in Three Perilla Varieties
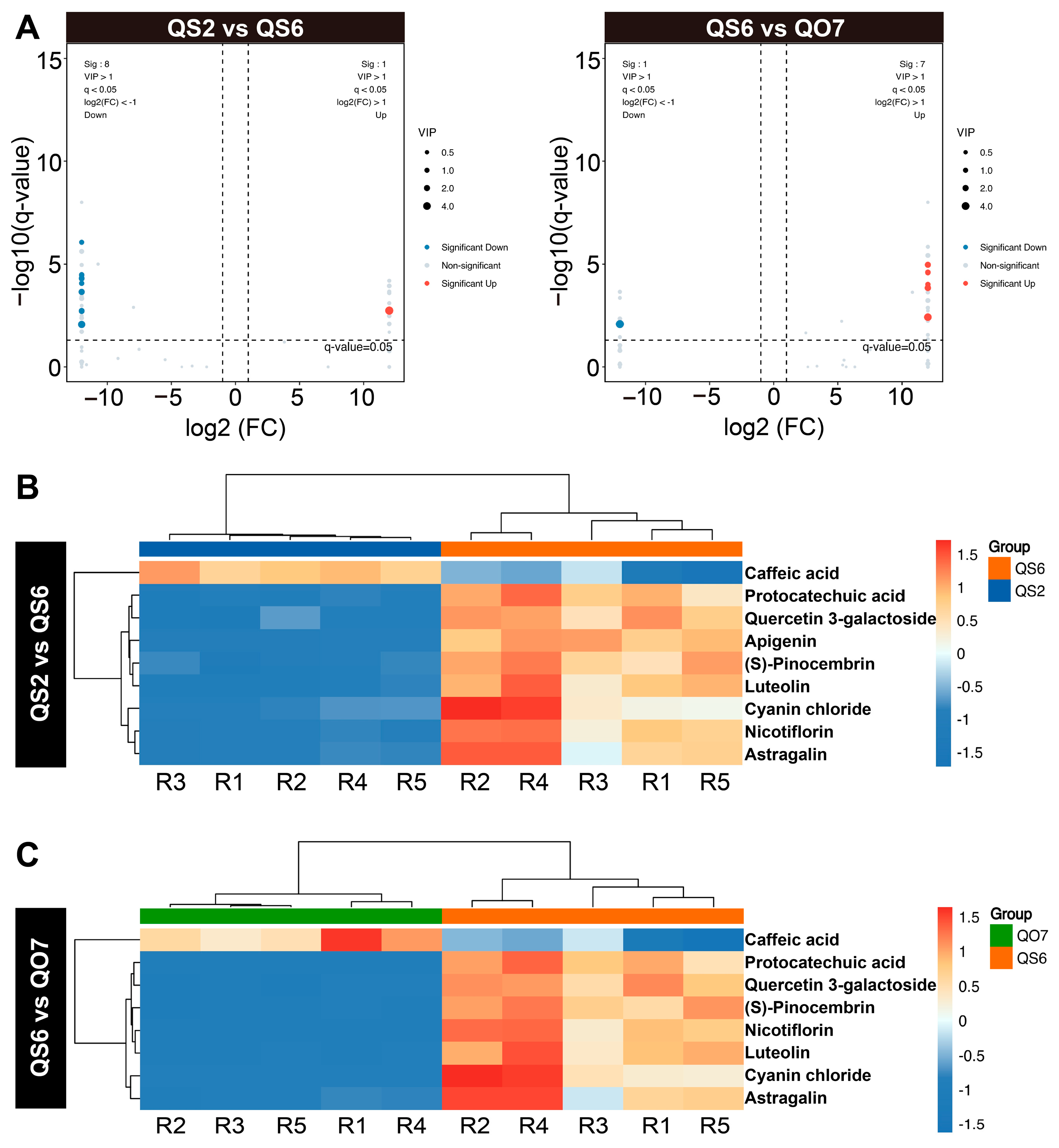
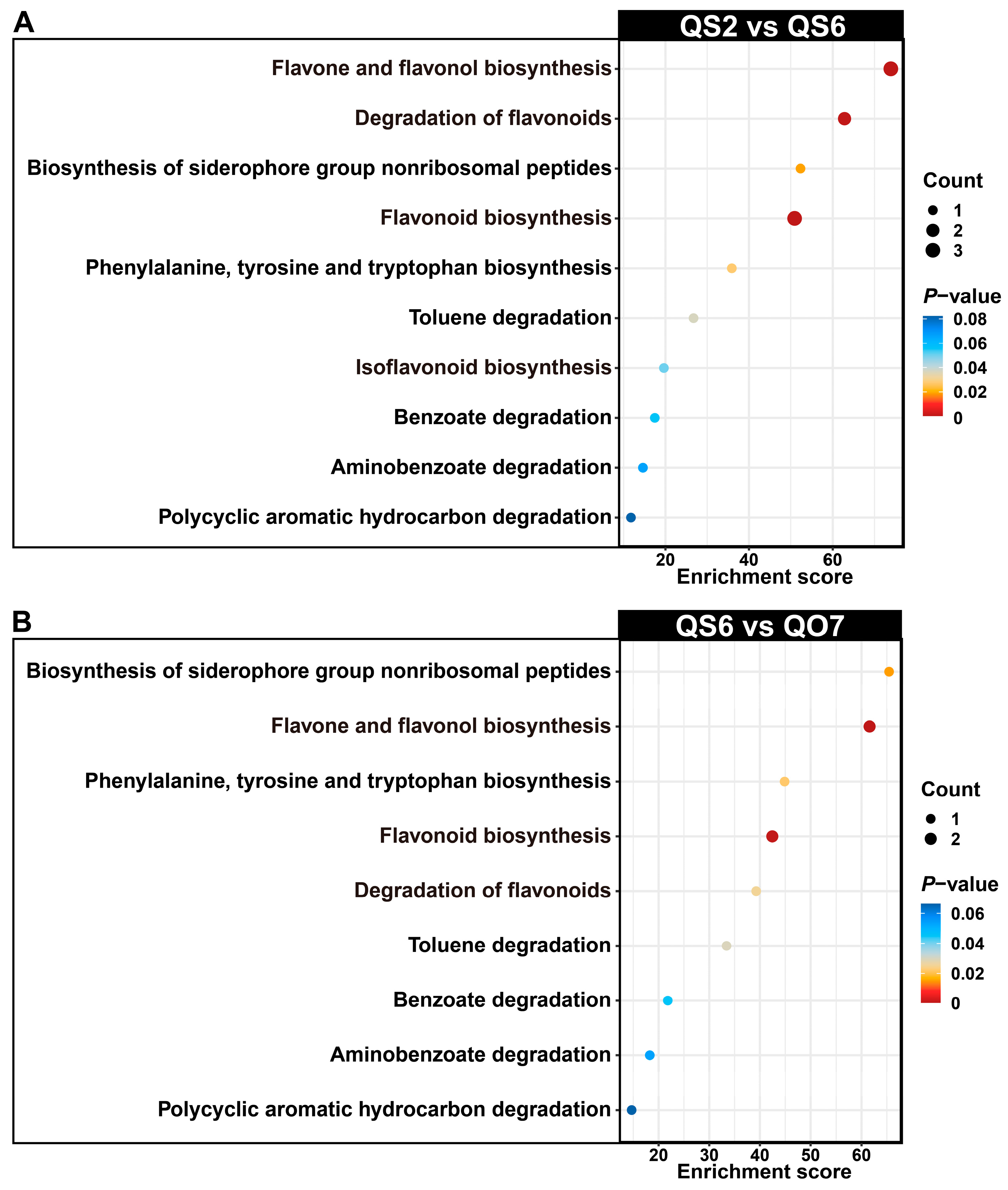
2.2. Isolation and Enrichment Analysis of Differentially Accumulated Phenolic Compounds
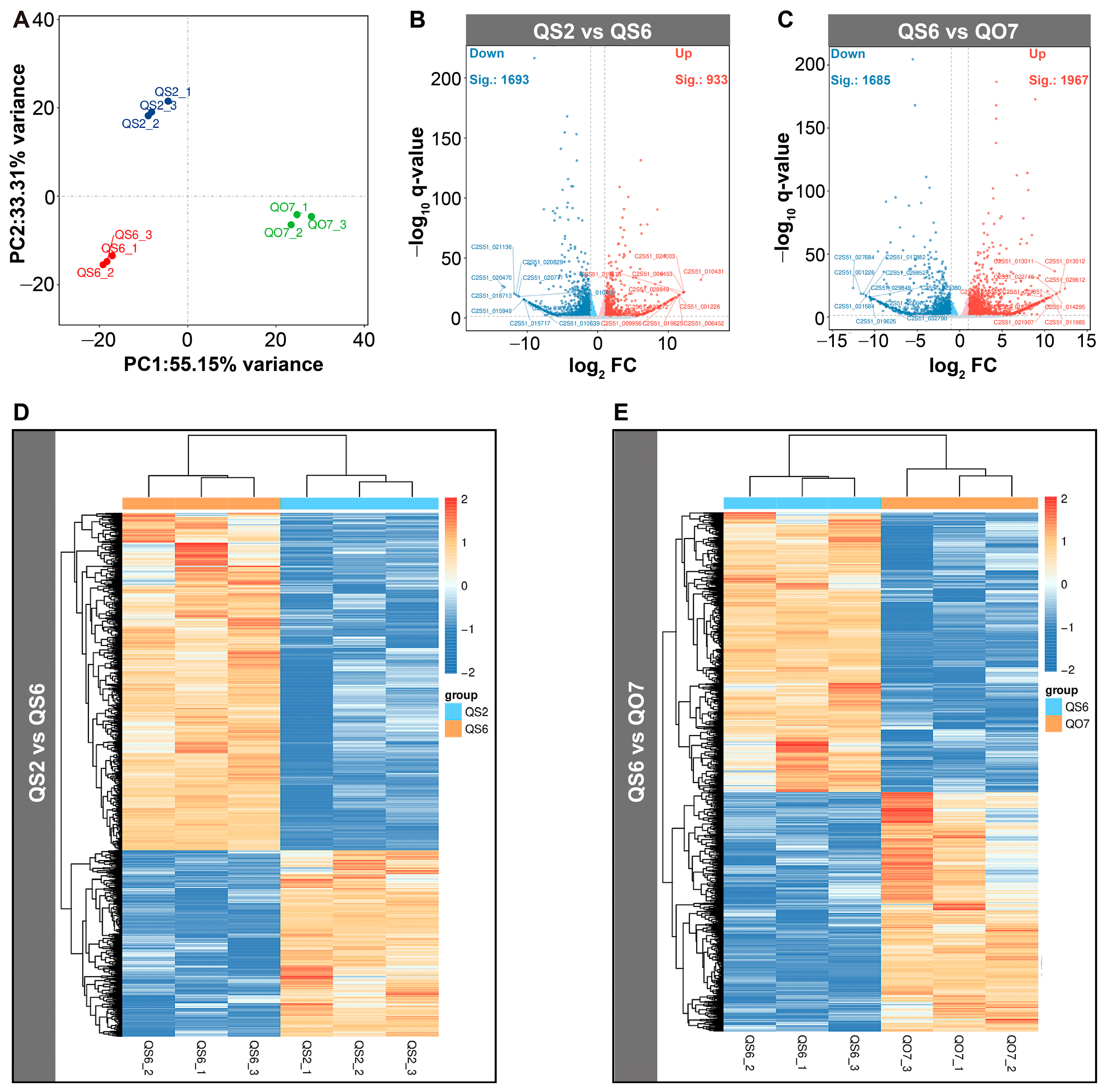
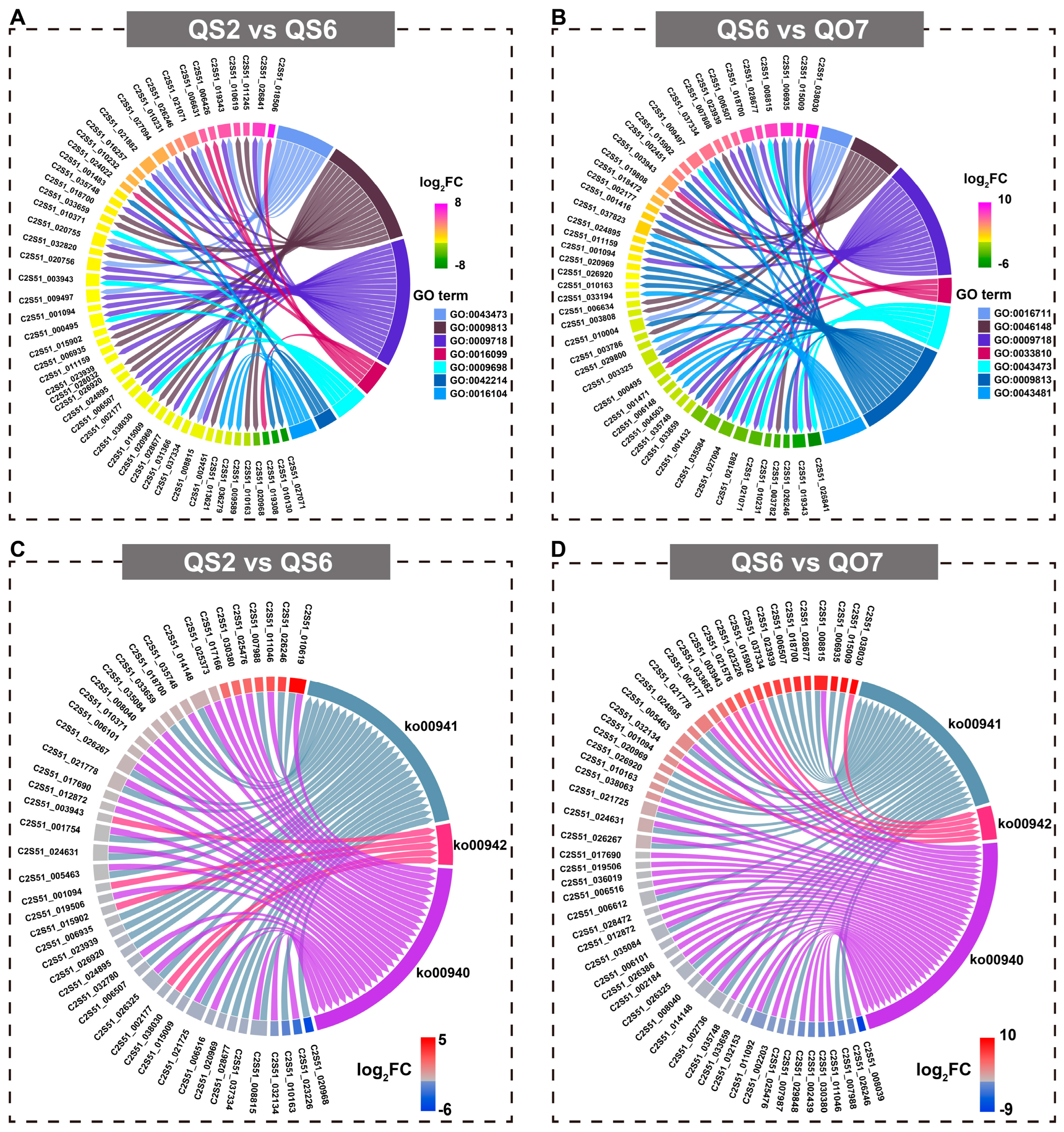
2.3. Comparative Transcriptome Analysis
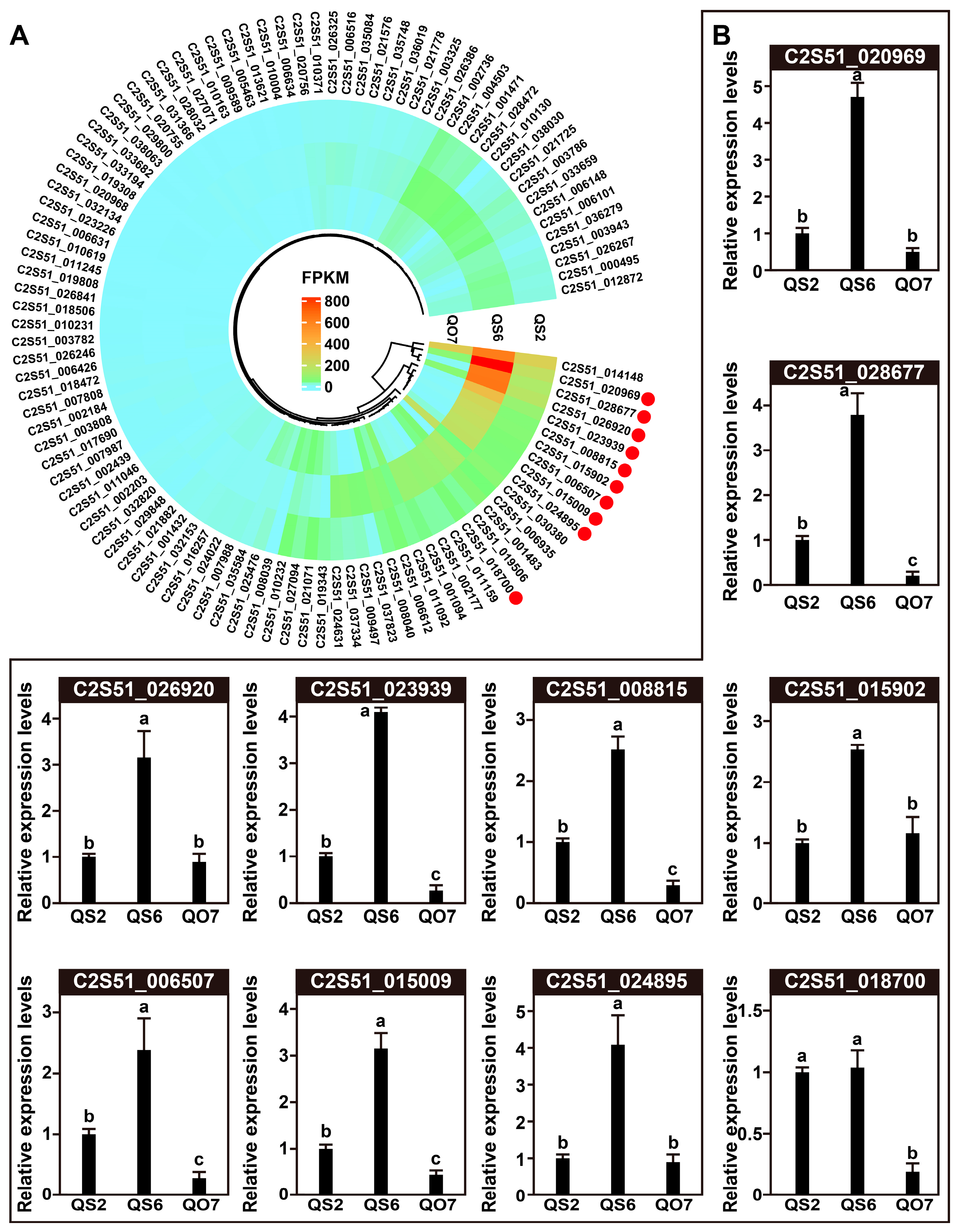
2.4. qRT-PCR Validation of DEGs
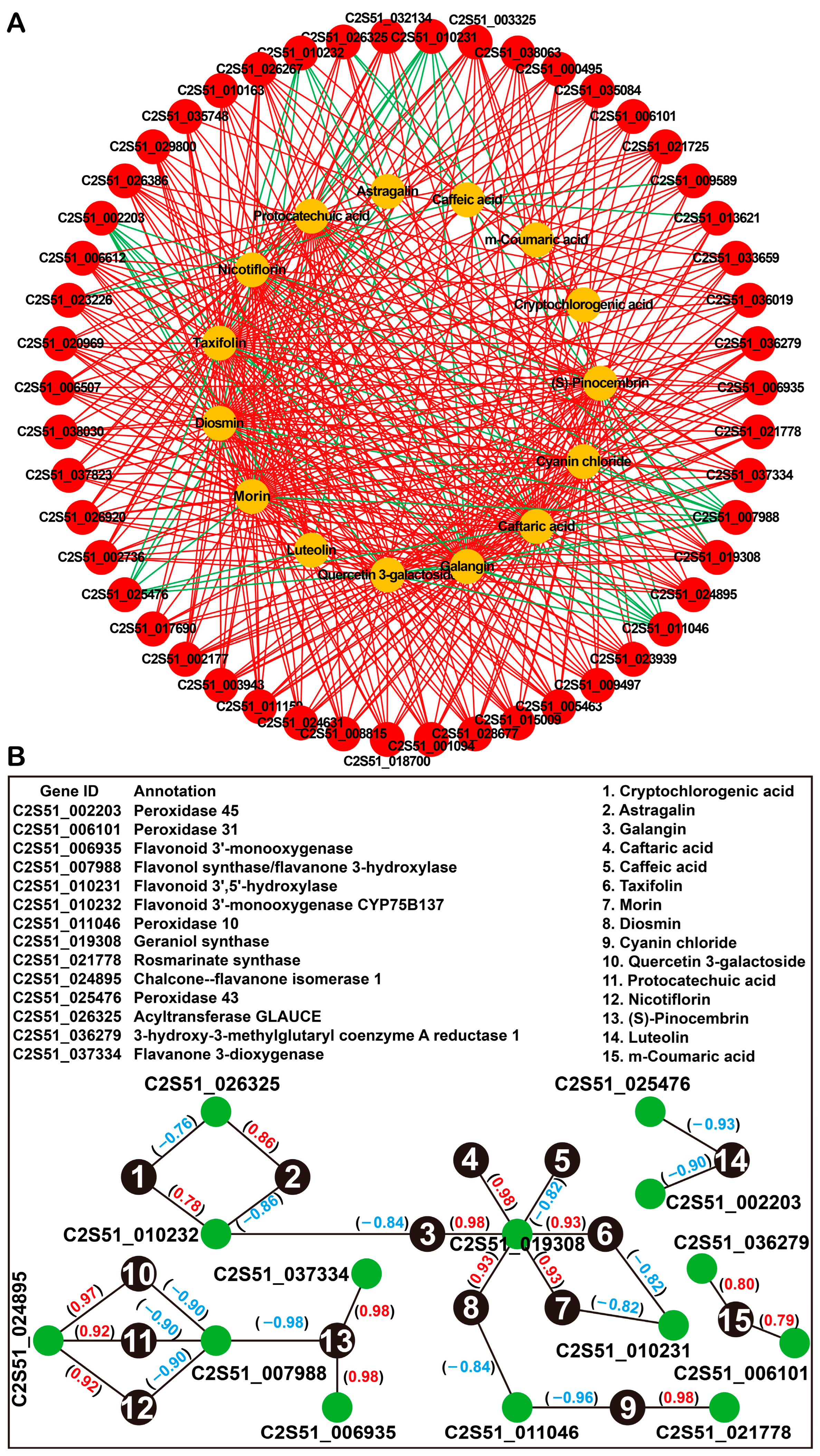
2.5. Integrated Analysis of Transcriptome and Metabolome Data
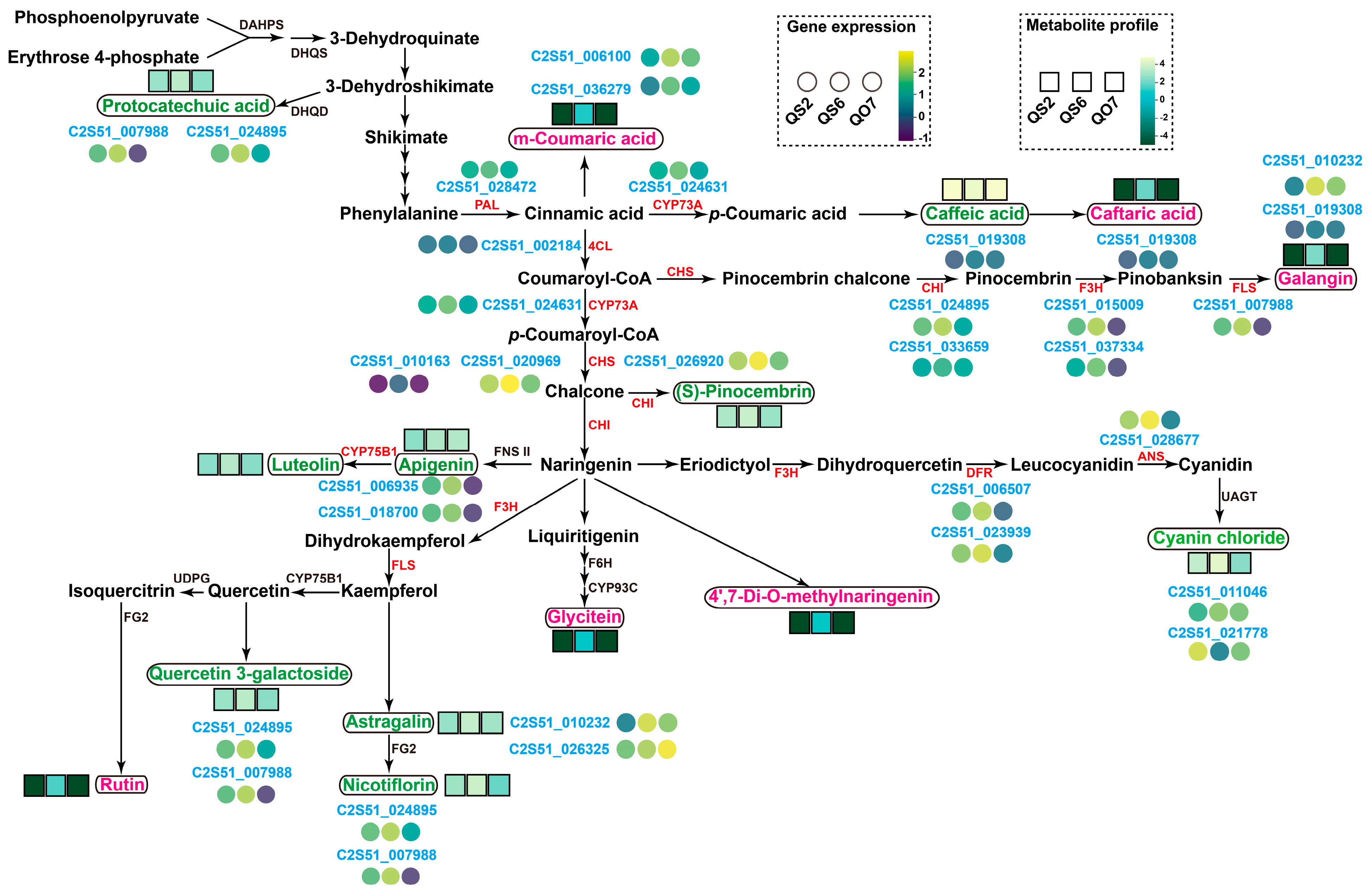
2.6. Profiles of Key Genes and Metabolites Associated with Phenolic Metabolisms in Perilla
3. Discussion
3.1. New Insights into the Constitution of Phenolic Compounds in Perilla Leaves
3.2. Candidate Genes Involved in Phenolic Compound Biosynthesis in Perilla
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Sample Collection
4.2. RNA Isolation and Library Preparation
4.3. RNA Sequencing and Differentially Expressed Genes Analysis
4.4. Quantitative Real-Time PCR Assay
4.5. Targeted Metabolome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 4CL | 4-coumarate-CoAligase |
| ANR | anthocyanidin reductase |
| ANS | anthocyanidin synthase |
| C4H | cinnamate 4- hydroxylase |
| cDNA | complementary DNA |
| CHI | chalcone isomerase |
| CHS | chalcone synthase |
| CYP73A | trans-cinnamate 4-monooxygenase |
| CYP75B1 | flavonoid 3′- monooxygenase |
| CYP93C | cytochrome P450 93C |
| DAHPS | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase |
| DAM | differentially accumulated metabolite |
| DEGs | differentially expressed genes |
| DFR | dihydrofavonol-4-reductase |
| DHQD | 3-Dehydroquinate dehydratase |
| DHQS | 3-Dehydroquinate synthase |
| F3H | naringenin 3-dioxygenase |
| F6H | feruloyl-CoA 6-hydroxylase |
| FG2 | flavonol-3-O-glucoside L-rhamnosyltransferase |
| FLS | flavonol synthase |
| FNS I | flavone synthase I |
| FPKM | fragments per kilobase per transcript per million mapped reads |
| GO | gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PAL | phenylalanine ammonia-lyase |
| PCA | principal component analysis |
| QC | quality control |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| RNA-Seq | RNA sequencing |
| UAGT | Anthocyanidin 3-O-glucosyltransferase |
| UDPG | UDP-glucosyltransferase |
| UGT | UDP-glycosyltransferases |
| UPLC-MS/MS | ultra-performance liquid chromatography/tandem mass spectrometry |
| VIP | variable importance of the projection |
References
- Hou, T.; Netala, V.R.; Zhang, H.; Xing, Y.; Li, H.; Zhang, Z. Perilla frutescens: A Rich Source of Pharmacological Active Compounds. Molecules 2022, 27, 3578. [Google Scholar] [CrossRef]
- Wu, X.; Dong, S.; Chen, H.; Guo, M.; Sun, Z.; Luo, H. Perilla frutescens: A traditional medicine and food homologous plant. Chin. Herb. Med. 2023, 15, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.M. Ethnomedicinal, phytochemical and pharmacological investigations of Perilla frutescens (L.) Britt. Molecules 2018, 24, 102. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Wang, J.; Liu, Y.M.; Zheng, Q.S.; Li, C.Y. Inhibitory effect of Malvidin on TNF-alpha-induced inflammatory response in endothelial cells. Eur. J. Pharmacol. 2014, 723, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Kuo, C.L.; Wang, J.P.; Cheng, J.S.; Huang, Z.W.; Chen, C.F. Growth inhibitory and apoptosis inducing effect of Perilla frutescens extract on human hepatoma HepG2 cells. J. Ethnopharmacol. 2007, 112, 557–567. [Google Scholar] [CrossRef]
- Saita, E.; Kishimoto, Y.; Tani, M.; Iizuka, M.; Toyozaki, M.; Sugihara, N.; Kondo, K. Antioxidant activities of Perilla frutescens against low density lipoprotein oxidation in vitro and in human subjects. J. Oleo Sci. 2012, 61, 113–120. [Google Scholar] [CrossRef]
- Yamamoto, H.; Ogawa, T. Antimicrobial activity of perilla seed polyphenols against oral pathogenic bacteria. Biosci. Biotechnol. Biochem. 2002, 66, 921–924. [Google Scholar] [CrossRef]
- Ghimire, B.K.; Yoo, J.H.; Yu, C.Y.; Chung, I.M. GC-MS analysis of volatile compounds of Perilla frutescens Britton var. Japonica accessions: Morphological and seasonal variability. Asian Pac. J. Trop. Med. 2017, 10, 643–651. [Google Scholar] [CrossRef]
- Peng, Y.; Ye, J.; Kong, J. Determination of phenolic compounds in Perilla frutescens L. by capillary electrophoresis with electrochemical detection. J. Agric. Food Chem. 2005, 53, 8141–8147. [Google Scholar] [CrossRef]
- Xie, G.; Zou, X.; Liang, Z.; Zhang, K.; Wu, D.; Jin, H.; Wang, H.; Shen, Q. GBF family member PfGBF3 and NAC family member PfNAC2 regulate rosmarinic acid biosynthesis under high light. Plant Physiol. 2004, 195, 1728–1744. [Google Scholar] [CrossRef]
- da Silva, A.P.G.; Sganzerla, W.G.; John, O.D.; Marchiosi, R. A comprehensive review of the classification, sources, biosynthesis, and biological properties of hydroxybenzoic and hydroxycinnamic acids. Phytochem. Rev. 2023. [Google Scholar] [CrossRef]
- Al Manari, H.H. Phenolic Compounds: Classification, Chemistry, and Updated Techniques of Analysis and Synthesis; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Marchiosi, R.; dos Santos, W.D.; Constantin, R.P.; de Lima, R.B.; Soares, A.R.; Finger-Teixeira, A.; Maota, T.R.; de Oliveira, D.M.; Foletto-Felipe, M.P.; Abrahão, J.; et al. Biosynthesis and metabolic actions of simple phenolic acids in plants. Phytochem. Rev. 2020, 19, 865–906. [Google Scholar] [CrossRef]
- Albuquerque, B.R.; Heleno, S.A.; Oliveira, M.B.P.P.; Barros, L.; Ferreira, I.C.F.R. Function phenolic compounds: Current industrial applications, limitations and future challenges. Food Funct. 2021, 12, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, H.B.; Negi, P.S. Phenolic acids from vegetables: A review on processing stability and health benefits. Food Res. Int. 2020, 136, 109298. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Sasaki, N.; Nakayama, T. Achievements and Perspectives in Biochemistry Concerning Anthocyanin Modification for Blue Flower Coloration. Plant Cell Physiol. 2015, 56, 28–40. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Wang, Z.L.; Wang, S.; Kuang, Y.; Hu, Z.M.; Qiao, X.; Ye, M. A comprehensive review on phytochemistry, pharmacology, and flavonoid biosynthesis of Scutellaria baicalensis. Pharm. Biol. 2018, 56, 465–484. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef]
- Qaderi, M.M.; Martel, A.B.; Strugnell, C.A. Environmental factors regulate plant secondary metabolites. Plants 2023, 12, 447. [Google Scholar] [CrossRef]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Carrington, Y.; Alber, A.; Ehlting, J. Molecular characterization of quinate and shikimate metabolism in populus trichocarpa. J. Biol. Chem. 2014, 289, 23846–23858. [Google Scholar] [CrossRef] [PubMed]
- Muir, R.M.; Ibáñez, A.M.; Uratsu, S.L.; Ingham, E.S.; Leslie, C.A.; McGranahan, G.H.; Batra, N.; Goyal, S.; Joseph, J.; Jemmis, E.D.; et al. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 2011, 75, 555–565. [Google Scholar] [CrossRef]
- Hong, E.; Park, K.H.; Kim, G.H. Phenolic-enriched fractions from perilla frutescens var. acuta: Determinating rosmarinic acid and antioxidant activity. J. Food Biochem. 2011, 35, 1637–1645. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, H.; Zhang, X.; Hu, X.; Wang, M. The effect of Perilla (Perilla frutescens) leaf extracts on the quality of surimi fish balls. Food Sci. Nutr. 2019, 7, 2083–2090. [Google Scholar] [CrossRef]
- Ghorbani, A. Mechanisms of antidiabetic effects of flavonoid rutin. Biomed. Pharmacother. 2017, 96, 305–312. [Google Scholar] [CrossRef]
- Thuan, N.H.; Shrestha, A.; Trung, N.T.; Tatipamula, V.B.; Van Cuong, D.; Canh, N.X.; Van Giang, N.; Kim, T.S.; Sohng, J.K.; Dhakal, D. Advances in biochemistry and the biotechnological production of taxifolin and its derivatives. Biotechnol. Appl. Biochem. 2022, 69, 848–861. [Google Scholar] [CrossRef]
- Wang, D.; Chen, J.; Pu, L.; Yu, L.; Xiong, F.; Sun, L.; Yu, Q.; Cao, X.; Chen, Y.; Peng, F.; et al. Galangin: A food-derived flavonoid with therapeutic potential against a wide spectrum of diseases. Phytother. Res. 2023, 37, 5700–5723. [Google Scholar] [CrossRef]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef]
- Quideau, S.; Deffieux, D.; Douat-Casassus, C.; Pouységu, L. Plant polyphenols: Chemical properties, biological activities, and synthesis. Angew. Chem. Int. Ed. Engl. 2011, 50, 586–621. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.M. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 1995, 107, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef]
- Kołton, A.; Długosz-Grochowska, O.; Wojciechowska, R.; Czaja, M. Biosynthesis regulation of folates and phenols in plants. Sci. Hortic. 2022, 291, 110561. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef]
- Song, J.; He, Y.; Luo, C.; Feng, B.; Ran, F.; Xu, H.; Ci, Z.; Xu, R.; Han, L.; Zhang, D. New progress in the pharmacology of protocatechuic acid: A compound ingested in daily foods and herbs frequently and heavily. Pharmacol. Res. 2020, 161, 105109. [Google Scholar] [CrossRef]
- Jiang, T.; Guo, K.; Liu, L.; Tian, W.; Xie, X.; Wen, S.; Wen, C. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 2020, 10, 16207. [Google Scholar] [CrossRef]
- Xie, G.; Zou, X.; Liang, Z.; Wu, D.; He, J.; Xie, K.; Jin, H.; Wang, H.; Shen, Q. Integrated metabolomic and transcriptomic analyses reveal molecular response of anthocyanins biosynthesis in perilla to light intensity. Front. Plant Sci. 2022, 13, 976449. [Google Scholar] [CrossRef]
- Ferreres, F.; Figueiredo, R.; Bettencourt, S.; Carqueijeiro, I.; Oliveira, J.; Gil-Izquierdo, A.; Pereira, D.M.; Valentão, P.; Andrade, P.B.; Duarte, P.; et al. Identification of phenolic compounds in isolated vacuoles of the medicinal plant Catharanthus roseus and their interaction with vacuolar class III peroxidase: An H2O2 affair? J. Exp. Bot. 2011, 62, 2841–2854. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Welinder, K.G.; Justesen, A.F.; Kjaersgård, I.V.; Jensen, R.B.; Rasmussen, S.K.; Jespersen, H.M.; Duroux, L. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002, 269, 6063–6081. [Google Scholar] [CrossRef]
- Sirikantaramas, S.; Yamazaki, M.; Saito, K. Mechanisms of resistance to self-produced toxic secondary metabolites in plants. Phytochem. Rev. 2008, 7, 467–477. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, H.; Mu, J.; Yang, T.; Shen, Q.; Wang, Y. Integrated Transcriptome and Metabolome Analyses Reveal Candidate Genes Associate with Phenolic Compound Biosynthesis in Different Varieties of Perilla frutescens. Int. J. Mol. Sci. 2025, 26, 2841. https://doi.org/10.3390/ijms26072841
Ye H, Mu J, Yang T, Shen Q, Wang Y. Integrated Transcriptome and Metabolome Analyses Reveal Candidate Genes Associate with Phenolic Compound Biosynthesis in Different Varieties of Perilla frutescens. International Journal of Molecular Sciences. 2025; 26(7):2841. https://doi.org/10.3390/ijms26072841
Chicago/Turabian StyleYe, Hong, Jiaxin Mu, Tong Yang, Qi Shen, and Yukun Wang. 2025. "Integrated Transcriptome and Metabolome Analyses Reveal Candidate Genes Associate with Phenolic Compound Biosynthesis in Different Varieties of Perilla frutescens" International Journal of Molecular Sciences 26, no. 7: 2841. https://doi.org/10.3390/ijms26072841
APA StyleYe, H., Mu, J., Yang, T., Shen, Q., & Wang, Y. (2025). Integrated Transcriptome and Metabolome Analyses Reveal Candidate Genes Associate with Phenolic Compound Biosynthesis in Different Varieties of Perilla frutescens. International Journal of Molecular Sciences, 26(7), 2841. https://doi.org/10.3390/ijms26072841





