Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max)
Abstract
1. Introduction
2. Results
2.1. Sequence Identification and Retrieval
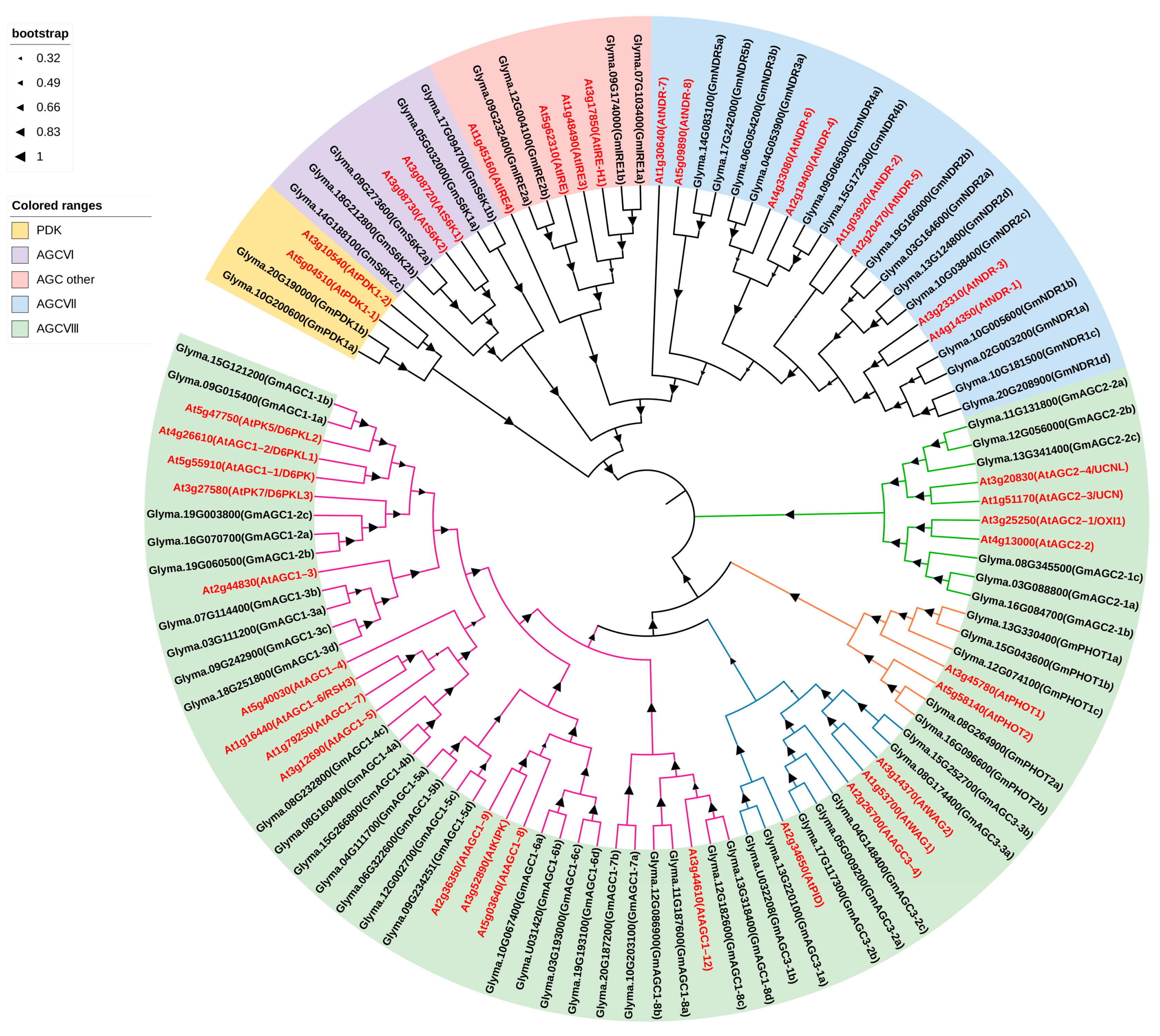
2.2. Sequence Alignment and Phylogeny
2.3. Physicochemical Properties of the GmAGC Kinase Family
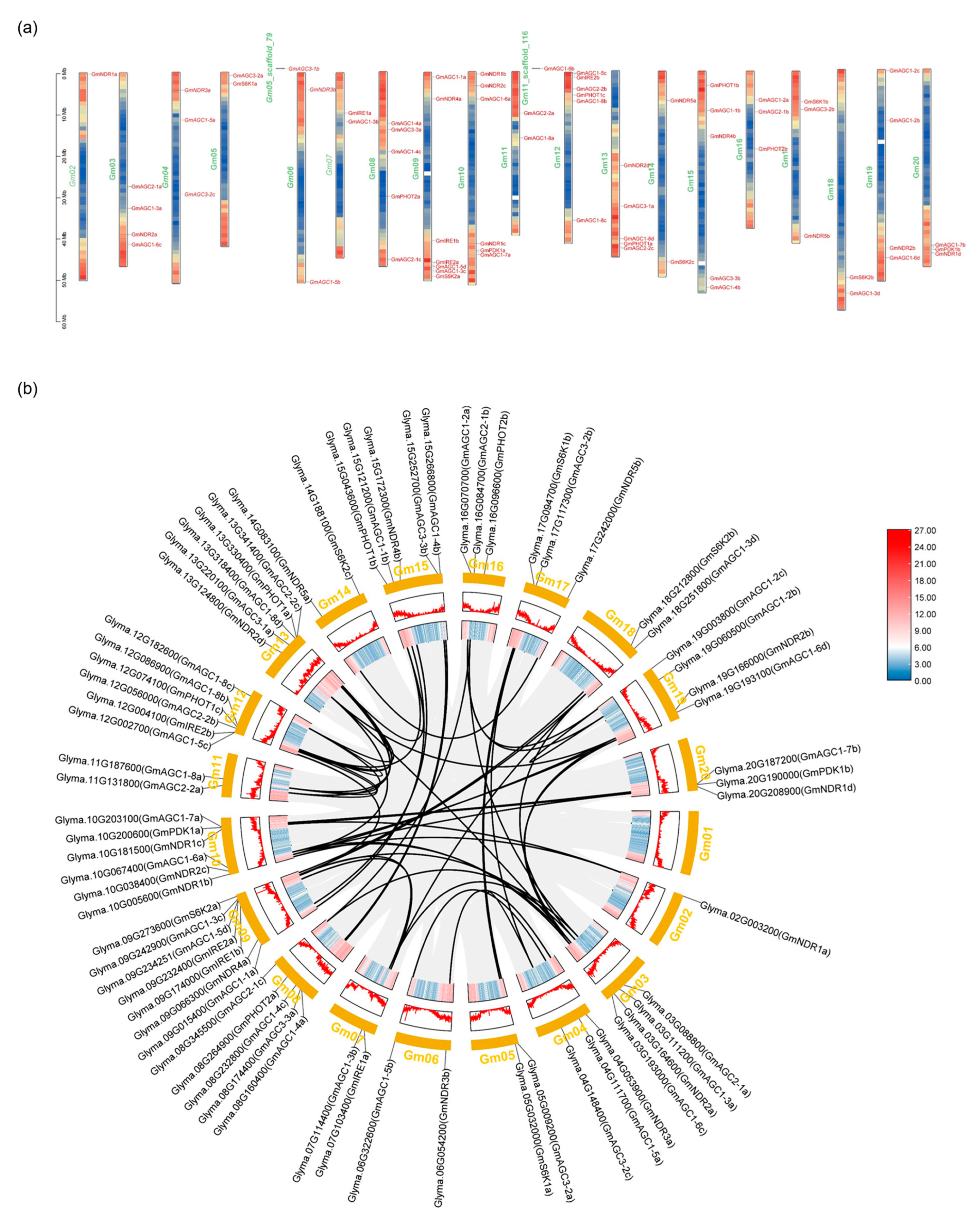
2.4. GmAGC Localization on Chromosomes, Collinearity Analysis, Gene Duplication, and Ka/Ks Ratio
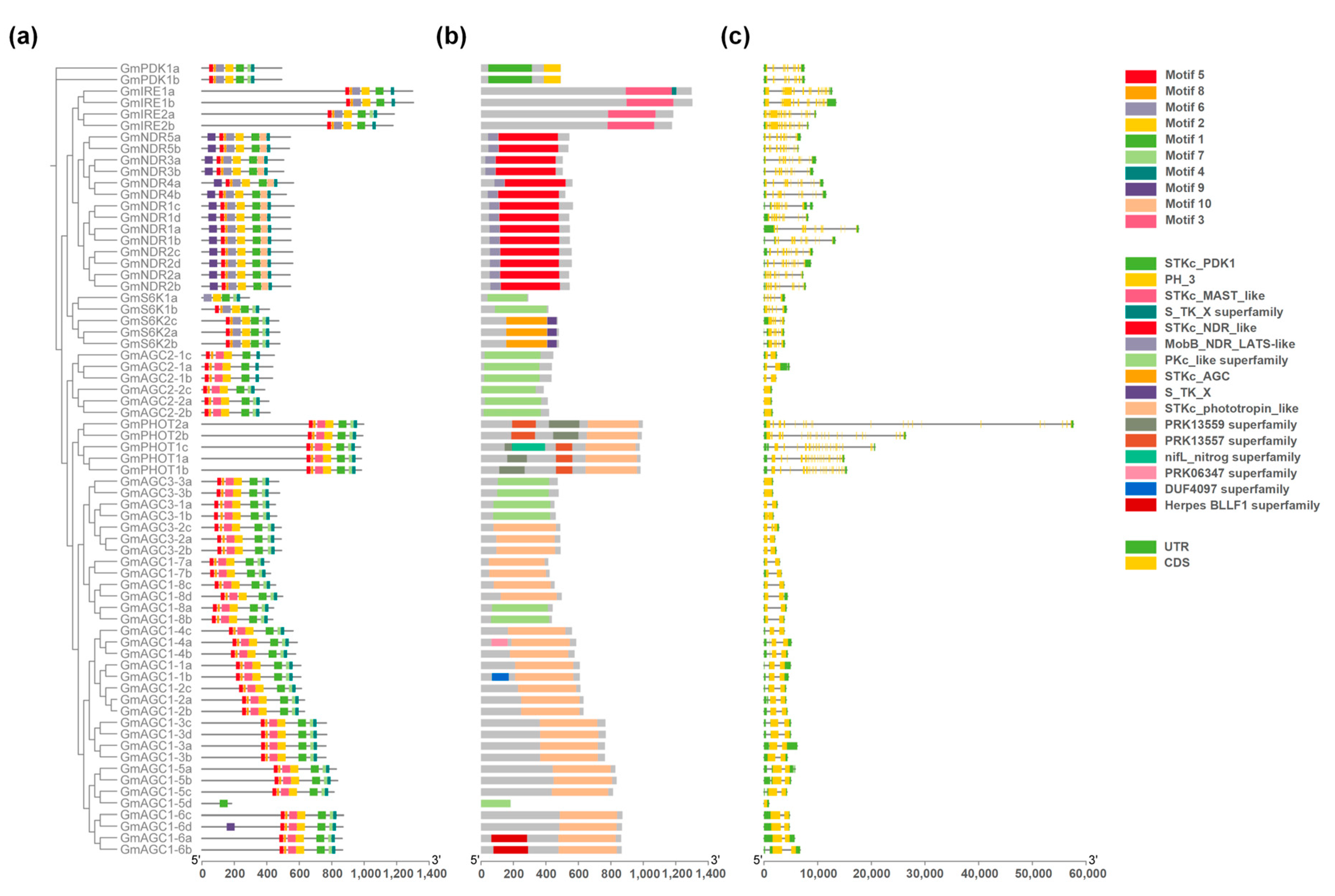
2.5. Motif Finding and Gene Structure Analysis of GmAGC Genes
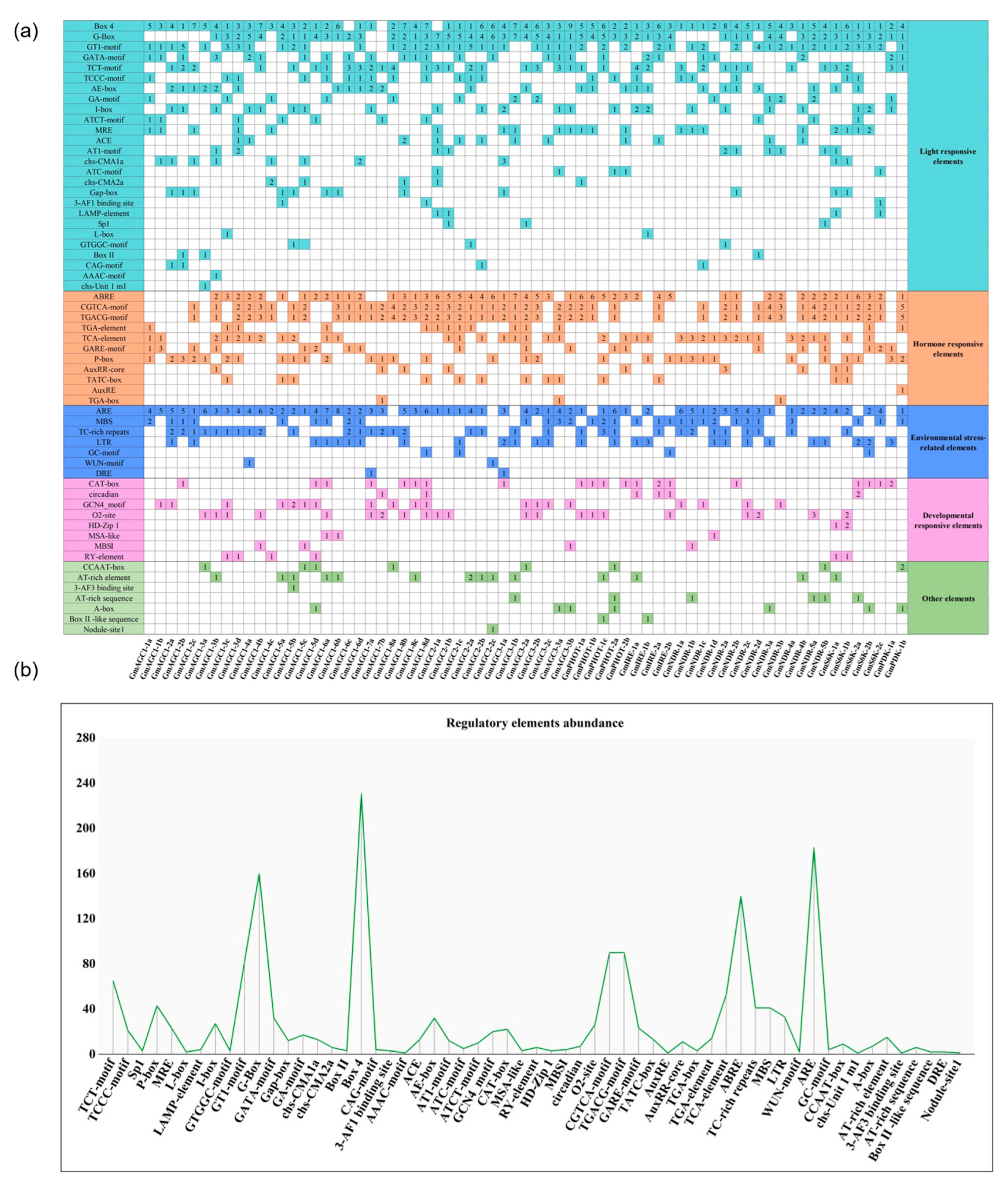
2.6. Analysis of Cis-Elements in the Promoters of GmAGC Genes
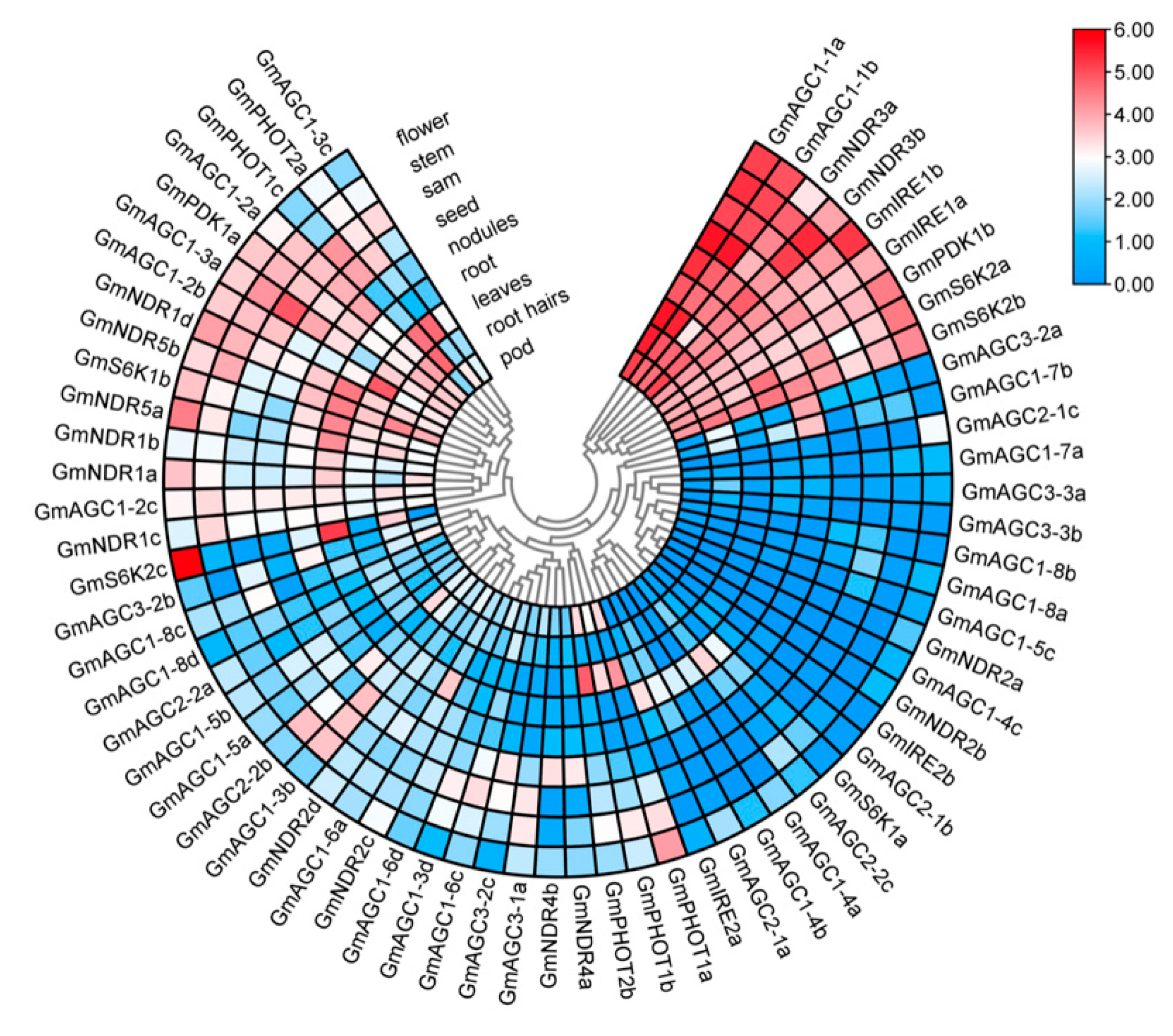
2.7. Expression Patterns of GmAGC Genes in Different Tissues
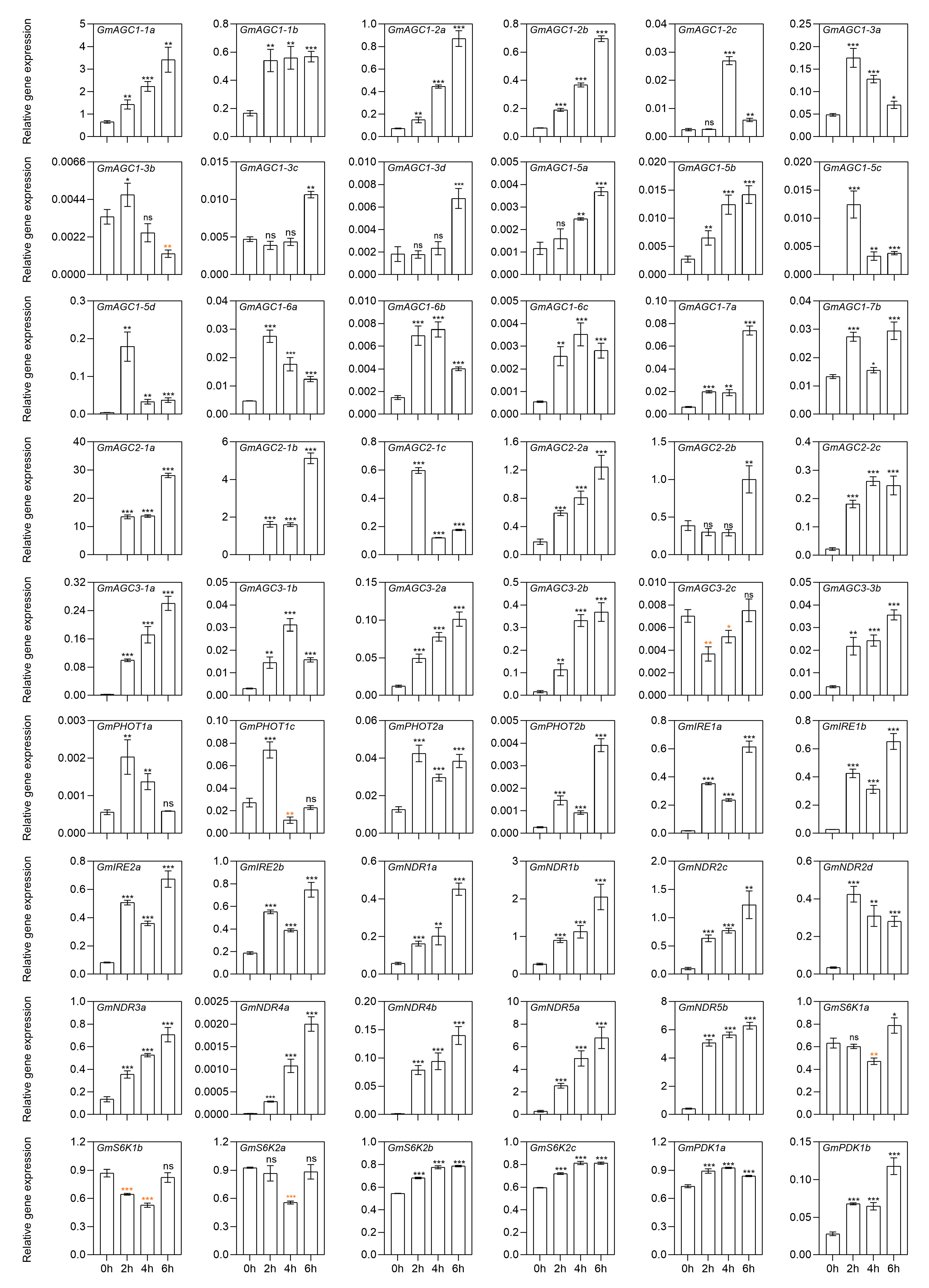
2.8. Gene Expression of GmAGC Members Under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval of AGC Protein Kinase Family Members in G. max
4.2. Phylogenetic Tree Construction, Gene Structure, and Conserved Motif Analysis of the GmAGC Gene Family
4.3. Collinearity Analysis of AGC Genes
4.4. Cis-Acting Elements Analysis of GmAGC Genes
4.5. Gene Expression Analysis of GmAGC Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, S.; Wei, M.; Wei, Y.; Jiang, Z. Insights into Plant Sensory Mechanisms under Abiotic Stresses. Plants 2024, 13, 1907. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein Kinases in Plant Responses to Drought, Salt, and Cold Stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Offringa, R. Evolutionary Adaptations of Plant AGC Kinases: From Light Signaling to Cell Polarity Regulation. Front. Plant Sci. 2012, 3, 250. [Google Scholar] [CrossRef]
- Liu, J.; Li, W.; Wu, G.; Ali, K. An Update on Evolutionary, Structural, and Functional Studies of Receptor-like Kinases in Plants. Front. Plant Sci. 2024, 15, 1305599. [Google Scholar] [CrossRef]
- Rehman, A.; Alwutayd, K.M.; Alshehri, D.; Alsudays, I.M.; Azeem, F.; Rahman, S.; Abid, M.; Shah, A.A. Regulatory Role of AGC Genes in Heat Stress Adaptation in Maize (Zea mays). Funct. Plant Biol. 2024, 51, FP23282. [Google Scholar] [CrossRef]
- Bögre, L. Growth Signalling Pathways in Arabidopsis and the AGC Protein Kinases. Trends Plant Sci. 2003, 8, 424–431. [Google Scholar] [CrossRef]
- Ek-Ramos, M.J.; Avila, J.; Cheng, C.; Martin, G.B.; Devarenne, T.P. The T-Loop Extension of the Tomato Protein Kinase AvrPto-Dependent Pto-Interacting Protein 3 (Adi3) Directs Nuclear Localization for Suppression of Plant Cell Death. J. Biol. Chem. 2010, 285, 17584–17594. [Google Scholar] [CrossRef]
- Haga, K.; Hayashi, K.; Sakai, T. PINOID AGC Kinases Are Necessary for Phytochrome-Mediated Enhancement of Hypocotyl Phototropism in Arabidopsis. Plant Physiol. 2014, 166, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Sakai, T. Photosensory Adaptation Mechanisms in Hypocotyl Phototropism: How Plants Recognize the Direction of a Light Source. J. Exp. Bot. 2023, 74, 1758–1769. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Katagiri, S.; Li, Y.; Yamashita, K.; Takase, H.; Umezawa, T. Hyperosmolarity-Induced Suppression of Group B1 Raf-like Protein Kinases Modulates Drought-Growth Trade-off in Arabidopsis. Proc. Natl. Acad. Sci. USA 2024, 121, e2419204121. [Google Scholar] [CrossRef]
- Wang, P.; Hsu, C.-C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.S.; Macho, A.P.; Tao, W.A.; et al. Mapping Proteome-Wide Targets of Protein Kinases in Plant Stress Responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling Salt Stress Signaling in Plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Garcia, A.V.; Al-Yousif, M.; Hirt, H. Role of AGC Kinases in Plant Growth and Stress Responses. Cell. Mol. Life Sci. 2012, 69, 3259–3267. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A.; et al. IRE1/bZIP60-Mediated Unfolded Protein Response Plays Distinct Roles in Plant Immunity and Abiotic Stress Responses. PLoS ONE 2012, 7, e31944. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, X.; Zhou, M.; Yang, J.; Ke, S.; Li, Y. Genome-Wide Identification of the AGC Protein Kinase Gene Family Related to Photosynthesis in Rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 12557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, K.; Wei, X.; Zhang, Q.; Rong, W.; Du, L.; Ye, X.; Qi, L.; Zhang, Z. The Wheat AGC Kinase TaAGC1 Is a Positive Contributor to Host Resistance to the Necrotrophic Pathogen Rhizoctonia cerealis. J. Exp. Bot. 2015, 66, 6591–6603. [Google Scholar] [CrossRef]
- Ahmad, F.; Abdullah, M.; Khan, Z.; Stępień, P.; Rehman, S.U.; Akram, U.; Rahman, M.H.U.; Ali, Z.; Ahmad, D.; Gulzar, R.M.A.; et al. Genome-Wide Analysis and Prediction of Chloroplast and Mitochondrial RNA Editing Sites of AGC Gene Family in Cotton (Gossypium hirsutum L.) for Abiotic Stress Tolerance. BMC Plant Biol. 2024, 24, 888. [Google Scholar] [CrossRef]
- Matsui, H.; Miyao, A.; Takahashi, A.; Hirochika, H. Pdk1 Kinase Regulates Basal Disease Resistance Through the OsOxi1–OsPti1a Phosphorylation Cascade in Rice. Plant Cell Physiol. 2010, 51, 2082–2091. [Google Scholar] [CrossRef]
- Wu, X.; Pan, L.; Guo, X.; Li, T.; Li, J.; Duan, Q.; Huang, J. Genome-Wide Identification and Expression Analysis of BrAGC Genes in Brassica Rapa Reveal Their Potential Roles in Sexual Reproduction and Abiotic Stress Tolerance. Front. Genet. 2022, 13, 1044853. [Google Scholar] [CrossRef]
- Li, X.; Yu, Q.; Hua, X.; He, J.; Liu, J.; Peng, L.; Wang, J.; Li, X.; Yang, Y. Phosphorylation of ADF7-Mediated by AGC1.7 Regulates Pollen Germination in Arabidopsis thaliana. Plant Cell Environ. 2025, 48, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Arenas, L.; Bayascas, J.R. Capturing Conformational Transitions of Full-Length PDK1 That Dictate Kinase Substrate Selectivity. Sci. Signal. 2023, 16, eadh5114. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.; Pleßmann, J.; Enugutti, B.; Hüttl, R.; Wassmer, K.; Schneitz, K. The AGC Protein Kinase UNICORN Controls Planar Growth by Attenuating PDK1 in Arabidopsis thaliana. PLOS Genet. 2019, 15, e1007927. [Google Scholar] [CrossRef]
- Tan, S.; Zhang, X.; Kong, W.; Yang, X.-L.; Molnár, G.; Vondráková, Z.; Filepová, R.; Petrášek, J.; Friml, J.; Xue, H.-W. The Lipid Code-Dependent Phosphoswitch PDK1–D6PK Activates PIN-Mediated Auxin Efflux in Arabidopsis. Nat. Plants 2020, 6, 556–569. [Google Scholar] [CrossRef]
- Xiao, Y.; Offringa, R. PDK1 Regulates Auxin Transport and Arabidopsis Vascular Development through AGC1 Kinase PAX. Nat. Plants 2020, 6, 544–555. [Google Scholar] [CrossRef]
- Ren, Q.W.; Liu, T.Y.; Lan, H.J.; Li, Z.C.; Huang, M.J.; Zhao, Y.T.; Chen, Y.; Liao, L.N.; Ma, X.H.; Liu, J.Z. Partially Knocking out NtPDK1a/1b/1c/1d Simultaneously in Nicotiana tabacum Using CRISPR/CAS9 Technology Results in Auxin-Related Developmental Defects. Plant Sci. 2024, 343, 112057. [Google Scholar] [CrossRef]
- Obomighie, I.; Lapenas, K.; Murphy, B.E.; Bowles, A.M.C.; Bechtold, U.; Prischi, F. The Role of Ribosomal Protein S6 Kinases in Plant Homeostasis. Front. Mol. Biosci. 2021, 8, 636560. [Google Scholar] [CrossRef]
- Yoon, H.S.; Fujino, K.; Liu, S.; Takano, T.; Tsugama, D. NDR/LATS-family Protein Kinase Genes Are Indispensable for Embryogenesis in Arabidopsis. FEBS Open Bio 2021, 11, 2600–2606. [Google Scholar] [CrossRef]
- Zhou, P.; Liang, Y.; Mei, J.; Liao, H.; Wang, P.; Hu, K.; Chen, L.; Zhang, X.; Ye, D. The Arabidopsis AGC Kinases NDR2/4/5 Interact with MOB1A/1B and Play Important Roles in Pollen Development and Germination. Plant J. 2021, 105, 1035–1052. [Google Scholar] [CrossRef]
- Yue, X.; Guo, Z.; Shi, T.; Song, L.; Cheng, Y. Arabidopsis AGC Protein Kinases IREH1 and IRE3 Control Root Skewing. J. Genet. Genom. 2019, 46, 259–267. [Google Scholar] [CrossRef]
- Pislariu, C.I.; Dickstein, R. The AGC Kinase MtIRE: A Link to Phospholipid Signaling During Nodulation? Plant Signal. Behav. 2007, 2, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, P.; Tong, C.; Li, X.; Yang, T.; Jiang, Y.; Zhao, B.; Dong, J.; Jiang, B.; Shen, J.; et al. CsIREH1 Phosphorylation Regulates DELLA Protein Affecting Plant Height in Cucumber (Cucumis sativus). New Phytol. 2024, 245, 1528–1546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; McCormick, S. AGCVIII Kinases: At the Crossroads of Cellular Signaling. Trends Plant Sci. 2009, 14, 689–695. [Google Scholar] [CrossRef]
- Galván-Ampudia, C.S.; Offringa, R. Plant Evolution: AGC Kinases Tell the Auxin Tale. Trends Plant Sci. 2007, 12, 541–547. [Google Scholar] [CrossRef]
- Zhang, Y. AGC Protein Kinase AGC1-4 Mediates Seed Size in Arabidopsis. Plant Cell Rep. 2020, 39, 825–837. [Google Scholar] [CrossRef]
- Han, J. AGC Kinases OXI1 and AGC2-2 Regulate Camalexin Secretion and Disease Resistance by Phosphorylating Transporter PDR6. Plant Physiol. 2024, 195, 1835–1850. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, X.; Li, M. PINOID-centered Genetic Interactions Mediate Auxin Action in Cotyledon Formation. Plant Direct 2024, 8, e587. [Google Scholar] [CrossRef]
- Wan, Y.L.; Eisinger, W.; Ehrhardt, D.; Kubitscheck, U.; Baluska, F.; Briggs, W. The Subcellular Localization and Blue-Light-Induced Movement of Phototropin 1-GFP in Etiolated Seedlings of Arabidopsis thaliana. Mol. Plant 2008, 1, 103–117. [Google Scholar] [CrossRef]
- Amanullah; Khan, J.A.; Yasir, M. Improving soybean yield and oil productivity: An integrated nutrient management approach for sustainable soybean production. BMC Plant Biol. 2025, 25, 293. [Google Scholar] [CrossRef]
- Graf, A.; Bassukas, A.E.L.; Xiao, Y.; Barbosa, I.C.R.; Mergner, J.; Grill, P.; Michalke, B.; Kuster, B.; Schwechheimer, C. D6PK plasma membrane polarity requires a repeated CXX(X)P motif and PDK1-dependent phosphorylation. Nat. Plants 2024, 10, 300–314. [Google Scholar] [CrossRef]
- Severin, A.J.; Cannon, S.B.; Graham, M.M.; Grant, D.; Shoemaker, R.C. Changes in Twelve Homoeologous Genomic Regions in Soybean Following Three Rounds of Polyploidy. Plant Cell 2011, 23, 3129–3136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, S.; Chang, X.; Wang, X.; Zhao, Y.; Xia, Y.; Trigiano, R.N.; Jiao, Y.; Chen, F. The Ancient Wave of Polyploidization Events in Flowering Plants and Their Facilitated Adaptation to Environmental Stress. Plant Cell Environ. 2020, 43, 2847–2856. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) Expansin Gene Superfamily Origins: Segmental and Tandem Duplication Events Followed by Divergent Selection among Subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Omer, S.; Harlow, T.J.; Gogarten, J.P. Does Sequence Conservation Provide Evidence for Biological Function? Trends Microbiol. 2017, 25, 11–18. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Tu, Z.; Shen, Y.; Wen, S.; Zong, Y.; Li, H. Alternative Splicing Enhances the Transcriptome Complexity of Liriodendron chinense. Front. Plant Sci. 2020, 11, 578100. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.K.; Sonah, H.; Singh, N.K. Intron Gain, a Dominant Evolutionary Process Supporting High Levels of Gene Expression in Rice. J. Plant Biochem. Biotechnol. 2016, 25, 142–146. [Google Scholar] [CrossRef]
- Pandey, S. Heterotrimeric G-Protein Signaling in Plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2019, 70, 213–238. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of Cis-Acting Regulatory Elements in Osmotic- and Cold-Stress-Responsive Promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-Induced Expression of the SCaM-4 Promoter Is Mediated in Part by a GT-1 Box That Interacts with a GT-1-Like Transcription Factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef]
- Xu, H.; Shi, X.; He, L.; Guo, Y.; Zang, D.; Li, H.; Zhang, W.; Wang, Y. Arabidopsis thaliana Trihelix Transcription Factor AST1 Mediates Salt and Osmotic Stress Tolerance by Binding to a Novel AGAG-Box and Some GT Motifs. Plant Cell Physiol. 2018, 59, 946–965. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis Myb Homolog Is Induced by Dehydration Stress and Its Gene Product Binds to the Conserved MYB Recognition Sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between Two Cis-acting Elements, ABRE and DRE, in ABA-dependent Expression of Arabidopsis rd29A Gene in Response to Dehydration and High-salinity Stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Qin, L.; Liu, W.; Wang, Y. ThERF1 Regulates Its Target Genes via Binding to a Novel Cis-acting Element in Response to Salt Stress. J. Integr. Plant Biol. 2015, 57, 838–847. [Google Scholar] [CrossRef]
- Zhu, C.; Schraut, D.; Hartung, W.; Schäffner, A.R. Differential Responses of Maize MIP Genes to Salt Stress and ABA. J. Exp. Bot. 2005, 56, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Du, J.; Tian, Z.; Sui, Y.; Zhao, M.; Song, Q.; Cannon, S.B.; Cregan, P.; Ma, J. Pericentromeric Effects Shape the Patterns of Divergence, Retention, and Expression of Duplicated Genes in the Paleopolyploid Soybean. Plant Cell 2012, 24, 21–32. [Google Scholar] [CrossRef]
| Sr.no. | Gene | Locus ID | Chr | Size (aa) | MW(Da) | pI | Subcellular Location | AGC Kinase Classification | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GmAGC1-1a | Glyma.09G015400 | 9 | 608 | 66,157.12 | 8.82 | Cytoplasm | AGC1 | AGCⅧ |
| 2 | GmAGC1-1b | Glyma.15G121200 | 15 | 608 | 66,112.15 | 8.51 | Cytoplasm | ||
| 3 | GmAGC1-2a | Glyma.16G070700 | 16 | 631 | 69,937.32 | 7.17 | Plasma membrane | ||
| 4 | GmAGC1-2b | Glyma.19G060500 | 19 | 631 | 69,931.22 | 7.98 | Plasma membrane | ||
| 5 | GmAGC1-2c | Glyma.19G003800 | 19 | 612 | 68,735.18 | 8.02 | Plasma membrane | ||
| 6 | GmAGC1-3a | Glyma.03G111200 | 3 | 763 | 83,526.82 | 6.20 | Extracellular | ||
| 7 | GmAGC1-3b | Glyma.07G114400 | 7 | 763 | 83,407.83 | 6.26 | Extracellular | ||
| 8 | GmAGC1-3c | Glyma.09G242900 | 9 | 766 | 83,736.08 | 6.99 | Extracellular | ||
| 9 | GmAGC1-3d | Glyma.18G251800 | 18 | 768 | 83,844.12 | 7.22 | Extracellular | ||
| 10 | GmAGC1-4a | Glyma.08G160400 | 8 | 586 | 64,793.02 | 8.89 | Cytoplasm | ||
| 11 | GmAGC1-4b | Glyma.15G266800 | 15 | 576 | 63,503.47 | 8.14 | Cytoplasm | ||
| 12 | GmAGC1-4c | Glyma.08G232800 | 8 | 560 | 62,559.09 | 9.23 | Cytoplasm | ||
| 13 | GmAGC1-5a | Glyma.04G111700 | 4 | 827 | 90,218.08 | 6.78 | Extracellular | ||
| 14 | GmAGC1-5b | Glyma.06G322600 | 6 | 835 | 90,986.97 | 6.72 | Extracellular | ||
| 15 | GmAGC1-5c | Glyma.12G002700 | 12 | 813 | 90,613.40 | 9.00 | Extracellular | ||
| 16 | GmAGC1-5d | Glyma.09G234251 | 9 | 182 | 19,986.92 | 5.76 | Plasma membrane | ||
| 17 | GmAGC1-6a | Glyma.10G067400 | 10 | 863 | 93,847.54 | 8.67 | Extracellular | ||
| 18 | GmAGC1-6b | Glyma.U031420 | un | 866 | 93,986.43 | 8.65 | Extracellular | ||
| 19 | GmAGC1-6c | Glyma.03G193000 | 3 | 871 | 95,373.38 | 8.83 | Extracellular | ||
| 20 | GmAGC1-6d | Glyma.19G193100 | 19 | 868 | 95,279.47 | 8.84 | Extracellular | ||
| 21 | GmAGC1-7a | Glyma.10G203100 | 10 | 414 | 45,918.41 | 6.40 | Plasma membrane | ||
| 22 | GmAGC1-7b | Glyma.20G187200 | 20 | 422 | 46,799.55 | 6.02 | Plasma membrane | ||
| 23 | GmAGC1-8a | Glyma.11G187600 | 11 | 441 | 48,991.94 | 6.44 | Plasma membrane | ||
| 24 | GmAGC1-8b | Glyma.12G086900 | 12 | 436 | 48,490.60 | 7.68 | Nuclear | ||
| 25 | GmAGC1-8c | Glyma.12G182600 | 12 | 453 | 50,636.74 | 6.66 | Cytoplasm | ||
| 26 | GmAGC1-8d | Glyma.13G318400 | 13 | 497 | 55,485.34 | 6.32 | Plasma membrane | ||
| 27 | GmAGC3-1a | Glyma.13G220100 | 13 | 452 | 50,551.26 | 8.29 | Plasma membrane | AGC3 | |
| 28 | GmAGC3-1b | Glyma.U032208 | un | 460 | 51,632.71 | 8.79 | Plasma membrane | ||
| 29 | GmAGC3-2a | Glyma.05G009200 | 5 | 488 | 55,368.00 | 9.28 | Plasma membrane | ||
| 30 | GmAGC3-2b | Glyma.17G117300 | 17 | 490 | 55,622.27 | 9.39 | Plasma membrane | ||
| 31 | GmAGC3-2c | Glyma.04G148400 | 4 | 488 | 54,931.86 | 9.09 | Plasma membrane | ||
| 32 | GmAGC3-3a | Glyma.08G174400 | 8 | 472 | 53,236.81 | 9.35 | Plasma membrane | ||
| 33 | GmAGC3-3b | Glyma.15G252700 | 15 | 478 | 54,469.23 | 9.44 | Plasma membrane | ||
| 34 | GmPHOT1a | Glyma.13G330400 | 13 | 982 | 110,192.34 | 7.29 | Plasma membrane | AGC4 | |
| 35 | GmPHOT1b | Glyma.15G043600 | 15 | 982 | 110,324.67 | 8.23 | Plasma membrane | ||
| 36 | GmPHOT1c | Glyma.12G074100 | 12 | 977 | 109,600.73 | 7.06 | Plasma membrane | ||
| 37 | GmPHOT2a | Glyma.08G264900 | 8 | 996 | 111,560.13 | 6.36 | Plasma membrane | ||
| 38 | GmPHOT2b | Glyma.16G096600 | 16 | 990 | 110,960.66 | 7.59 | Plasma membrane | ||
| 39 | GmAGC2-1a | Glyma.03G088800 | 3 | 436 | 49,579.15 | 6.62 | Plasma membrane | AGC2 | |
| 40 | GmAGC2-1b | Glyma.16G084700 | 16 | 434 | 49,826.72 | 7.69 | Extracellular | ||
| 41 | GmAGC2-1c | Glyma.08G345500 | 8 | 445 | 50,615.27 | 7.21 | Extracellular | ||
| 42 | GmAGC2-2a | Glyma.11G131800 | 11 | 411 | 46,524.23 | 9.53 | Plasma membrane | ||
| 43 | GmAGC2-2b | Glyma.12G056000 | 12 | 419 | 47,344.31 | 9.71 | Plasma membrane | ||
| 44 | GmAGC2-2c | Glyma.13G341400 | 13 | 386 | 43,528.03 | 9.65 | Plasma membrane | ||
| 45 | GmIRE1a | Glyma.07G103400 | 7 | 1297 | 142,803.63 | 5.81 | Nuclear | AGC other | |
| 46 | GmIRE1b | Glyma.09G174000 | 9 | 1302 | 143,144.25 | 5.64 | Nuclear | ||
| 47 | GmIRE2a | Glyma.09G232400 | 9 | 1184 | 132,490.49 | 5.41 | Nuclear | ||
| 48 | GmIRE2b | Glyma.12G004100 | 12 | 1176 | 131,889.99 | 5.49 | Nuclear | ||
| 49 | GmNDR1a | Glyma.02G003200 | 2 | 547 | 63,207.30 | 7.20 | Nuclear | AGCⅦ | |
| 50 | GmNDR1b | Glyma.10G005600 | 10 | 547 | 63,353.58 | 8.15 | Nuclear | ||
| 51 | GmNDR1c | Glyma.10G181500 | 10 | 566 | 66,294.51 | 7.85 | Nuclear | ||
| 52 | GmNDR1d | Glyma.20G208900 | 20 | 543 | 62,958.90 | 8.45 | Nuclear | ||
| 53 | GmNDR2a | Glyma.03G164600 | 3 | 542 | 62,867.10 | 5.84 | Cytoplasm | ||
| 54 | GmNDR2b | Glyma.19G166000 | 19 | 546 | 63,507.89 | 5.88 | Nuclear | ||
| 55 | GmNDR2c | Glyma.10G038400 | 10 | 558 | 64,301.02 | 6.37 | Cytoplasm | ||
| 56 | GmNDR2d | Glyma.13G124800 | 13 | 559 | 64,545.18 | 6.21 | Cytoplasm | ||
| 57 | GmNDR3a | Glyma.04G053900 | 4 | 503 | 58,226.48 | 5.80 | Cytoplasm | ||
| 58 | GmNDR3b | Glyma.06G054200 | 6 | 503 | 58,254.45 | 5.80 | Cytoplasm | ||
| 59 | GmNDR4a | Glyma.09G066300 | 9 | 563 | 64,847.96 | 6.50 | Extracellular | ||
| 60 | GmNDR4b | Glyma.15G172300 | 15 | 519 | 59,615.93 | 6.14 | Cytoplasm | ||
| 61 | GmNDR5a | Glyma.14G083100 | 14 | 544 | 62,311.44 | 6.69 | Extracellular | ||
| 62 | GmNDR5b | Glyma.17G242000 | 17 | 538 | 61,522.02 | 5.77 | Extracellular | ||
| 63 | GmS6K1a | Glyma.05G032000 | 5 | 291 | 32,603.31 | 7.69 | Plasma membrane | AGCⅥ | |
| 64 | GmS6K1b | Glyma.17G094700 | 17 | 415 | 46,873.57 | 7.28 | Nuclear | ||
| 65 | GmS6K2a | Glyma.09G273600 | 9 | 1005 | 112,107.84 | 7.34 | Plasma membrane | ||
| 66 | GmS6K2b | Glyma.18G212800 | 18 | 479 | 54,328.16 | 6.21 | Plasma membrane | ||
| 67 | GmS6K2c | Glyma.14G188100 | 14 | 472 | 53,323.71 | 7.24 | Nuclear | ||
| 68 | GmPDK1a | Glyma.10G200600 | 10 | 491 | 54,802.36 | 6.73 | Cytoplasm | PDK1 | |
| 69 | GmPDK1b | Glyma.20G190000 | 20 | 491 | 54,731.24 | 6.73 | Cytoplasm | ||
| Gene Pairs | Ka | Ks | Ka/Ks | Date (Mya) | Type of Selection | Type of Duplication |
|---|---|---|---|---|---|---|
| GmAGC1-1a/GmAGC1-1b | 0.02 | 0.15 | 0.13 | 12.45 | Purifying | Segmental/WGD |
| GmAGC1-2a/GmAGC1-2b | 0.09 | 0.65 | 0.13 | 53.34 | Purifying | Segmental/WGD |
| GmAGC1-2a/GmAGC1-2c | 0.02 | 0.17 | 0.09 | 14.34 | Purifying | Segmental/WGD |
| GmAGC1-2c/GmAGC1-2b | 0.08 | 0.67 | 0.12 | 54.95 | Purifying | Segmental/WGD |
| GmAGC1-3a/GmAGC1-3b | 0.02 | 0.13 | 0.13 | 11.02 | Purifying | Segmental/WGD |
| GmAGC1-3a/GmAGC1-3c | 0.08 | 0.62 | 0.13 | 50.53 | Purifying | Segmental/WGD |
| GmAGC1-3a/GmAGC1-3d | 0.08 | 0.56 | 0.15 | 45.67 | Purifying | Segmental/WGD |
| GmAGC1-3b/GmAGC1-3c | 0.08 | 0.62 | 0.14 | 50.44 | Purifying | Segmental/WGD |
| GmAGC1-3b/GmAGC1-3d | 0.09 | 0.56 | 0.16 | 46.02 | Purifying | Segmental/WGD |
| GmAGC1-3c/GmAGC1-3d | 0.02 | 0.14 | 0.12 | 11.78 | Purifying | Segmental/WGD |
| GmAGC1-4a/GmAGC1-4b | 0.02 | 0.16 | 0.15 | 13.17 | Purifying | Segmental/WGD |
| GmAGC1-5a/GmAGC1-5b | 0.02 | 0.09 | 0.25 | 7.74 | Purifying | Segmental/WGD |
| GmAGC1-5d/GmAGC1-5c | 0.05 | 0.12 | 0.41 | 10.04 | Purifying | Segmental/WGD |
| GmAGC1-6a/GmAGC1-6d | 0.13 | 0.45 | 0.29 | 36.89 | Purifying | Segmental/WGD |
| GmAGC1-6c/GmAGC1-6a | 0.13 | 0.43 | 0.31 | 35.39 | Purifying | Segmental/WGD |
| GmAGC1-6c/GmAGC1-6d | 0.02 | 0.10 | 0.25 | 7.83 | Purifying | Segmental/WGD |
| GmAGC1-7a/GmAGC1-7b | 0.04 | 0.22 | 0.17 | 17.69 | Purifying | Segmental/WGD |
| GmAGC1-8a/GmAGC1-8b | 0.04 | 0.23 | 0.18 | 19.15 | Purifying | Segmental/WGD |
| GmAGC1-8a/GmAGC1-8c | 0.17 | 0.79 | 0.22 | 64.52 | Purifying | Segmental/WGD |
| GmAGC1-8a/GmAGC1-8d | 0.17 | 0.85 | 0.20 | 69.56 | Purifying | Segmental/WGD |
| GmAGC1-8b/GmAGC1-8c | 0.18 | 0.79 | 0.23 | 64.59 | Purifying | Segmental/WGD |
| GmAGC1-8b/GmAGC1-8d | 0.16 | 0.88 | 0.19 | 72.14 | Purifying | Segmental/WGD |
| GmAGC1-8c/GmAGC1-8d | 0.02 | 0.17 | 0.11 | 14.16 | Purifying | Segmental/WGD |
| GmAGC2-1a/GmAGC2-1b | 0.28 | 1.53 | 0.18 | 125.04 | Purifying | Segmental/WGD |
| GmAGC2-1a/GmAGC2-1c | 0.09 | 0.35 | 0.26 | 28.69 | Purifying | Segmental/WGD |
| GmAGC2-1c/GmAGC2-1b | 0.30 | 1.54 | 0.20 | 126.41 | Purifying | Segmental/WGD |
| GmAGC2-2a/GmAGC2-2b | 0.05 | 0.20 | 0.24 | 16.69 | Purifying | Segmental/WGD |
| GmAGC2-2a/GmAGC2-2c | 0.21 | 0.84 | 0.25 | 68.60 | Purifying | Segmental/WGD |
| GmAGC2-2b/GmAGC2-2c | 0.20 | 0.93 | 0.22 | 76.50 | Purifying | Segmental/WGD |
| GmAGC3-2a/GmAGC3-2b | 0.03 | 0.18 | 0.17 | 14.35 | Purifying | Segmental/WGD |
| GmAGC3-2c/GmAGC3-2a | 0.17 | 1.00 | 0.17 | 82.04 | Purifying | Segmental/WGD |
| GmAGC3-2c/GmAGC3-2b | 0.15 | 0.90 | 0.17 | 73.81 | Purifying | Segmental/WGD |
| GmAGC3-3a/GmAGC3-3b | 0.06 | 0.27 | 0.23 | 21.76 | Purifying | Segmental/WGD |
| GmIRE1a/GmIRE1b | 0.01 | 0.09 | 0.16 | 7.49 | Purifying | Segmental/WGD |
| GmIRE2a/GmIRE2b | 0.03 | 0.10 | 0.27 | 8.06 | Purifying | Segmental/WGD |
| GmNDR1a/GmNDR1b | 0.01 | 0.08 | 0.15 | 6.91 | Purifying | Segmental/WGD |
| GmNDR1a/GmNDR1c | 0.09 | 0.45 | 0.19 | 36.99 | Purifying | Segmental/WGD |
| GmNDR1c/GmNDR1d | 0.06 | 0.14 | 0.45 | 11.09 | Purifying | Segmental/WGD |
| GmNDR2a/GmNDR2b | 0.08 | 0.64 | 0.12 | 52.49 | Purifying | Segmental/WGD |
| GmNDR2a/GmNDR2c | 0.08 | 0.63 | 0.13 | 51.47 | Purifying | Segmental/WGD |
| GmNDR2a/GmNDR2d | 0.04 | 0.18 | 0.22 | 14.96 | Purifying | Segmental/WGD |
| GmNDR2c/GmNDR1d | 0.11 | 2.16 | 0.05 | 176.75 | Purifying | Segmental/WGD |
| GmNDR2c/GmNDR2b | 0.08 | 0.69 | 0.11 | 56.93 | Purifying | Segmental/WGD |
| GmNDR2c/GmNDR2d | 0.02 | 0.09 | 0.19 | 6.97 | Purifying | Segmental/WGD |
| GmNDR2d/GmNDR2b | 0.08 | 0.64 | 0.13 | 52.77 | Purifying | Segmental/WGD |
| GmNDR3a/GmNDR3b | 0.01 | 0.10 | 0.05 | 8.25 | Purifying | Segmental/WGD |
| GmNDR4a/GmNDR4b | 0.02 | 0.10 | 0.18 | 8.02 | Purifying | Segmental/WGD |
| GmNDR5a/GmNDR5b | 0.03 | 0.15 | 0.22 | 12.65 | Purifying | Segmental/WGD |
| GmPDK1a/GmPDK1b | 0.01 | 0.07 | 0.15 | 5.37 | Purifying | Segmental/WGD |
| GmPHOT1a/GmPHOT1b | 0.02 | 0.15 | 0.16 | 12.22 | Purifying | Segmental/WGD |
| GmPHOT1c/GmPHOT1a | 0.12 | 0.66 | 0.19 | 54.33 | Purifying | Segmental/WGD |
| GmPHOT1c/GmPHOT1b | 0.12 | 0.68 | 0.18 | 55.45 | Purifying | Segmental/WGD |
| GmS6K1a/GmS6K1b | 0.07 | 0.17 | 0.44 | 13.55 | Purifying | Segmental/WGD |
| GmS6K2a/GmS6K2b | 0.02 | 0.14 | 0.17 | 11.25 | Purifying | Segmental/WGD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yang, S.; Chen, Y.; Ye, S.; Lin, W.; Lin, X.; Tang, Y.; Liu, B. Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max). Int. J. Mol. Sci. 2025, 26, 2588. https://doi.org/10.3390/ijms26062588
Liu W, Yang S, Chen Y, Ye S, Lin W, Lin X, Tang Y, Liu B. Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max). International Journal of Molecular Sciences. 2025; 26(6):2588. https://doi.org/10.3390/ijms26062588
Chicago/Turabian StyleLiu, Wenmin, Shuichan Yang, Yi Chen, Sujun Ye, Wenmin Lin, Xiaoya Lin, Yang Tang, and Baohui Liu. 2025. "Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max)" International Journal of Molecular Sciences 26, no. 6: 2588. https://doi.org/10.3390/ijms26062588
APA StyleLiu, W., Yang, S., Chen, Y., Ye, S., Lin, W., Lin, X., Tang, Y., & Liu, B. (2025). Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max). International Journal of Molecular Sciences, 26(6), 2588. https://doi.org/10.3390/ijms26062588






