Genome-Wide Dissection of MATE Gene Family in Cultivated Peanuts and Unveiling Their Expression Profiles Under Aluminum Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of AhMATEs
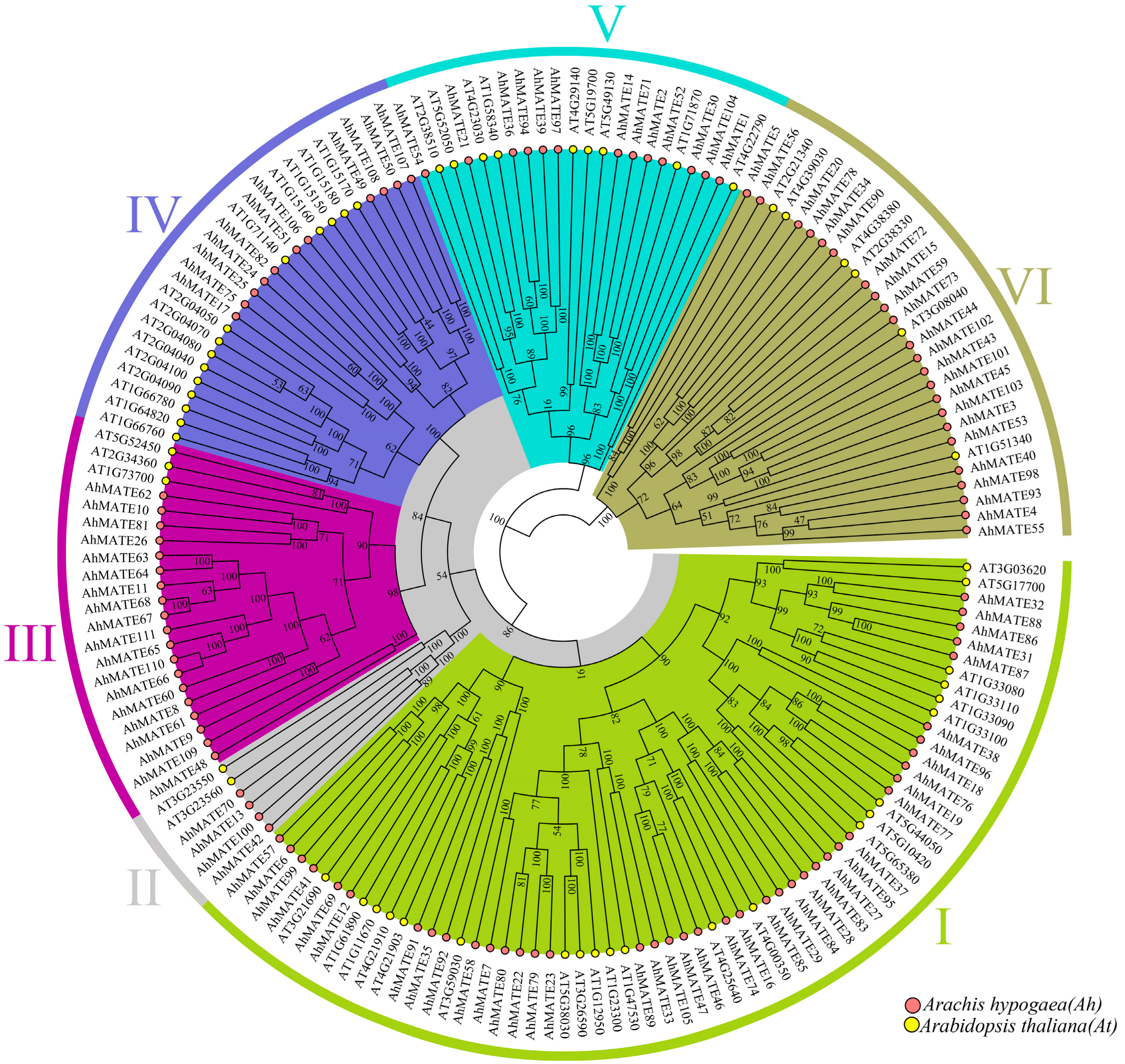
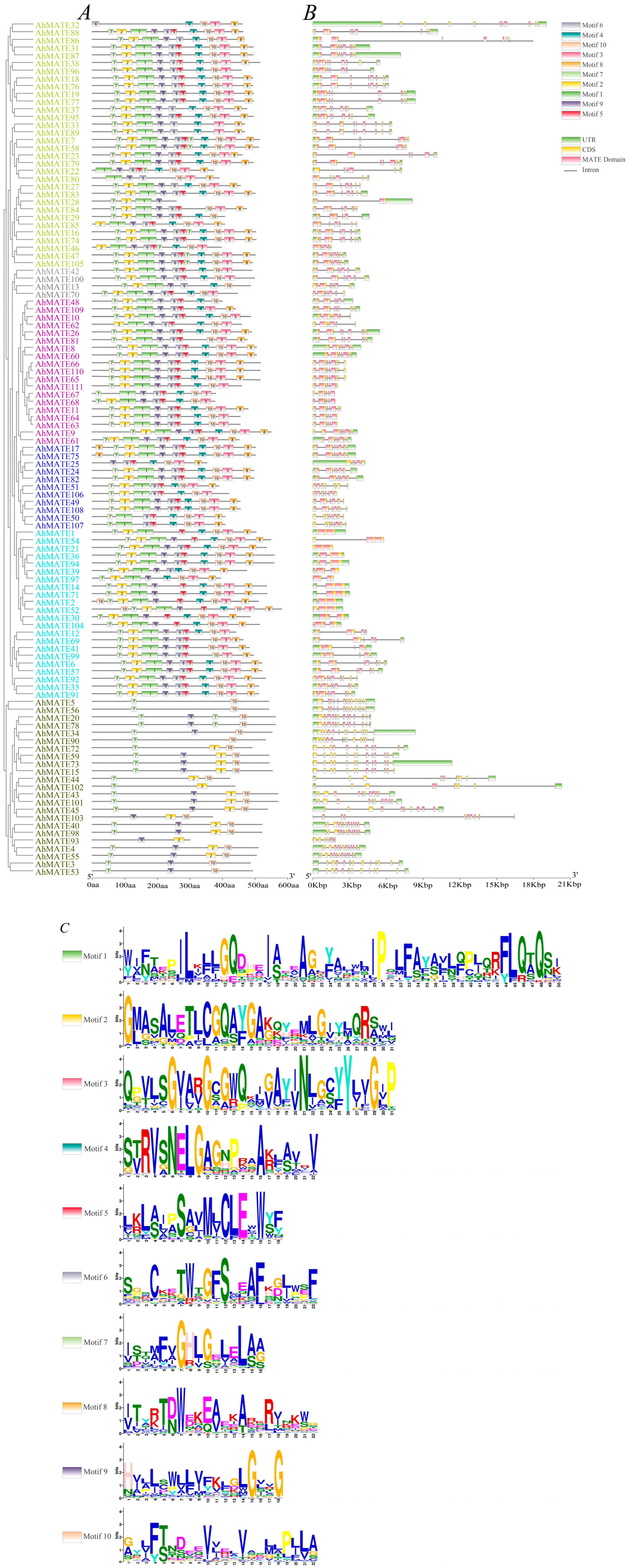
2.2. Phylogenetic Relationship, Conserved Motifs, and Gene Structure Analysis of AhMATEs
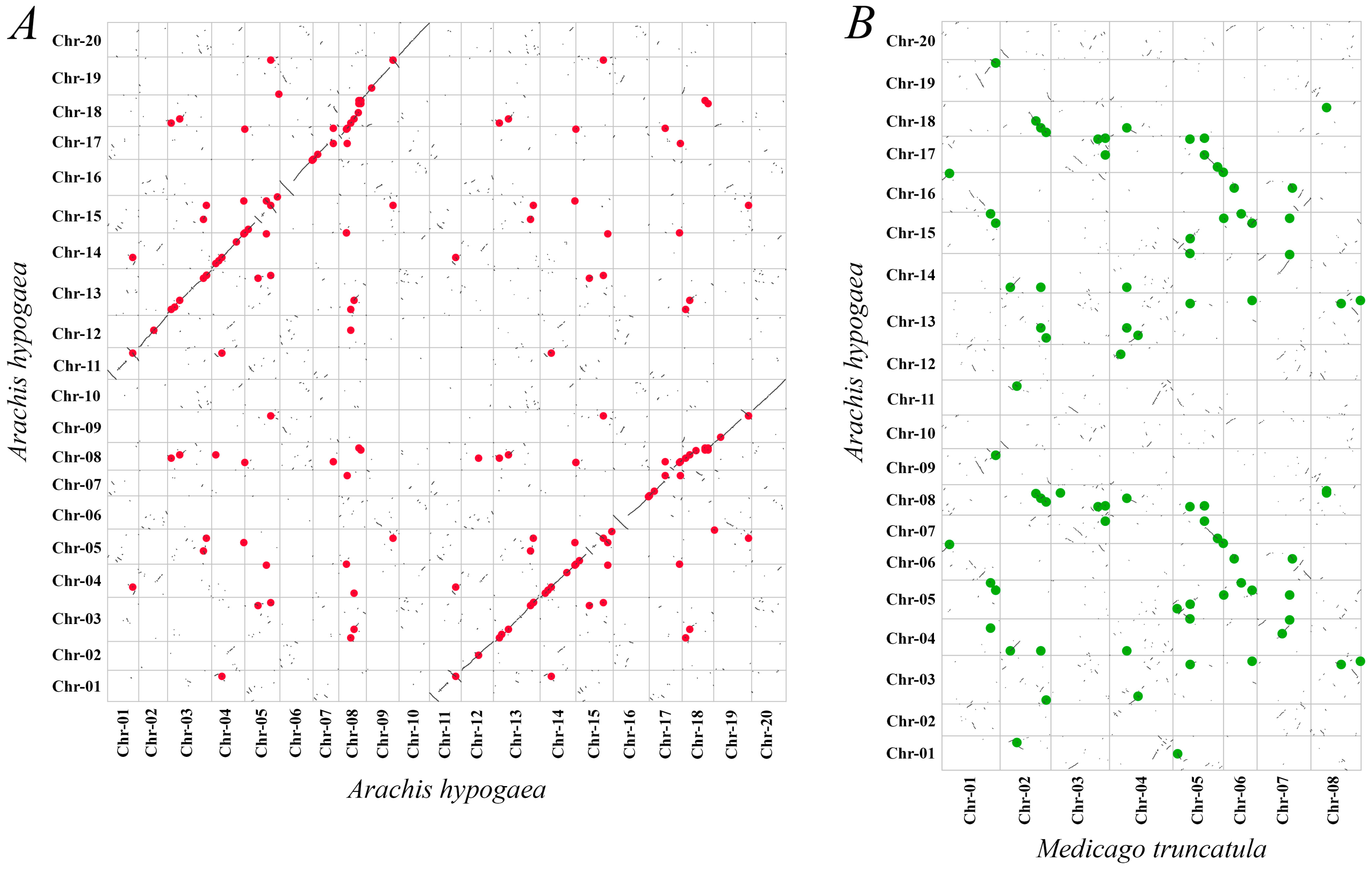
2.3. Gene Duplication and Synteny Analysis
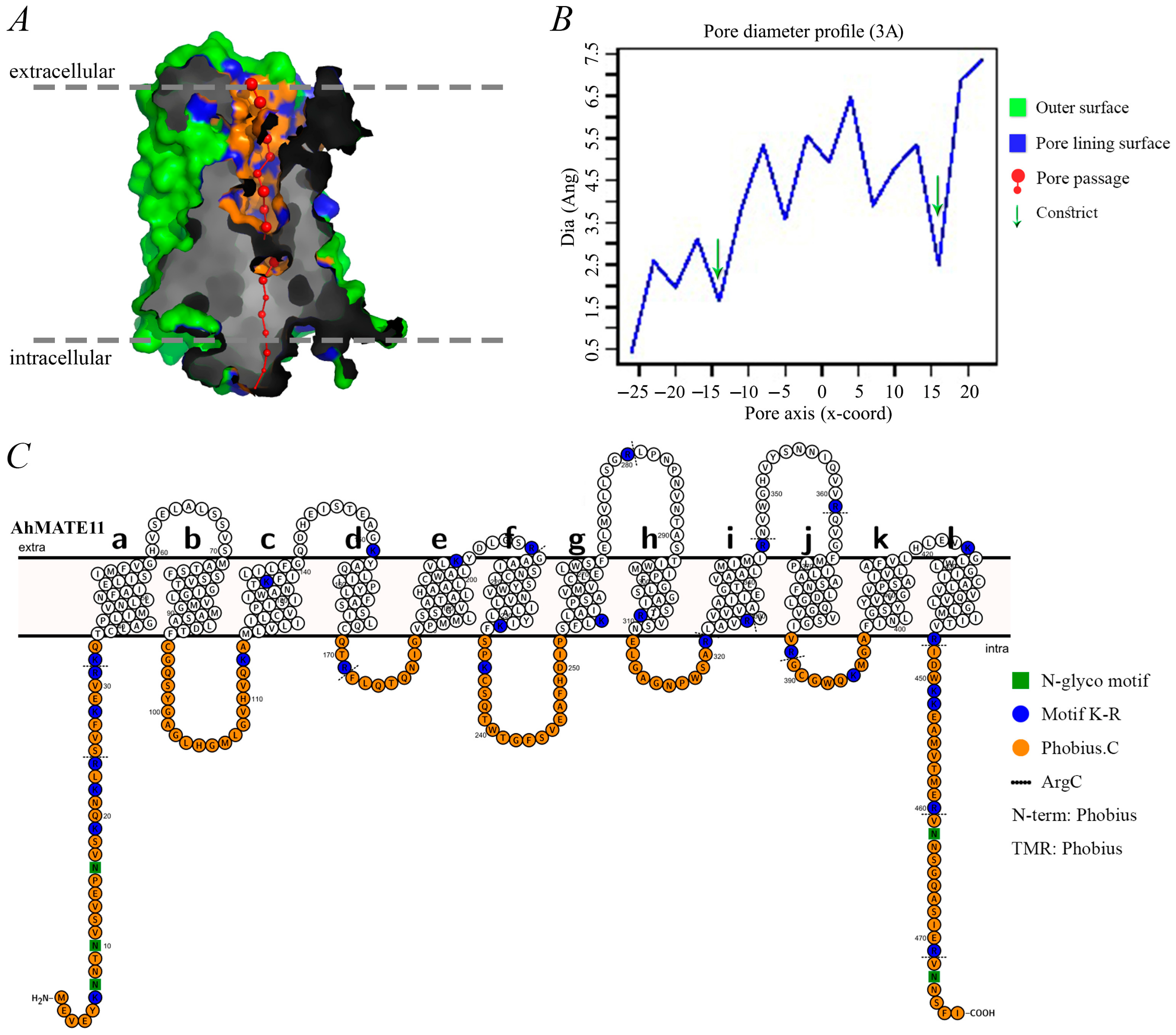
2.4. Pore Morphology, Dimensions, and Protein Topology
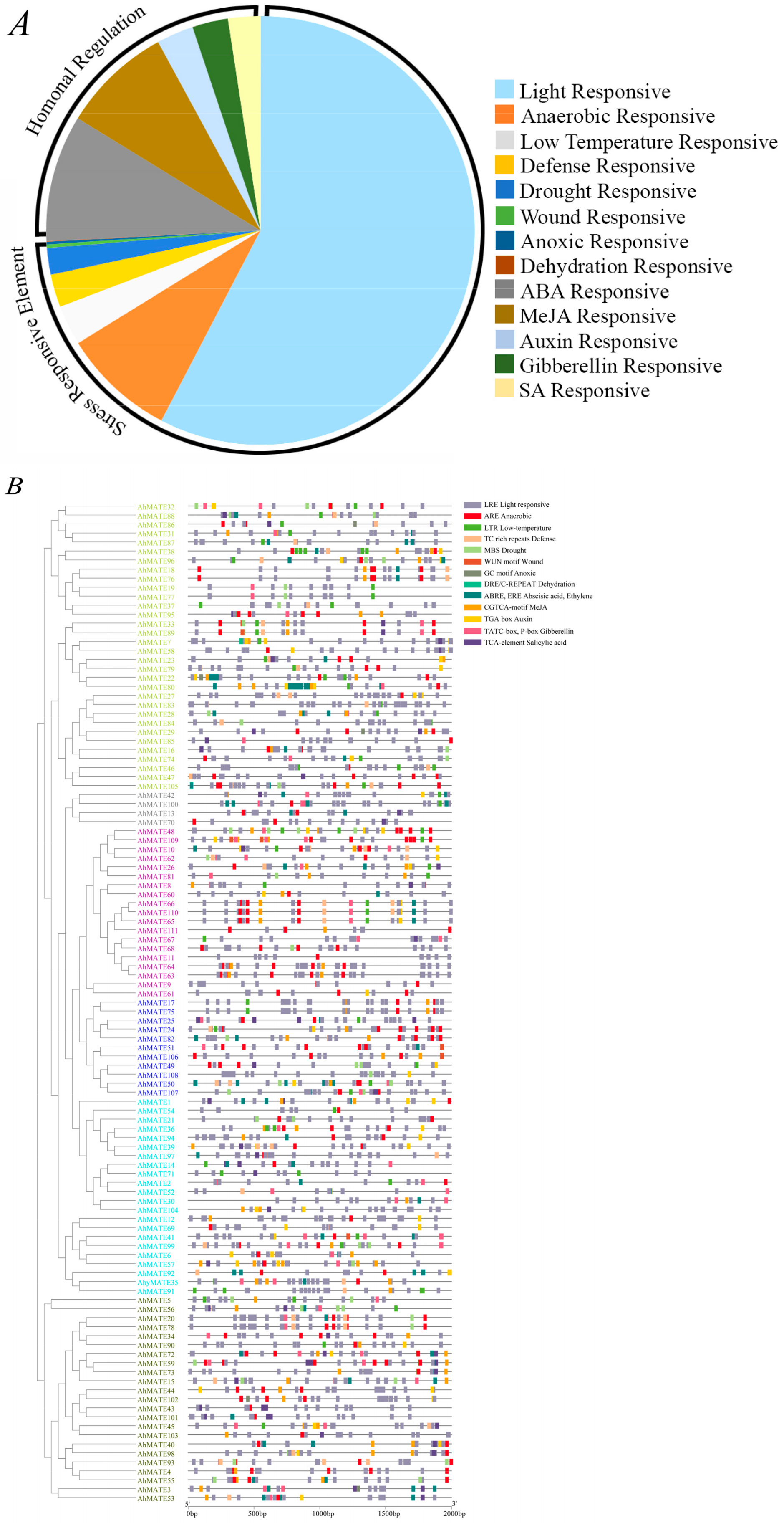
2.5. Cis-Acting Regulatory Elements (CAREs) Detection
2.6. Expression Profiles of AhMATEs Under Al Stress
3. Discussion
4. Materials and Methods
4.1. Identification of MATE Family Genes in Peanut
4.2. Phylogenetic Tree, Motifs, and Domains Analysis
4.3. Duplication Mode and Synteny Analysis
4.4. Cis-Acting Regulatory Elements Analysis
4.5. Protein Tertiary Structure Prediction
4.6. Expression Analysis of AhMATEs
4.7. Plant Growth, Al Treatment, and RNA Isolation
4.8. Quantitative Real-Time PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Al | aluminum |
| aa | amino acids |
| bp | base pair |
| CARE | cis-acting regulatory element |
| CDS | coding sequence |
| TMs | transmembrane segments |
| UTR | untranslated region |
| ZH2 | Zhonghua No.2 |
References
- Çiftçi, S.; Suna, G.G. Functional components of peanuts (Arachis hypogaea L.) and health benefits: A review. Future Foods 2022, 5, 100140. [Google Scholar] [CrossRef]
- Zhao, S.C.; LÜ, J.L.; Xu, X.P.; Lin, X.M.; Luiz, M.R.; Qiu, S.J.; Ciampitti, I.; He, P. Peanut yield, nutrient uptake and nutrient requirements in different regions of China. J. Integr. Agric. 2021, 20, 2502–2511. [Google Scholar] [CrossRef]
- Zhao, X.; Pan, X.; Ma, H.; Dong, X.; Che, J.; Wang, C.; Shi, Y.; Liu, K.; Shen, R. Scientific Issues and Strategies of Acid Soil Use in China. Acta Pedol. Sin. 2023, 60, 1248–1263. [Google Scholar] [CrossRef]
- Chauhan, D.K.; Yadav, V.; Vaculík, M.; Gassmann, W.; Pike, S.; Arif, N.; Singh, V.P.; Deshmukh, R.; Sahi, S.; Tripathi, D.K. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Bojórquez-Quintal, E.; Escalante-Magaña, C.; Echevarría-Machado, I.; Martínez-Estévez, M. Aluminum, a friend or foe of higher plants in acid soils. Front. Plant Sci. 2017, 8, 1767. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. Molecular mechanisms for coping with Al toxicity in plants. Int. J. Mol. Sci 2019, 20, 1551. [Google Scholar] [CrossRef]
- Brown, M.H.; Paulsen, I.T.; Skurray, R.A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 1999, 31, 394–395. [Google Scholar] [CrossRef]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782. [Google Scholar] [CrossRef]
- Morita, Y.; Kataoka, A.; Shiota, S.; Mizushima, T.; Tsuchiya, T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 2000, 182, 6694–6697. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Brown, M.H.; Skurray, R.A. Proton-dependent multidrug efflux systems. Microbiol. Rev. 1996, 60, 575–608. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Yasuda, M.; Morita, Y.; Otsuka, C.; Tsuchiya, T.; Omote, H.; Moriyama, Y. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 2005, 187, 1552–1558. [Google Scholar] [CrossRef] [PubMed]
- Kobara, A.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Omote, H.; Moriyama, Y. A novel variant of mouse MATE-1 H+/organic cation antiporter with a long hydrophobic tail. Arch. Biochem. Biophys. 2008, 469, 195–199. [Google Scholar] [CrossRef]
- Omote, H.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Moriyama, Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006, 27, 587–593. [Google Scholar] [CrossRef]
- Masuda, S.; Terada, T.; Yonezawa, A.; Tanihara, Y.; Kishimoto, K.; Katsura, T.; Ogawa, O.; Inui, K. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 2006, 17, 2127–2135. [Google Scholar] [CrossRef]
- Wang, L.; Bei, X.; Gao, J.; Li, Y.; Yan, Y.; Hu, Y. The similar and different evolutionary trends of MATE family occurred between rice and Arabidopsis thaliana. BMC Plant Biol. 2016, 16, 207. [Google Scholar] [CrossRef]
- Wang, J.; Hou, Q.; Li, P.; Yang, L.; Sun, X.; Benedito, V.A.; Wen, J.; Chen, B.; Mysore, K.S.; Zhao, J. Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula. Plant J. 2017, 90, 79–95. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 223. [Google Scholar] [CrossRef]
- Zhu, H.; Wu, J.; Jiang, Y.; Jin, J.; Zhou, W.E.I.; Wang, Y.U.; Han, G.; Zhao, Y.; Cheng, B. Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J. Genet. 2016, 95, 691–704. [Google Scholar] [CrossRef]
- Huang, J.J.; AN, W.J.; Wang, K.J.; Jiang, T.H.; Ren, Q.; Liang, W.H.; Wang, H.H. Expression profile analysis of MATE gene family in rice. Biol. Plant. 2019, 63, 556–564. [Google Scholar] [CrossRef]
- dos Santos, A.L.; Chaves-Silva, S.; Yang, L.; Maia, L.G.S.; Chalfun-Júnior, A.; Sinharoy, S.; Zhao, J.; Benedito, V.A. Global analysis of the MATE gene family of metabolite transporters in tomato. BMC Plant Biol. 2017, 17, 185. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Meng, H.; Xing, H.; Liang, L.; Zhao, X.; Luo, K. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus. J. Exp. Bot. 2017, 68, 5669–5683. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Magwanga, R.O.; Guo, X.; Kirungu, J.N.; Lu, H.; Cai, X.; Zhou, Z.; Wei, Y.; Wang, X.; Zhang, Z.; et al. Genome-wide analysis of multidrug and toxic compound extrusion (MATE) family in Gossypium raimondii and Gossypium arboreum and its expression analysis under salt, cadmium, and drought stress. G3 Genes Genom. Genet. 2018, 8, 2483–2500. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, H.; He, L.F. Genome-wide analysis of the MATE gene family in potato. Mol. Biol. Rep. 2019, 46, 403–414. [Google Scholar] [CrossRef]
- Dong, B.; Niu, L.; Meng, D.; Song, Z.; Wang, L.; Jian, Y.; Fan, X.; Dong, M.; Yang, Q.; Fu, Y. Genome-wide analysis of MATE transporters and response to metal stress in Cajanus cajan. J. Plant Interact. 2019, 14, 265–275. [Google Scholar] [CrossRef]
- Zhang, X.; Weir, B.; Wei, H.; Deng, Z.; Zhang, X.; Zhang, Y.; Xu, X.; Zhao, C.; Berger, J.D.; Vance, W.; et al. Genome-wide identification and transcriptional analyses of MATE transporter genes in root tips of wild Cicer spp. under aluminium stress. bioRxiv 2020, 2020-04. [Google Scholar] [CrossRef]
- Singh, D.; Tripathi, A.; Mitra, R.; Bhati, J.; Rani, V.; Taunk, J.; Singh, D.; Yadav, R.K.; Siddiqui, M.H.; Pal, M. Genome-wide identification of MATE and ALMT genes and their expression profiling in mungbean (Vigna radiata L.) under aluminium stress. Ecotoxicol. Environ. Saf. 2024, 280, 116558. [Google Scholar] [CrossRef]
- Tripathi, A.; Singh, D.; Bhati, J.; Singh, D.; Tanuk, J.; Alkahtani, J.; Alhasshmi, A.; Singh, M.P. Genome wide identification of MATE and ALMT gene family in lentil (Lens culinaris Medikus) and expression profiling under Al stress condition. BMC Plant Biol. 2025, 25, 88. [Google Scholar] [CrossRef]
- Diener, A.C.; Gaxiola, R.A.; Fink, G.R. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 2001, 13, 1625–1638. [Google Scholar] [CrossRef]
- Li, L.; He, Z.; Pandey, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 2002, 277, 5360–5368. [Google Scholar] [CrossRef]
- Furukawa, J.; Yamaji, N.; Wang, H.; Mitani, N.; Murata, Y.; Sato, K.; Katsuhara, M.; Takeda, K.; Ma, J.F. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 2007, 48, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.V.; Liu, J.; Guimarães, C.T.; Lana, U.G.P.; Alves, V.M.C.; Wang, Y.H.; Schaffert, R.E.; Hoekenga, O.A.; Piñeros, M.A.; Shaff, J.E.; et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Yokosho, K.; Yamaji, N.; Saisho, D.; Yamane, M.; Takahashi, H.; Sato, K.; Nakazono, M.; Ma, J.F. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat. Commun. 2012, 3, 713. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Delhaize, E.; Zhou, M.; Ryan, P.R. The barley MATE gene, HvAACT1, increases citrate efflux and Al3+ tolerance when expressed in wheat and barley. Ann. Bot. 2013, 112, 603–612. [Google Scholar] [CrossRef]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.X.; Zhang, Q.; Chang, G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef]
- Gani, U.; Sharma, P.; Tiwari, H.; Nautiyal, A.K.; Kundan, M.; Wajid, M.A.; Kesari, R.; Nargotra, A.; Misra, P. Comprehensive genome-wide identification, characterization, and expression profiling of MATE gene family in Nicotiana tabacum. Gene 2021, 783, 145554. [Google Scholar] [CrossRef]
- Nagy, L.G.; Merényi, Z.; Hegedüs, B.; Bálint, B. Novel phylogenetic methods are needed for understanding gene function in the era of mega-scale genome sequencing. Nucleic Acids Res. 2020, 48, 2209–2219. [Google Scholar] [CrossRef]
- Miyauchi, H.; Moriyama, S.; Kusakizako, T.; Kumazaki, K.; Nakane, T.; Yamashita, K.; Hirata, K.; Dohmae, N.; Nishizawa, T.; Ito, K.; et al. Structural basis for xenobiotic extrusion by eukaryotic MATE transporter. Nat. Commun. 2017, 8, 1633. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Pan, Y.; Jia, J.; Zhang, H.; Bai, F.; Zhang, P.; Zhu, H.; He, Y.; et al. A molecular pathway for CO2 response in Arabidopsis guard cells. Nat. Commun. 2015, 6, 6057. [Google Scholar] [CrossRef]
- Wang, R.; Liu, X.; Liang, S.; Ge, Q.; Li, Y.; Shao, J.; Qi, Y.; An, L.; Yu, F. A subgroup of MATE transporter genes regulates hypocotyl cell elongation in arabidopsis. J. Exp. Bot. 2015, 66, 6327–6343. [Google Scholar] [CrossRef]
- Ren, H.; Wang, Z.; Shang, X.; Zhang, X.; Ma, L.; Bian, Y.; Wang, D.; Liu, W. Involvement of GA3-oxidase in inhibitory effect of nitric oxide on primary root growth in Arabidopsis. Plant Biol. 2024, 26, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, X.; Shaff, J.; Liang, C.; Jia, X.; Li, Z.; Magalhaes, J.; Kochian, L.V. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J. 2012, 71, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Dingus, A.; Roslund, M.I.; Brauner, S.; Sinkkonen, A.; Weidenhamer, J.D. Arabidopsis response to copper is mediated by density and root exudates: Evidence that plant density and toxic soils can shape plant communities. Am. J. Bot. 2024, 111, e16285. [Google Scholar] [CrossRef]
- Qiu, W.; Wang, N.; Dai, J.; Wang, T.; Kochian, L.V.; Liu, J.; Zuo, Y. AhFRDL1-mediated citrate secretion contributes to adaptation to iron deficiency and aluminum stress in peanuts. J. Exp. Bot. 2019, 70, 2873–2886. [Google Scholar] [CrossRef]
- Jo, B.S.; Choi, S.S. Introns: The Functional Benefits of Introns in Genomes. Genom. Inform. 2015, 13, 112–118. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Lu, M. Structures of multidrug and toxic compound extrusion transporters and their mechanistic implications. Channels 2016, 2, 88–100. [Google Scholar] [CrossRef]
- Vogel, H.J. A numerical experiment on pore size, pore connectivity, water retention, permeability, and solute transport using network models. Eur. J. Soil Sci. 2000, 51, 99–105. [Google Scholar] [CrossRef]
- Lee, J.K.; Kozono, D.; Remis, J.; Kitagawa, Y.; Agre, P.; Stroud, R.M. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc. Natl. Acad. Sci. USA 2005, 102, 18932–18937. [Google Scholar] [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 3, 151–159. [Google Scholar] [CrossRef]
- Tanaka, Y.; Iwaki, S.; Tsukazaki, T. Crystal Structure of a Plant Multidrug and Toxic Compound Extrusion Family Protein. Structure 2017, 9, 1455–1460.e2. [Google Scholar] [CrossRef] [PubMed]
- Claxton, D.P.; Jagessar, K.L.; McHaourab, H.S. Principles of Alternating Access in Multidrug and Toxin Extrusion (MATE) Transporters. J. Mol. Biol. 2021, 16, 166959. [Google Scholar] [CrossRef] [PubMed]
- Kusakizako, T.; Claxton, D.P.; Tanaka, Y.; Maturana, A.D.; Kuroda, T.; Ishitani, R.; McHaourab, H.S.; Nureki, O. Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter. Structure 2019, 27, 293–301.e293. [Google Scholar] [CrossRef]
- Qiao, C.; Yang, J.; Wan, Y.; Xiang, S.; Guan, M.; Du, H.; Tang, Z.; Lu, K.; Li, J.; Qu, C. A genome-wide survey of MATE transporters in Brassicaceae and unveiling their expression profiles under abiotic stress in rapeseed. Plants 2020, 9, 1072. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Zhou, P.; Silverstein, K.A.T.; Ramaraj, T.; Guhlin, J.; Denny, R.; Liu, J.; Farmer, A.D.; Steele, K.P.; Stupar, R.M.; Miller, J.R.; et al. Exploring structural variation and gene family architecture with De Novo assemblies of 15 Medicago genomes. BMC Genom. 2017, 18, 261. [Google Scholar] [CrossRef]
- Kumar, G.M.; Mamidala, P.; Podile, A.R. Regulation of polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics 2009, 2, 238–249. [Google Scholar]
- Ibraheem, O.; Botha, C.E.J.; Bradley, G. In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 2010, 34, 268–283. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.N.; Yao, J.F.; Ren, Y.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene. Plant Soil 2017, 418, 255–266. [Google Scholar] [CrossRef]
- Arbelaez, J.; Maron, L.; Jobe, T.; Piñeros, M.; Famoso, A.; Rebelo, A.R.; Singh, N.; Ma, Q.; Fei, Z.; Kochian, L.; et al. Aluminum Resistance Transcription Factor 1 (ART1) contributes to natural variation in aluminum resistance in diverse genetic backgrounds of rice (O. sativa). Plant Direct 2017, 1, 14. [Google Scholar] [CrossRef]
- Xiao, D.; Li, X.; Zhou, Y.Y.; Wei, L.; Keovongkod, C.; He, H.; Zhan, J.; Wang, A.Q.; He, L.F. Transcriptome analysis reveals significant difference in gene expression and pathways between two peanut cultivars under Al stress. Gene 2021, 781, 145535. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.E.; Guerinot, M.L. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 2002, 14, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 1, 1023–1031. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro-Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 15, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2019, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2012, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hameed, S.; Li, X.; Zhou, Y.; Zhan, J.; Wang, A.; Han, Z.; Xiao, D.; He, L. Genome-Wide Dissection of MATE Gene Family in Cultivated Peanuts and Unveiling Their Expression Profiles Under Aluminum Stress. Int. J. Mol. Sci. 2025, 26, 2707. https://doi.org/10.3390/ijms26062707
Hameed S, Li X, Zhou Y, Zhan J, Wang A, Han Z, Xiao D, He L. Genome-Wide Dissection of MATE Gene Family in Cultivated Peanuts and Unveiling Their Expression Profiles Under Aluminum Stress. International Journal of Molecular Sciences. 2025; 26(6):2707. https://doi.org/10.3390/ijms26062707
Chicago/Turabian StyleHameed, Saba, Xia Li, Yunyi Zhou, Jie Zhan, Aiqin Wang, Zhuqiang Han, Dong Xiao, and Longfei He. 2025. "Genome-Wide Dissection of MATE Gene Family in Cultivated Peanuts and Unveiling Their Expression Profiles Under Aluminum Stress" International Journal of Molecular Sciences 26, no. 6: 2707. https://doi.org/10.3390/ijms26062707
APA StyleHameed, S., Li, X., Zhou, Y., Zhan, J., Wang, A., Han, Z., Xiao, D., & He, L. (2025). Genome-Wide Dissection of MATE Gene Family in Cultivated Peanuts and Unveiling Their Expression Profiles Under Aluminum Stress. International Journal of Molecular Sciences, 26(6), 2707. https://doi.org/10.3390/ijms26062707





