Abstract
Wheat powdery mildew disease caused by the obligate biotrophic fungal pathogen Blumeria graminis forma specialis tritici (B.g. tritici) seriously threatens global wheat production. Although improved powdery mildew resistance is an aim in wheat breeding, the regulatory mechanism underlying the wheat–B.g. tritici interaction remains poorly understood. In this study, the wheat chromatin remodeling protein TaSWP73 was identified as a negative regulator of post-penetration resistance against B.g. tritici. The transient overexpression of TaSWP73 attenuates wheat post-penetration resistance against B.g. tritici, while the silencing of TaSWP73 potentiates salicylic acid (SA) biosynthesis and activates post-penetration resistance against B.g. tritici. Importantly, chromatin in the promoter regions of TaSARD1, an activator gene of SA biosynthesis, is marked by high nucleosome occupancy in the TaSWP73-silenced wheat leaves. The silencing of TaSARD1 could suppress SA biosynthesis and attenuate post-penetration resistance against B.g. tritici with a lack of TaSWP73. In addition, TaICS1 was characterized as an essential component of wheat SA biosynthetic machinery. Potentiated SA biosynthesis and increased post-penetration resistance against B.g. tritici with a lack of TaSWP73 could be suppressed by the silencing of TaICS1 expression. These results collectively support the hypothesis that the wheat chromatin remodeling protein TaSWP73 contributes to the compatible wheat–powdery mildew interaction presumably via the suppression of the TaSARD1-TaICS1-SA pathway.
1. Introduction
As one of the most widely cultivated staple crops, allohexaploid bread wheat (Triticum aestivum L.) provides about one-fifth of the calories in human food [1,2]. The increasing global population drives the demand for wheat grains, but wheat growth and yields are threatened by host-adapted pathogens and pests (P&Ps) [3]. The biotrophic fungal pathogen Blumeria graminis forma specialis tritici (B.g. tritici) is the causal agent of the devastating powdery mildew disease, which results in a wheat yield loss of 10–40% [4,5]. During the infection, B.g. tritici conidia landing on the wheat host epidermal cells firstly germinate and then produce appressorium, enabling penetration through the plant cell wall [4,5]. In the post-penetration stages, a feeding structure of B.g. tritici, the haustorium, is developed to absorb nutrients from wheat cells, and finally, microcolonies are formed to disperse conidia [4,5]. Cultivating B.g. tritici-resistant wheat varieties is one of the most effective and economical ways to control this epidemic [4,5]. To this end, it is vital to decipher the regulatory mechanism underlying the wheat–B.g. tritici interaction.
The recognition of invading pathogens such as B.g. tritici by host plants like bread wheat could trigger induced defenses like pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) to cope with pathogen infections [6,7,8,9,10,11,12,13,14,15,16]. Defense-related phytohormone salicylic acid (SA) plays a key role in initiating the intertwined PTI and ETI, and the activation of PTI and ETI usually culminates in massive transcriptomic reprogramming [17,18,19]. In the dicot model plant Arabidopsis thaliana, the major route for SA biosynthesis takes place in the chloroplast, and isochorismate synthase AtICS1 (isochorismate synthase 1) is one of the key enzymes in the SA biosynthetic machinery [20]. Arabidopsis calmodulin-binding protein SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (AtSARD1) was identified as a key regulator of AtICS1 induction and SA biosynthesis [21]. In addition, epigenetic modulators like histone acetylases and chromatin remodeling proteins were characterized as key regulators of plant defense in A. thaliana [22]. For instance, the expression of AtSARD1 and SA biosynthesis was epigenetically suppressed by histone deacetylase AtHDA6 [23]. In addition, Arabidopsis switch/sucrose non-fermentable (SWI/SNF)-associated protein AtSWP73A epigenetically suppresses the plant immune receptor [24]. However, the potential regulation of the wheat–B.g. tritici interaction by the chromatin remodeling protein SWP73 remains unknown.
Herein, the wheat chromatin remodeling protein TaSWP73 was identified as an epigenetic suppressor of post-penetration resistance against B.g. tritici. The transient overexpression of TaSWP73 attenuates wheat post-penetration resistance against B.g. tritici, while the silencing of TaSWP73 potentiates SA accumulation and activates post-penetration resistance against B.g. tritici. Importantly, chromatin in the promoter regions of TaSARD1 is marked by high nucleosome occupancy in TaSWP73-silenced wheat leaves, suggesting that the chromatin remodeling protein TaSWP73 suppresses TaSARD1 transcription at the epigenetic level. The silencing of TaSARD1 and isochorismate synthase gene TaICS1 could suppress SA biosynthesis and attenuate post-penetration resistance against B.g. tritici with a lack of TaSWP73. These results collectively support the hypothesis that the wheat chromatin remodeling protein TaSWP73 contributes to the compatible wheat–powdery mildew interaction presumably via the epigenetic suppression of TaSARD1 transcription and the attenuation of the TaSARD1-TaICS1-SA pathway. These findings shed novel light on the epigenetic mechanism underlying wheat–B.g. tritici interactions and provide valuable information for genetic improvement in wheat resistance against devastating powdery mildew disease.
2. Results
2.1. Characterization of TaSWP73 Genes in Regulation of Compatible Wheat–B.g. tritici Interaction
In this study, we are interested in examining the function of wheat SWP73 homologs in the wheat–B.g. tritici interaction. To this end, amino acid sequences of Arabidopsis AtSWP73A (At3g01890) and AtSWP73B (At5g14170) were used as queries to search the reference genome of the hexaploid bread wheat (http://plants.ensembl.org/). TaSWP73 was identified as the wheat homolog of AtSWP73A and AtSWP73B. Three TaSWP73 genes separately located on wheat chromosomes 2A, 2B, and 2D were designated as TaSWP73-2A (TraesCS2A02G281500), TaSWP73-2B (TraesCS2B02G298800), and TaSWP73-2D (TraesCS2D02G280300) (Figure S1). As shown in Figure 1A, these predicted TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D proteins shared more than 57% of their identity with Arabidopsis AtSWP73A and AtSWP73B. Phylogenetic analysis validated that wheat TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D proteins are homologs of Brachypodium BdSWP73, maize ZmSWP73, rice OsSWP73, and Arabidopsis TaSWP73A and TaSWP73B (Figure 1B). As shown in Figure 1C,D, a conserved SWIB-like domain was identified in all TaSWP73 proteins, and the coding regions of TaSWP73 genomic sequences all contained two exons and one intron.
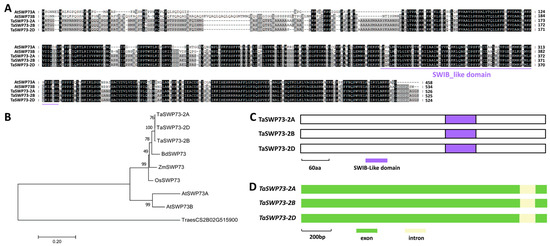
Figure 1.
Identification of wheat TaSWP73 based on homology with Arabidopsis AtSWP73A and AtSWP73B. (A) Protein sequence alignments of Arabidopsis AtSWP73A, AtSWP73B, wheat TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D. Conserved residues among 5 protein sequences are shaded in dark, while residues conserved in at least 3 of the 5 proteins are shaded in gray. (B) Phylogenetic relationships of the SWP73 proteins from Arabidopsis, Brachypodium, maize, rice, and wheat. Two-letter genus species prefixes: At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Os, Oryza sativa; Ta, Triticum aestivum; Zm, Zea mays. (C) Domain structures of wheat TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D proteins. (D) Gene architectures of wheat TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D genes.
To analyze the function of the TaSWP73 gene in the regulation of the wheat–B.g. tritici interaction, transient gene expression assays were performed to overexpress the TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D genes in the wheat leaf epidermal cells. These bombarded wheat leaves were inoculated with B.g. tritici conidia, and the B.g. tritici haustorium index (HI%) was analyzed. As shown in Figure 2A, the B.g. tritici HI% increased from 54.6% for the empty vector (OE-EV) control to above 69.7% for wheat cells overexpressing the TaSWP73-2A, TaSWP73-2B, or TaSWP73-2D gene, suggesting that the TaSWP73 gene negatively regulates wheat post-penetration resistance against powdery mildew and positively contributes to the B.g. tritici post-penetration event haustorial formation. Thereafter, we employed barley stripe mosaic virus (BSMV)-induced gene silencing (BSMV-VIGS) to silence all endogenous TaSWP73 genes in the wheat leaves. As shown in Figure 2B, the reverse transcription–quantitative polymerase chain reaction (RT-qPCR) assay demonstrated that the accumulation level of TaSWP73 gene transcripts decreased significantly in wheat leaves silencing the TaSWP73 gene. These BSMV-VIGS wheat leaves were inoculated with B.g. tritici conidia, and the B.g. tritici microcolony index (MI%) was analyzed. As shown in Figure 2C, the B.g. tritici MI% decreased from 59.3% for the control plants (BSMV-γ) to 31.7% for TaSWP73-silenced plants (BSMV-TaSWP73as), confirming that the TaSWP73 gene negatively regulates wheat post-penetration resistance against B.g. tritici and positively contributes to the B.g. tritici post-penetration event microcolony formation. We measured the accumulation of defense-related phytohormone SA in TaSWP73-silenced wheat leaves infected by B.g. tritici. As shown in Figure 2D, the SA level was remarkably elevated in the TaSWP73-silenced wheat leaves, compared with that of the BSMV-γ control plants, suggesting that TaSWP73 suppresses SA accumulation in bread wheat. We analyzed the transcript levels of SA signaling marker genes TaPR1 and TaPR2 in TaSWP73-silenced wheat leaves. As shown in Figure 2E, the accumulation levels of the TaPR1 and TaPR2 transcripts were greatly enhanced by the silencing of TaSWP73. These results suggested that the chromatin remodeling protein TaSWP73 negatively regulates SA accumulation and contributes to the compatible wheat–powdery mildew interaction.

Figure 2.
Functional characterization of TaSWP73 gene in wheat–B.g. tritici interaction. (A) Statistical analysis of B.g. tritici haustorial formation in wheat epidermal cells transiently overexpressing TaSWP73 (OE-TaSWP73). Wheat epidermal cells bombarded with empty vector (OE-EV) were set as a control. More than 50 wheat cells were analyzed for each experiment. (B) RT-qPCR analysis of TaSWP73 transcript accumulation in the wheat leaves silencing TaSWP73 (BSMV-TaSWP73as). (C) Statistical analysis of B.g. tritici microcolony formation in wheat leaves silencing TaSWP73. (D) Measurement of SA content in wheat leaves silencing TaSWP73. (E) RT-qPCR analysis of TaPR1 and TaPR2 transcript accumulation in wheat leaves silencing TaSWP73. For (B–E), leaves of wheat plants infected with BSMV-γ were employed as the negative control. For (A–E), three technical replicates per treatment were statistically analyzed, and data are presented as the mean ± SE (Student’s t-test; ** p < 0.01), and these assays were repeated in three independent biological replicates with similar results.
2.2. Epigenetic Regulation of TaSARD1 Genes by TaSWP73
In the dicot model plant A. thaliana, the transcription of the AtSARD1 gene is tightly regulated at epigenetic levels [23]. TaSARD1 genes were identified as key regulators of wheat post-penetration resistance against B.g. tritici [25]. We ask whether the wheat chromatin remodeling protein TaSWP73 is involved in the epigenetic regulation of the TaSARD1 gene. To examine the potential regulation of the chromatin structure in promoter regions of TaSARD1 genes by the wheat chromatin remodeling protein TaSWP73, the nucleosome occupancy micrococcal nuclease (MNase) assay was performed (Figure 3A). As shown in Figure 3A, the MNase assay showed significantly reduced nucleosome occupancy in TaSARD1 promoters in the wheat leaves silencing the TaSWP73 gene compared with the BSMV-γ control, suggesting that the wheat chromatin remodeling protein TaSWP73 might function as an epigenetic repressor of the TaSARD1 gene. Consistent with this, nuclear run-on and RT-qPCR assays demonstrated that the silencing of the TaSWP73 gene resulted in a significant enhancement in the transcription rates and transcript accumulation of the TaSARD1 gene (Figure 3B,C). These results suggested that the chromatin remodeling protein TaSWP73 contributes to the establishment of a repressive chromatin state in the TaSARD1 gene.

Figure 3.
Characterization of nucleosomal occupancy and gene transcription at TaSARD1 loci in TaSWP73-silenced wheat leaves. (A) MNase analysis of nucleosome occupancy in TaSARD1 promoters in wheat leaves silencing TaSWP73 (BSMV-TaSWP73as). The nucleosome occupancy levels in wheat leaves infected with the BSMV-γ empty vector (negative control) were set to 1.0. Transcription rates (B) and expression levels (C) of the TaSARD1 gene in wheat leaves silencing TaSWP73 were measured by nuclear run-on and RT-qPCR assays, respectively. For (A–C), leaves of wheat plants infected with BSMV-γ were employed as the negative control. For (A–C), three technical replicates per treatment were statistically analyzed, and data are presented as the mean ± SE (Student’s t-test; ** p < 0.01), and these assays were repeated in three independent biological replicates with similar results.
2.3. Functional Analysis of TaSARD1 Genes in the TaSWP73-Mediated Suppression of SA Accumulation and Compatible Wheat–B.g. tritici Interaction
Having already demonstrated that TaSARD1 gene expression and wheat post-penetration resistance against B.g. tritici are negatively regulated by the chromatin remodeling protein TaSWP73, we next ask whether the chromatin remodeling protein TaSWP73 negatively regulates wheat post-penetration resistance against B.g. tritici via the suppression of TaSARD1 gene expression. To test this hypothesis, we simultaneously silenced the TaSWP73 and TaSARD1 genes and analyzed the B.g. tritici MI%. As shown in Figure 4A, the accumulation levels of the TaSWP73 or TaSARD1 gene transcript decreased remarkably in wheat leaves co-silencing TaSWP73 and TaSARD1 genes, compared with the BSMV-γ control. The B.g. tritici MI% decreased from 58.9% for the control plants (BSMV-γ) to 30.2% for the TaSWP73-silenced (BSMV-TaSWP73as) plants but increased to above 77.2% for wheat leaves co-silencing TaSWP73 with the TaSARD1 gene (Figure 4B). Under B.g. tritici infection, the SA level showed a significant increase in the wheat leaves silencing the TaSWP73 gene but a remarkable reduction in the wheat leaves co-silencing TaSWP73 with the TaSARD1 gene (Figure 4C). The RT-qPCR assay further demonstrated that the accumulation levels of TaPR1 and TaPR2 transcripts significantly increased in the wheat leaves silencing the TaSWP73 gene but remarkably decreased in the wheat leaves co-silencing the TaSWP73 and TaSARD1 genes, compared with the BSMV-γ control (Figure 4D). The above results indicated that potentiated SA biosynthesis and increased post-penetration resistance against B.g. tritici with a lack of TaSWP73 could be attenuated by the silencing of TaSARD1 expression, suggesting that the epigenetic suppression of TaSARD1 by the chromatin remodeling protein TaSWP73 might contribute to the negative regulation of TaSWP73 in SA biosynthesis and a positive contribution to the compatible wheat–powdery mildew interaction.

Figure 4.
Characterization of the genetic interplay between TaSWP73 and TaSARD1 in the wheat–B.g. tritici interaction. (A) RT-qPCR analysis of TaSWP73 and TaSARD1 transcript accumulation in wheat leaves silencing TaSWP73 (BSMV-TaSWP73as) or TaSARD1 (BSMV-TaSARD1as) or co-silencing TaSWP73 and TaSARD1 (BSMV-TaSWP73as + BSMV-TaSARD1as). (B) Statistical analysis of B.g. tritici microcolony formation in wheat leaves silencing TaSWP73 or TaSARD1 or co-silencing TaSWP73 and TaSARD1. (C) Measurement of SA content in wheat leaves silencing TaSWP73 or TaSARD1 or co-silencing TaSWP73 and TaSARD1. (D) RT-qPCR analysis of TaPR1 and TaPR2 transcript accumulation in wheat leaves silencing TaSWP73 or TaSARD1 or co-silencing TaSWP73 and TaSARD1. For (A–D), leaves of wheat plants infected with BSMV-γ were employed as the negative control. For (A–D), three technical replicates per treatment were statistically analyzed, and data are presented as the mean ± SE (Student’s t-test; ** p < 0.01), and these assays were repeated in three independent biological replicates with similar results.
2.4. Functional Analysis of TaICS1 Genes in the TaSWP73-Mediated Suppression of SA Accumulation and Wheat–B.g. tritici Interaction
In the dicot model plant A. thaliana, AtICS1 is involved in SA biosynthesis [20]. Herein, we are interested in examining the function of wheat ICS1 homologs in the wheat–B.g. tritici interaction. To this end, we first searched the reference genome of the hexaploid bread wheat using the amino acid sequence of Arabidopsis AtICS1 (At1g74710) as a query and identified TaICS1 as a wheat homolog of AtICS1. Three TaICS1 genes separately located on wheat chromosomes 5A, 5B, and 5D were designated as TaICS1-5A (TraesCS5A02G193800), TaICS1-5B (TraesCS5B02G189100), and TaICS1-5D (TraesCS5D02G196200). As shown in Figure 5A, these predicted TaICS1-5A, TaICS1-5B, and TaICS1-5D proteins shared more than 48% of their identity with Arabidopsis AtICS1. Phylogenetic analysis validated that wheat TaICS1-5A, TaICS1-5B, and TaICS1-5D proteins are homologs of Brachypodium BdICS1, maize ZmICS1, rice OsICS1, and Arabidopsis TaICS1 (Figure 5B). As shown in Figure 5C,D, a conserved chorismate-binding enzyme (Chorismate_bind) domain was identified in the C-terminal parts of all TaICS1 proteins, and the coding regions of the TaICS1 genomic sequences all contained 14 exons and 13 introns.

Figure 5.
Identification of wheat TaICS1 based on homology with Arabidopsis AtICS1. (A) Protein sequence alignments of Arabidopsis AtICS1, wheat TaICS1-5A, TaICS1-5B, and TaICS1-5D. Conserved residues among 4 protein sequences are shaded in dark, while residues conserved in at least 2 of the 4 proteins are shaded in gray. (B) Phylogenetic relationships of the ICS1 proteins from Arabidopsis, Brachypodium, maize, rice, and wheat. Two-letter genus species prefixes: At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Os, Oryza sativa; Ta, Triticum aestivum; Zm, Zea mays. (C) Domain structures of wheat TaICS1-5A, TaICS1-5B, and TaICS1-5D proteins. (D) Gene architectures of wheat TaICS1-5A, TaICS1-5B, and TaICS1-5D genes.
To examine the function of the TaICS1 genes in the TaSWP73-mediated suppression of SA accumulation and wheat post-penetration resistance against B.g. tritici, we simultaneously silenced TaSWP73 and TaICS1 genes and analyzed the B.g. tritici MI%. As shown in Figure 6A, the accumulation levels of the TaSWP73 or TaICS1 gene transcripts decreased remarkably in wheat leaves co-silencing TaSWP73 and TaICS1 genes, compared with the BSMV-γ control. The B.g. tritici MI% decreased from 56.8% for the control plants (BSMV-γ) to 29.7% for TaSWP73-silenced (BSMV-TaSWP73as) leaves but increased to above 76.9% for wheat leaves co-silencing TaSWP73 with the TaICS1 gene (Figure 6B). Under B.g. tritici infection, the SA level showed a significant increase in the wheat leaves silencing the TaSWP73 gene but a remarkable reduction in the wheat leaves co-silencing TaSWP73 with the TaICS1 gene (Figure 6C). The RT-qPCR assay further demonstrated that the accumulation levels of TaPR1 and TaPR2 gene transcripts significantly increased in the wheat leaves silencing the TaSWP73 gene but remarkably decreased in the wheat leaves co-silencing TaSWP73 with the TaICS1 gene, compared with the BSMV-γ control (Figure 6D). The above results indicated that potentiated SA biosynthesis and increased post-penetration resistance against B.g. tritici with a lack of TaSWP73 could be attenuated by the silencing of TaICS1 expression. These findings collectively suggested that the wheat chromatin remodeling protein TaSWP73 contributes to the compatible wheat–powdery mildew interaction probably via the suppression of the TaSARD1-TaICS1-SA pathway.

Figure 6.
Characterization of the genetic interplay between TaSWP73 and TaICS1 in the wheat–B.g. tritici interaction. (A) RT-qPCR analysis of TaSWP73 and TaICS1 transcript accumulation in wheat leaves silencing TaSWP73 (BSMV-TaSWP73as) or TaICS1 (BSMV-TaICS1as) or co-silencing TaSWP73 and TaICS1 (BSMV-TaSWP73as + BSMV-TaICS1as). (B) Statistical analysis of B.g. tritici microcolony formation in wheat leaves silencing TaSWP73 or TaICS1 or co-silencing TaSWP73 and TaICS1. (C) Measurement of SA content in wheat leaves silencing TaSWP73 or TaICS1 or co-silencing TaSWP73 and TaICS1. (D) RT-qPCR analysis of TaPR1 and TaPR2 transcript accumulation in wheat leaves silencing TaSWP73 or TaICS1 or co-silencing TaSWP73 and TaICS1. For (A–D), leaves of wheat plants infected with BSMV-γ were employed as the negative control. For (A–D), three technical replicates per treatment were statistically analyzed, and data are presented as the mean ± SE (Student’s t-test; ** p < 0.01), and these assays were repeated in three independent biological replicates with similar results.
3. Discussion
3.1. Wheat Chromatin Remodeling Protein TaSWP73 Positively Contributes to the Compatible Wheat–B.g. tritici Interaction
In this study, three TaSWP73 genes (TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D) were separately identified from wheat chromosomes 2A, 2B and 2D. Sequence alignment and phylogenetic analysis demonstrated that the highly homologous TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D proteins are the closest homologs of Brachypodium BdSWP73, maize ZmSWP73, rice OsSWP73, and Arabidopsis TaSWP73A and TaSWP73B. The overexpression of TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D resulted in a significantly increased B.g. tritici haustorium index (HI%), while silencing TaSWP73 led to a remarkably decreased B.g. tritici microcolony index (MI%), suggesting that the wheat chromatin remodeling protein TaSWP73 negatively regulates post-penetration resistance against powdery mildew and positively contributes to B.g. tritici post-penetration events like haustorium development and microcolony formation. Interestingly, SA accumulation and the expression levels of SA signaling-related defense marker genes TaPR1 and TaPR2 were potentiated in the TaSWP73-silenced wheat leaves, suggesting that TaSWP73 negatively regulates SA accumulation. Interestingly, AtSWP73A was revealed to suppress the Arabidopsis defense response against the bacterium Pseudomonas syringae pv. tomato (Pst) strain carrying the effector AvrRpt2 [24]. These studies imply that SWP73 homologs negatively regulate plant resistance against (hemi)biotrophic pathogenic bacteria and fungi in the dicot Arabidopsis and monocot bread wheat. Interestingly, RT-qPCR demonstrated that the accumulation level of TaSWP73 gene transcripts was not significantly changed by B.g. tritici inoculation (Figure S2), suggesting that the suppression of wheat defense by TaSWP73 is maintained during B.g. tritici infection.
3.2. Isochorismate Synthase TaICS1 Positively Contributes to SA Biosynthesis and Wheat Post-Penetration Resistance Against B.g. tritici
Herein, three TaICS1 genes (TaICS1-5A, TaICS1-5B, and TaICS1-5D) were separately identified from wheat chromosomes 5A, 5B, and 5D. Sequence alignment and phylogenetic analysis demonstrated that the highly homologous TaICS1-5A, TaICS1-5B, and TaICS1-5D proteins are the closest homologs of Brachypodium BdICS1, maize ZmICS1, rice OsICS1, and Arabidopsis AtICS1. The B.g. tritici haustorium index (HI%) and microcolony index (MI%) were enhanced by the silencing of the TaICS1 gene, suggesting that wheat isochorismate synthase TaICS1 positively regulates post-penetration resistance against powdery mildew and suppresses B.g. tritici post-penetration events like haustorium development and microcolony formation. Notably, the knockdown of the TaICS1 gene attenuated the SA accumulation in wheat leaves under B.g. tritici infection. In contrast, SA accumulation in the absence of B.g. tritici infection was not significantly affected by the silencing of the TaICS1 gene, indicating that wheat isochorismate synthase TaICS1 specifically contributes to SA biosynthesis under B.g. tritici infection. Consistent with this study, Zhang et al. reported that the mutation of one or two TaICS1 homoeoalleles in wheat reduced the SA levels under ultraviolet treatment and Fusarium graminearum infection, further enhancing susceptibility to Fusarium head blight (FHB) [26]. These studies suggested that wheat TaICS1 mainly governs the endogenous SA levels under infection with pathogens like B.g. tritici and F. graminearum. Interestingly, isochorismate synthase OsICS1 was reported to be required for phylloquinone biosynthesis in rice, and it would be intriguing to examine the potential regulation of phylloquinone biosynthesis by TaICS1 in future research [27].
3.3. Wheat Chromatin Remodeling Protein TaSWP73 Positively Contributes to the Compatible Wheat–Powdery Mildew Interaction Presumably via Suppression of the TaSARD1-TaICS1-SA Pathway
In this study, the MNase assay demonstrated that the wheat chromatin remodeling protein TaSWP73 facilitates chromatin assembly in promoter regions of the TaSARD1 gene. The silencing of the TaSWP73 gene resulted in reduced nucleosomal occupancy in TaSARD1 promoters and potentiated TaSARD1 transcription. The knockdown of TaSARD1 and its downstream gene TaICS1 could attenuate SA biosynthesis and post-penetration resistance against B.g. tritici with a lack of TaSWP73, suggesting that the epigenetic suppression of TaSARD1 by chromatin remodeling protein TaSWP73 might contribute to the negative regulation of SA biosynthesis and post-penetration resistance against B.g. tritici by TaSWP73. Based on these results, we propose a model of how the chromatin remodeling protein TaSWP73 functions in the regulation of the wheat–B.g. tritici interaction. In this model, depicted in Figure 7, the wheat chromatin remodeling protein TaSWP73 facilitates chromatin assembly in promoter regions of the TaSARD1 gene and maintains the TaSARD1 gene transcription in the resting state. Other elusive chromatin remodeling proteins might be involved in the chromatin disassembly in the TaSARD1 promoter to keep the basal expression of the TaSARD1 gene. As a result, the expression level of the SA biosynthesis gene TaICS1 is low, and SA accumulation is maintained at a basal level, leading to a fully compatible wheat–B.g. tritici interaction. In the absence of the chromatin remodeling protein TaSWP73, chromatin in TaSARD1 promoters remains in an activated state marked by reduced nucleosomal occupancy, leading to the epigenetic activation of the TaSARD1 gene. As a result, the expression of the SA biosynthesis gene TaICS1 is up-regulated and SA accumulation is increased, leading to an attenuated compatible wheat–B.g. tritici interaction.

Figure 7.
Proposed model for regulation of the compatible wheat–powdery mildew interaction by the chromatin remodeling protein TaSWP73. In the wild-type wheat plants (A), the wheat chromatin remodeling protein TaSWP73 facilitates chromatin assembly in promoter regions of the TaSARD1 gene and maintains the TaSARD1 gene transcription in the resting state. Other elusive chromatin remodeling protein might be involved in chromatin disassembly in the TaSARD1 promoter to keep the basal expression of the TaSARD1 gene. As a result, the expression level of the SA biosynthesis gene TaICS1 is low, and SA accumulation is maintained at a basal level, leading to a fully compatible wheat–B.g. tritici interaction. In the absence of chromatin remodeling protein TaSWP73 (B), chromatin at TaSARD1 promoters remains in an activated state marked by reduced nucleosomal occupancy, leading to the epigenetic activation of the TaSARD1 gene. As a result, the expression of the SA biosynthesis gene TaICS1 is up-regulated and SA accumulation is increased, leading to an attenuated compatible wheat–B.g. tritici interaction.
Wheat histone deacetylase TaHDA6 and TaHDT701 were previously identified as epigenetic suppressors of post-penetration resistance against B.g. tritici [28,29]. As discussed by prior reviews, different types of epigenetic modulators like chromatin remodeling proteins and histone deacetylases could function in concert to regulate plant development and environmental adaptation [30]. Therefore, it is intriguing to examine the potential interplays between the chromatin remodeling protein TaSWP73 and histone deacetylases TaHDA6 and TaHDT701 in the epigenetic suppression of SA biosynthesis and post-penetration resistance against B.g. tritici. In addition, calmodulin-binding transcription activators (CAMTAs) TaCAMTA2 and TaCAMTA3 were previously identified as suppressors of wheat post-penetration resistance against powdery mildew [25]. Interestingly, the expressions of TaSARD1 and TaEDS1 were potentiated by the silencing of the TaCAMTA2 and TaCAMTA3 genes, suggesting that transcription factors TaCAMTA2 and TaCAMTA3 negatively regulate the expression of the TaSARD1 and TaEDS1 genes [25]. Analyzing the potential association of the chromatin remodeling protein TaSWP73 with transcription factors TaCAMTA2 and TaCAMTA3 might shed novel light on the molecular mechanism underlying TaSWP73 function in SA biosynthesis and the wheat–B.g. tritici interaction in future research.
Herein, the wheat TaSWP73 gene was identified as a susceptibility (S) gene contributing to the establishment of a compatible wheat–B.g. tritici interaction [31,32,33]. As summarized by previous reviews, a plethora of S genes such as TaMLO, TaEDR1, TaPOD70, and TaDND1/2 have been identified [34,35,36,37,38]. Inactivating the S genes TaMLO and TaEDR1 via newly developed genome editing and targeting induced local lesions in genomes (TILLING) techniques could reduce wheat compatibility with B.g. tritici and confer durable resistance [39,40,41,42,43,44,45,46]. Therefore, genetically manipulating the TaSWP73 gene by TILLING or genome editing approaches like transcription activator-like effector nucleases (TALENs) and CRISPR (clustered regularly interspaced short palindromic repeats)–Cas 9 (CRISPR-associated 9) might provide a new avenue for breeding new wheat varieties with improved powdery mildew resistance.
4. Materials and Methods
4.1. Plant and Fungal Materials
B.g. tritici-susceptible wheat cultivar Yannong 999 and virulent B.g. tritici isolate E09 were employed for wheat–B.g. tritici interaction characterization in this study. Wheat seedlings were grown in growth chambers under 16 h light/8 h dark with a light intensity of 150 μmol photons s−1m−2, a 20 °C/18 °C day/night cycle, and 70% relative humidity (RH). B.g. tritici isolate E09 was maintained on the plants of wheat cultivar Yannong 999 and kept at 70% RH and a 20 °C day/18 °C night cycle.
4.2. Gene Expression Analysis
Reverse transcription–quantitative polymerase chain reaction (RT-qPCR) and nuclear run-on assays were performed to analyze the transcript accumulation and transcription rates of the TaSWP73, TaSARD1, and TaICS1 genes. The newly grown wheat leaves (n = 5, randomly chosen) with virus symptoms about two weeks post BSMV infection were harvested for the RT-qPCR and nuclear run-on assays. For the RT-qPCR assay, the total RNA was extracted from the wheat leaves using TRizol solution and treated with RNase-free DNase I to remove potential DNA contamination. The first-strand cDNA was synthesized using 1 μg of the total RNA and used in RT-qPCR as a template to detect the expression of the indicated wheat gene. The RT-qPCR assay was performed using the ABI step-one real-time PCR system with the GoTaq qPCR Master Mix. The expression of TaEF1 was set as the internal control, and the expression levels of TaSWP73, TaSARD1, TaICS1, TaPR1, or TaPR2 were measured by qPCR using the qPCR Master Mix (Invitrogen, Waltham, MA, USA) under the following programs: 95 °C for 3 min, 40 cycles at 95 °C for 20 s, 56 °C for 30 s, and 72 °C for 15 s, followed by 72 °C for 1 min. For the nuclear run-on assay, wheat cell nuclei were isolated and mixed with reaction buffer (25 mM biotin-16-UTP and 0.75 mM of ATP, CTP, and GTP) for the transcription reaction. After RNA extraction, the nascent RNA was enriched by streptavidin magnetic beads and subjected to the RT-qPCR assay. In the RT-qPCR and nuclear run-on assays, TaEF1, TaSWP73, TaSARD1, TaICS1, TaPR1, and TaPR2 were analyzed using the primers 5′CAGGACGTTTACAAGATTG3′/5′CAAAACCACGCTTCAGATC3′, 5′CTTATAAGGCTGCTAACTC3′/5′GGGACGGTGGTGTCTTGAG3′, 5′GCGAGTAATGAAAGCAT3′/5′TTAATCAACTTGATCCC3′, 5′CCACAAGGAGCAGTGGGAG3′/5′TGTGGAACAACGAAGTAGA3′, 5′GAGAATGCAGACGCCCAAG3′/5′TGGAGCTTGCAGTCGTTGATC3′, and 5′AGGATGTTGCTTCCATGTTTG3′/5′AGTAGATGCGCATGCCGTTG3′. For the RT-qPCR and nuclear run-on assays, three technical replicates using replicate samples were statistically analyzed, and the data are presented as the mean ± SE (Student’s t-test; ** p < 0.01). All RT-qPCR and nuclear run-on assays were repeated in three biological replicates using independently prepared samples with similar results.
4.3. BSMV-Mediated Gene Silencing and B.g. tritici Microcolony Formation Analysis
Barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) was employed to silence the TaSWP73, TaSARD1, and TaICS1 genes in the Yannong 999 plants. For the BSMV-VIGS assay, about 200bp antisense (as) fragments of TaSWP73, TaSARD1, or TaICS1 were amplified using the primers 5′AAGGAAGTTTAACACCAGCACCAGGGCCATC3′/5′AACCACCACCACCGTGATCCATGAGCATCGTAGG3′, 5′AAGGAAGTTTAATGGTTCTAGTATCTATAAG3′/5′AACCACCACCACCGTGTTTGGAACCAGTTATTCG3′, and 5′AAGGAAGTTTAATCAATGTCCCCATGTTTCC3′/5′AACCACCACCACCGTCTGTTGGTTGGTTTGGTGG3′. PCR products were cloned into the pCa-γbLIC vector through the ligation-independent cloning technique to create the Agrobacterium-mediated BSMV-VIGS constructs BSMV-TaSWP73as, BSMV-TaSARD1as, and BSMV-TaICS1as. The BSMV-VIGS assay silencing the indicated genes was performed as previously described [47]. Briefly, the construct DNA of pCaBS-α, pCaBS-β, and pCa-γbLIC derivatives (BSMV-γ, BSMV-TaSWP73as, BSMV-TaSARD1as, or BSMV-TaICS1as) was separately transformed into the Agrobacterium tumefaciens strain GV3101. Agrobacteria grown in LB liquid media were harvested and resuspended in infiltration buffer (10 mM MgCl2, 100 µM acetosyringone, and 10 mM MES). Equal amounts of cell suspension harboring pCaBS-α, pCaBS-β, and pCa-γbLIC derivatives were mixed for the agroinfiltration of Nicotiana benthamiana leaves. After maintenance in a growth chamber for 12 days post infiltration, the infiltrated N. benthamiana leaves were ground, and the sap was inoculated onto the two-leaf stages of wheat plants. The newly grown wheat leaves (n = 5) with virus symptoms about two weeks post BSMV infection were randomly collected for further experiments like gene expression analysis, wheat–B.g. tritici interaction characterization, and nucleosomal occupancy analysis. For the B.g. tritici microcolony formation analysis, the newly grown upper leaves with virus symptoms were collected and subjected to inoculation with B.g. tritici strain E09 conidia. About 72 h post B.g. tritici inoculation, leaf samples were fixed in an ethanol–acetic acid solution (1:1, v/v) and kept in a destaining solution (lactic acid–glycerol–water, 1:1:1, v/v/v). Thereafter, B.g. tritici-infected leaves were stained with 0.1% (w/v) Coomassie brilliant blue R250 to visualize the fungal epiphytic structure under a microscope. About 2000 B.g. tritici–wheat interaction sites (randomly chosen) were analyzed in one experiment. For the B.g. tritici microcolony index (MI%) analysis, three technical replicates using replicate samples were statistically analyzed, and the data are presented as the mean ± SE (Student’s t-test; ** p < 0.01). All the B.g. tritici microcolony index (MI%) analyses were repeated in three biological replicates using independently prepared samples with similar results.
4.4. Single-Cell Transient Gene Overexpression Assay and B.g. tritici Haustorium Formation Analysis
For the single-cell transient gene overexpression assay, the coding regions of TaSWP73-2A, TaSWP73-2B, and TaSWP73-2D were amplified using the primers 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTTC ATGGCCACCGGTGGCAACC3′/5′GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAGAACCACCAGCACCA3′, 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCCACCGGCGGCAACC3′/5′GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAGAACCACCAGCACCA3′, and 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCCACCGGCGGCAACC3′/5′GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAGAACCACCAGCACCA3′. PCR products were cloned into pIPKb001, an expression vector driven by the maize ubiquitin promoter, to create the pIPKb001-TaSWP73-2A (for OE-TaSWP73-2A), pIPKb001-TaSWP73-2B (for OE-TaSWP73-2B), and pIPKb001-TaSWP73-2D (for OE-TaSWP73-2D) constructs using GATEWAY cloning technology (Invitrogen). The single-cell transient gene overexpression assay was performed as previously described [38]. The β-glucuronidase (GUS) reporter gene was co-delivered into the wheat epidermal cell to mark the transformed cells and better visualize the fungal haustorium in these cells. The pIPKb001 overexpression constructs were mixed with the GUS reporter vector at a 1:1 molar ratio before coating the DNA microcarrier. The exogenous DNA on the microcarrier was delivered into the wheat epidermal cell through a particle inflow gun (Bio-Rad, Hercules, CA, USA). The inoculation of B.g. tritici conidia spores was performed at least 16 h post bombardment. The leaf segments were stained for GUS activity 48 h post B.g. tritici spore inoculation and kept in a destaining solution. Before mounting for microscopy, the B.g. tritici-infected wheat leaves were stained with Coomassie blue to visualize the fungal epiphytic structure. About 50 B.g. tritici-infected wheat epidermal cells (randomly chosen) were analyzed in one experiment. For the B.g. tritici haustorium index (HI%) analysis, three technical replicates using replicate samples were statistically analyzed, and the data are presented as the mean ± SE (Student’s t-test; ** p < 0.01). All the B.g. tritici haustorium index (HI%) analyses were repeated in three biological replicates using independently prepared samples with similar results.
4.5. SA Measurement
Free SA was analyzed using High-Performance Liquid Chromatography (HPLC), as previously described [48]. Briefly, the newly grown wheat leaves (n = 5) with virus symptoms about two weeks post BSMV infection were randomly collected and ground with liquid nitrogen into powder and then homogenized in 70% ethanol (v/v) containing the internal standard ortho-anisic acid. After centrifugation, the supernatant was collected, and the pellet was homogenized with 90% v/v methanol. After centrifugation, both supernatants were pooled and evaporated under vacuum. Then, 5% trichloroacetic acid was added to the remaining aqueous solution. After centrifugation, the supernatant was collected and mixed with ethyl acetate/cyclohexane. After centrifugation, the upper organic phase was collected. For SA quantification, organic phases were resuspended in HPLC starting solvent (methanol 40%, water 60%, acetic acid 1%) and analyzed by a reverse-phase HPLC column. The free SA amount was calculated in ng mg−1 fresh weight (FW) with reference to the amount of internal standard. For the free SA measurement, three technical replicates using replicate samples were statistically analyzed, and the data are presented as the mean ± SE (Student’s t-test; ** p < 0.01). All the free SA measurements were repeated in three biological replicates using independently prepared samples with similar results.
4.6. Nucleosomal Occupancy Analysis
The nucleosome occupancy micrococcal nuclease (MNase) assay was conducted to analyze the chromatin assembly structure in the TaSARD1 promoter regions as previously described [29]. Briefly, the newly grown wheat leaves (n = 5) with virus symptoms about two weeks post BSMV infection were randomly collected and cross-linked and then subjected to nuclear isolation and MNase digestion. Genomic DNA was then recovered and underwent qPCR analysis to analyze the TaSARD1 promoter regions using the primers 5′CTGTGACTTCATGCTCAAG3′/5′CCAAATCATCTAACTTTCC3′, 5′ATGTACACTGAAATTAATC3′/5′GATGCAGGTAGAAAGCAGG3′, 5′TGAATTGTCAAATGTCTCT3′/5′GTTGGTAGCGTCTCTTATC3′, 5′TGGTGCGTGCACTGAAATC3′/5′AGCTGCAGGCAGCTAGGGA3′, 5′ACGGGCTGCCCTGACACTC3′/5′GAGCTCCTGAAGCAGCTGG3′, 5′CACCCGACATCAAAACAAC3′/5′GCCGTTAGTTTAGGACAGG3′, 5′GCTTTGCAAAGCAACTTGG3′/5′CTGGCGTAATGATAAGAAG3′, 5′ACAAATAACCATCGACCCA3′/5′ATTAGTTGTTTATTTAATT3′, 5′TACAAAGCGATGAATGCCA3′/5′TACTCTGTTGCTATGTTAG3′, and 5′AACCATCGACCACCTATTG3′/5′CAAGGCTTCGAGCTCCCAA3′. Nuclei without MNase digestion treatment were employed as the input control. For the MNase assay, three technical replicates using replicate samples were statistically analyzed, and the data are presented as the mean ± SE (Student’s t-test; ** p < 0.01). All MNase assays were repeated in three biological replicates using independently prepared samples with similar results.
4.7. Phylogenetic Tree Reconstruction
SWP73 and ICS1 homologs from Arabidopsis, Brachypodium, maize, rice, and wheat were subjected to protein alignment with Clustal W, and phylogenetic trees were reconstructed using the Neighbor-Joining method with 1000 bootstraps by MEGA 7 (Molecular Evolutionary Genetics Analysis) software.
4.8. Statistical Analysis
For the statistical analysis of gene expression, SA measurement, nucleosomal occupancy in gene promoter regions, B.g. tritici microcolony formation, and B.g. tritici haustorium formation, at least three independent experiments were performed for each assay, and at least 5 wheat leaves (for the RT-qPCR, SA measurement, nuclear run-on, and MNase assay), 50 B.g. tritici-infected wheat epidermal cells (for the B.g. tritici haustorium index (HI%) analysis), and 2000 B.g. tritici–wheat interaction sites (for the B.g. tritici microcolony index (MI%) analysis) were analyzed in one experiment or were randomly chosen for each group. Three technical replicates per assay were analyzed using Student’s t-test, and the value represents the mean ± standard deviation (n. s. p > 0.05, * 0.01 < p < 0.05, ** p < 0.01; n. s. represents no significant difference). These assays were repeated in three independent biological replicates using independently prepared samples with similar results.
5. Conclusions
Herein, we characterized the function of the wheat chromatin remodeling protein TaSWP73 in regulating the wheat–B.g. tritici interaction and demonstrated that TaSWP73 negatively regulates wheat post-penetration resistance against B.g. tritici. The overexpression of TaSWP73 attenuates wheat post-penetration resistance against B.g. tritici, while the silencing of TaSWP73 potentiates SA biosynthesis and activates post-penetration resistance against B.g. tritici. Furthermore, we found that chromatin in the promoter regions of TaSARD1, an activator gene of SA biosynthesis, is marked by high nucleosome occupancy in the TaSWP73-silenced wheat leaves. The silencing of TaSARD1 could suppress SA biosynthesis and attenuate post-penetration resistance against B.g. tritici with a lack of TaSWP73. In addition, we identified TaICS1 as an essential component of wheat SA biosynthetic machinery and found that potentiated SA biosynthesis and increased post-penetration resistance against B.g. tritici with a lack of TaSWP73 could be suppressed by the silencing of TaICS1 expression. These results collectively suggest that the wheat chromatin remodeling protein TaSWP73 negatively regulates post-penetration resistance against B.g. tritici probably via the suppression of the TaSARD1-TaICS1-SA pathway. These findings shed novel light on the epigenetic mechanism underlying wheat–B.g. tritici interactions, and genetically manipulating the TaSWP73, TaSARD1, and TaICS1 genes characterized in this study might provide a promising new avenue to improve wheat post-penetration resistance against powdery mildew disease in future research.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26062590/s1.
Author Contributions
Y.F., Z.Y., J.L., X.W., H.L., P.Z. and C.C. planned and designed this research; Y.F., Z.Y., J.L., X.W., H.L. and P.Z. performed experiments; Y.F., Z.Y. and C.C. analyzed the data and wrote this manuscript with contributions from J.L., X.W., H.L. and P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Shandong Province (ZR2022MC008, ZR2017BC109), the Qingdao Science and Technology Bureau Fund (17-1-1-50-jch), and the Qingdao University Fund (DC1900005385).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data presented here are available on request through correspondence.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Levy, A.A.; Feldman, M. Evolution and origin of bread wheat. Plant Cell 2022, 34, 2549–2567. [Google Scholar] [CrossRef] [PubMed]
- Lee, R. The outlook for population growth. Science 2011, 333, 569–573. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Qian, J.; Loos, A.; Kümmel, F.; Spanu, P.D.; Panstruga, R. Long-term and rapid evolution in powdery mildew fungi. Mol. Ecol. 2023, 33, e16909. [Google Scholar] [CrossRef]
- Mapuranga, J.; Chang, J.; Yang, W. Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat. Front. Plant Sci. 2022, 13, 1102908. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- van der Burgh, A.M.; Joosten, M.H.A.J. Plant immunity: Thinking outside and inside the box. Trends Plant Sci. 2019, 24, 587–601. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Gust, A.A.; Nürnberger, T. Plant immunity unified. Nat. Plants 2021, 7, 382–383. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J.M. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, M.; Pimprikar, P.; Nürnberger, T.; Zipfel, C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 2021, 7, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, 6316. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–503. [Google Scholar] [CrossRef]
- Adachi, H.; Tsuda, K. Convergence of cell-surface and intracellular immune receptor signalling. New Phytol. 2019, 221, 1676–1678. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [PubMed]
- Zhi, P.; Chang, C. Exploiting epigenetic variations for crop disease resistance improvement. Front. Plant Sci. 2021, 12, 692328. [Google Scholar] [CrossRef]
- Wu, Z.; He, L.; Jin, Y.; Chen, J.; Shi, H.; Wang, Y.; Yang, W. HISTONE DEACETYLASE 6 suppresses salicylic acid biosynthesis to repress autoimmunity. Plant Physiol. 2021, 187, 2592–2607. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Rangel, D.S.; Qin, X.; Bui, C.; Li, R.; Jia, Z.; Cui, X.; Jin, H. The chromatin-remodeling protein BAF60/SWP73A regulates the plant immune receptor NLRs. Cell Host Microbe 2021, 29, 425–434. [Google Scholar] [CrossRef]
- Li, M.; Yang, Z.; Liu, J.; Chang, C. Wheat susceptibility genes TaCAMTA2 and TaCAMTA3 negatively regulate post-penetration resistance against Blumeria graminis forma specialis tritici. Int. J. Mol. Sci. 2023, 24, 10224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Man, J.; Xu, D.; Wen, L.; Li, Y.H.; Deng, M.; Jiang, Q.T.; Xu, Q.; Chen, G.Y.; Wei, Y.M. Investigating the mechanisms of isochorismate synthase: An approach to improve salicylic acid synthesis and increase resistance to Fusarium head blight in wheat. Crop. J. 2024, 12, 1054–1063. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, G.; Zhang, D.; Li, G.; Qiu, J.L.; Wu, J. Isochorismate synthase is required for phylloquinone, but not salicylic acid biosynthesis in rice. aBIOTECH 2024, 5, 488–496. [Google Scholar] [CrossRef]
- Liu, J.; Zhi, P.; Wang, X.; Fan, Q.; Chang, C. Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f.sp. tritici. J. Exp. Bot. 2019, 70, 255–268. [Google Scholar] [CrossRef]
- Zhi, P.; Kong, L.; Liu, J.; Zhang, X.; Wang, X.; Li, H.; Sun, M.; Li, Y.; Chang, C. Histone deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f.sp. tritici. Int. J. Mol. Sci. 2020, 21, 2640. [Google Scholar] [CrossRef]
- Liu, J.; Chang, C. Concerto on chromatin: Interplays of different epigenetic mechanisms in plant development and environmental adaptation. Plants 2021, 10, 2766. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef] [PubMed]
- van Schie, C.C.; Takken, F.L. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, Z.; Chang, C. Susceptibility is new resistance: Wheat susceptibility genes and exploitation in resistance breeding. Agriculture 2022, 12, 1419. [Google Scholar] [CrossRef]
- Koseoglou, E.; van der Wolf, J.M.; Visser, R.; Bai, Y. Susceptibility reversed: Modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 2022, 27, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, E.; Giczey, G.; Burgyán, J. Virus-induced gene silencing of MLO genes induces powdery mildew resistance in Triticum aestivum. Arch. Virol. 2012, 157, 1345–1350. [Google Scholar] [CrossRef]
- Li, R.; Zhang, X.; Zhao, B.; Song, P.; Zhang, X.; Wang, B.; Li, Q. Wheat Class III Peroxidase TaPOD70 is a potential susceptibility factor negatively regulating wheat resistance to Blumeria graminis f. sp. tritici. Phytopathology 2023, 113, 873–883. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Zhi, P.; Gao, R.; Chen, W.; Chang, C. Wheat transcriptional corepressor TaTPR1 suppresses susceptibility genes TaDND1/2 and potentiates post-penetration resistance against Blumeria gramin is forma specialis tritici. Int. J. Mol. Sci. 2024, 25, 1695. [Google Scholar] [CrossRef]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000, 123, 439–442. [Google Scholar] [CrossRef]
- Kurowska, M.; Daszkowska-Golec, A.; Gruszka, D.; Marzec, M.; Szurman, M.; Szarejko, I.; Maluszynski, M. TILLING: A shortcut in functional genomics. J. Appl. Genet. 2011, 52, 371–390. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Schenke, D.; Cai, D. Applications of CRISPR/Cas to improve crop disease resistance: Beyond inactivation of susceptibility factors. iScience 2020, 23, 101478. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Yuan, C.; Li, C.; Yan, L.; Jackson, A.O.; Liu, Z.; Han, C.; Yu, J.; Li, D. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 2011, 6, e26468. [Google Scholar] [CrossRef]
- Fragnière, C.; Serrano, M.; Abou-Mansour, E.; Métraux, J.P.; L’Haridon, F. Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 2011, 585, 1847–1852. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).