Molecular Interplay Between Non-Coding RNAs and Connexins and Its Possible Role in Cancer
Abstract
:1. Introduction
2. Selection of Articles
3. ncRNAs in Cancer
4. Connexins in Cancer
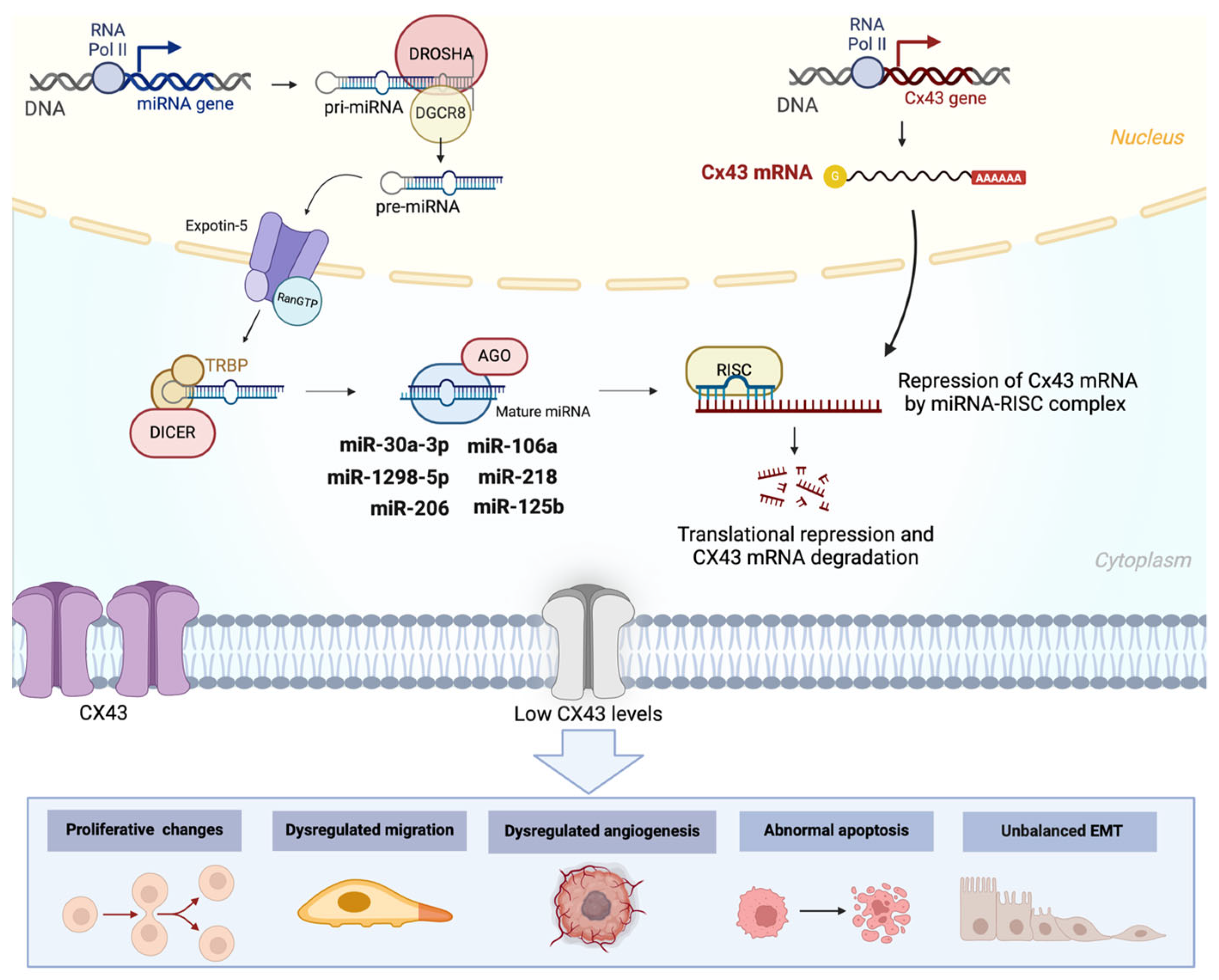
5. Interplay Between ncRNAs and Cxs in Cancer
5.1. Pancreatic Cancer
5.2. Bladder Cancer
5.3. Breast Cancer
5.4. Epithelial Ovarian Cancer
5.5. Melanoma
5.6. Glioblastoma
5.7. Other Types of Cancers
6. Potential Biomedical Applications of ncRNAs and Cxs
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| CCA | choriocarcinoma |
| circRNA | circular RNA |
| CRC | colorectal cancer |
| EMT | epithelial–mesenchymal transition |
| EOC | epithelial ovarian cancer |
| GC | gastric cancer |
| GBM | glioblastoma multiforme |
| GJC | gap junction channel |
| HNSCC | head and neck squamous cell carcinoma |
| lncRNA | long non-coding RNA |
| miRNA | microRNA |
| MIBC | muscle-invasive bladder cancer |
| ncRNA | non-coding RNA |
| NEAT1 | Nuclear-Enriched Abundant Transcript 1 |
| NPC | nasopharyngeal carcinoma |
| PC | prostate cancer |
| PDAC | pancreatic ductal adenocarcinoma |
| piRNA | PIWI-interacting RNA |
| PFS | progression-free survival |
| rRNA | ribosomal RNA |
| siRNA | small interfering RNA |
| snoRNA | small nucleolar RNA |
| snRNA | small nuclear RNA |
| TNM | Tumor Node Metastasis |
| CCA | choriocarcinoma |
| circRNA | circular RNA |
References
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.; Morton, C.C. Identification and function of long non-coding RNA. Front. Cell. Neurosci. 2013, 7, 168. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Saez, J.C.; Berthoud, V.M.; Branes, M.C.; Martinez, A.D.; Beyer, E.C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 2003, 83, 1359–1400. [Google Scholar] [CrossRef]
- Dbouk, H.A.; Mroue, R.M.; El-Sabban, M.E.; Talhouk, R.S. Connexins: A myriad of functions extending beyond assembly of gap junction channels. Cell Commun. Signal. 2009, 7, 4. [Google Scholar] [CrossRef]
- Sáez, J.C.; Leybaert, L. Hunting for connexin hemichannels. FEBS Lett. 2014, 588, 1205–1211. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Chen, N. Perspective and Therapeutic Potential of the Noncoding RNA-Connexin Axis. Int. J. Mol. Sci. 2024, 25, 6146. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Leithe, E.; Graham, S.V.; Kameritsch, P.; Mayán, M.D.; Mesnil, M.; Pogoda, K.; Tabernero, A. Connexins in cancer: Bridging the gap to the clinic. Oncogene 2019, 38, 4429–4451. [Google Scholar] [CrossRef]
- Muñoz, J.P.; Pérez-Moreno, P.; Pérez, Y.; Calaf, G.M. The Role of MicroRNAs in Breast Cancer and the Challenges of Their Clinical Application. Diagnostics 2023, 13, 3072. [Google Scholar] [CrossRef]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef]
- Rhim, J.; Baek, W.; Seo, Y.; Kim, J.H. From Molecular Mechanisms to Therapeutics: Understanding MicroRNA-21 in Cancer. Cells 2022, 11, 2791. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Noori, M.; Sarrafzadeh, S.; Tamehri Zadeh, S.S.; Nemati, M.; Chatrabnous, N.; Jafarzadeh, S.; Hamblin, M.R.; Jafari Najaf Abadi, M.H.; Mirzaei, H. MicroRNA-383: A tumor suppressor miRNA in human cancer. Front. Cell Dev. Biol. 2022, 10, 955486. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell. Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- He, Y.; Xu, Y.; Yu, X.; Sun, Z.; Guo, W. The Vital Roles of LINC00662 in Human Cancers. Front. Cell Dev. Biol. 2021, 9, 711352. [Google Scholar] [CrossRef]
- Pérez-Moreno, P.; Riquelme, I.; Bizama, C.; Vergara-Gómez, L.; Tapia, J.C.; Brebi, P.; García, P.; Roa, J.C. LINC00662 Promotes Aggressive Traits by Modulating OCT4 Expression through miR-335-5p in Gallbladder Cancer Cells. Int. J. Mol. Sci. 2024, 25, 6740. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Q.; Hann, S.S. The functions and oncogenic roles of CCAT1 in human cancer. Biomed. Pharmacother. 2019, 115, 108943. [Google Scholar] [CrossRef]
- Ma, M.Z.; Chu, B.F.; Zhang, Y.; Weng, M.Z.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef]
- Dong, L.; Ding, H.; Li, Y.; Xue, D.; Liu, Y. LncRNA TINCR is associated with clinical progression and serves as tumor suppressive role in prostate cancer. Cancer Manag. Res. 2018, 10, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Zhu, C.; Wang, K. A review of current evidence about lncRNA MEG3: A tumor suppressor in multiple cancers. Front. Cell Dev. Biol. 2022, 10, 997633. [Google Scholar] [CrossRef]
- Yu, X.; Liu, H.; Chang, N.; Fu, W.; Guo, Z.; Wang, Y. Circular RNAs: New players involved in the regulation of cognition and cognitive diseases. Front. Neurosci. 2023, 17, 1097878. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Riquelme, I.; Pérez-Moreno, P.; Letelier, P.; Brebi, P.; Roa, J.C. The Emerging Role of PIWI-Interacting RNAs (piRNAs) in Gastrointestinal Cancers: An Updated Perspective. Cancers 2021, 14, 202. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jiang, X.Y.; Qi, W.; Ji, C.G.; Xie, X.L.; Zhang, D.X.; Cui, Z.J.; Wang, C.K.; Bai, Y.; Wang, J.; et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Xu, B.; Ye, M.H.; Lv, S.G.; Wang, Q.X.; Wu, M.J.; Xiao, B.; Kang, C.S.; Zhu, X.G. SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget 2017, 8, 43953–43966. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, J.; Ju, S.; Cui, M.; Jing, R. Disorders and roles of tsRNA, snoRNA, snRNA and piRNA in cancer. J. Med. Genet. 2022, 59, 623–631. [Google Scholar] [CrossRef]
- Oh, J.M.; Venters, C.C.; Di, C.; Pinto, A.M.; Wan, L.; Younis, I.; Cai, Z.; Arai, C.; So, B.R.; Duan, J.; et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 2020, 11, 1. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, Z.; Zhang, S.; Han, X.; Li, Y.; Xu, Q.; Zhou, L.; Xu, H.; Bai, Y.; Xu, C.; et al. Down-regulation of small nuclear RNA (snRNA) RNU5E-1 in hepatocellular carcinoma presents with vital clinical significance. J. Gastrointest. Oncol. 2020, 11, 738–746. [Google Scholar] [CrossRef]
- López, J.; Blanco, S. Exploring the role of ribosomal RNA modifications in cancer. Curr. Opin. Genet. Dev. 2024, 86, 102204. [Google Scholar] [CrossRef]
- Babaian, A.; Rothe, K.; Girodat, D.; Minia, I.; Djondovic, S.; Milek, M.; Spencer Miko, S.E.; Wieden, H.J.; Landthaler, M.; Morin, G.B.; et al. Loss of m1acp3Ψ Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020, 31, 107611. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, B.; Gan, C.; Sun, H.; Feng, L. A Comprehensive Review of Small Interfering RNAs (siRNAs): Mechanism, Therapeutic Targets, and Delivery Strategies for Cancer Therapy. Int. J. Nanomed. 2023, 18, 7605–7635. [Google Scholar] [CrossRef]
- Ubby, I.; Krueger, C.; Rosato, R.; Qian, W.; Chang, J.; Sabapathy, K. Cancer therapeutic targeting using mutant-p53-specific siRNAs. Oncogene 2019, 38, 3415–3427. [Google Scholar] [CrossRef]
- Zhou, M.; Zheng, M.; Zhou, X.; Tian, S.; Yang, X.; Ning, Y.; Li, Y.; Zhang, S. The roles of connexins and gap junctions in the progression of cancer. Cell Commun. Signal. 2023, 21, 8. [Google Scholar] [CrossRef]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef]
- Raza, A.; Ghoshal, A.; Chockalingam, S.; Ghosh, S.S. Connexin-43 enhances tumor suppressing activity of artesunate via gap junction-dependent as well as independent pathways in human breast cancer cells. Sci. Rep. 2017, 7, 7580. [Google Scholar] [CrossRef]
- Sirnes, S.; Bruun, J.; Kolberg, M.; Kjenseth, A.; Lind, G.E.; Svindland, A.; Brech, A.; Nesbakken, A.; Lothe, R.A.; Leithe, E.; et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int. J. Cancer 2012, 131, 570–581. [Google Scholar] [CrossRef]
- Han, Y.; Wang, H.; Chen, H.; Tan, T.; Wang, Y.; Yang, H.; Ding, Y.; Wang, S. CX43 down-regulation promotes cell aggressiveness and 5-fluorouracil-resistance by attenuating cell stiffness in colorectal carcinoma. Cancer Biol. Ther. 2023, 24, 2221879. [Google Scholar] [CrossRef]
- Kazan, J.M.; El-Saghir, J.; Saliba, J.; Shaito, A.; Jalaleddine, N.; El-Hajjar, L.; Al-Ghadban, S.; Yehia, L.; Zibara, K.; El-Sabban, M. Cx43 Expression Correlates with Breast Cancer Metastasis in MDA-MB-231 Cells In Vitro, In a Mouse Xenograft Model and in Human Breast Cancer Tissues. Cancers 2019, 11, 460. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, X.; He, Q.; Gu, Z.; Jia, X.; Zhuang, Z. Connexin 43 controls metastatic behavior in triple negative breast cancer through TGFβ1-Smad3-intergin αV signaling axis Based on optical image diagnosis. SLAS Technol. 2024, 29, 100190. [Google Scholar] [CrossRef]
- Zhang, A.; Hitomi, M.; Bar-Shain, N.; Dalimov, Z.; Ellis, L.; Velpula, K.K.; Fraizer, G.C.; Gourdie, R.G.; Lathia, J.D. Connexin 43 expression is associated with increased malignancy in prostate cancer cell lines and functions to promote migration. Oncotarget 2015, 6, 11640–11651. [Google Scholar] [CrossRef]
- McCutcheon, S.; Spray, D.C. Glioblastoma-Astrocyte Connexin 43 Gap Junctions Promote Tumor Invasion. Mol. Cancer Res. 2022, 20, 319–331. [Google Scholar] [CrossRef]
- Acuña, R.A.; Varas-Godoy, M.; Herrera-Sepulveda, D.; Retamal, M.A. Connexin46 Expression Enhances Cancer Stem Cell and Epithelial-to-Mesenchymal Transition Characteristics of Human Breast Cancer MCF-7 Cells. Int. J. Mol. Sci. 2021, 22, 12604. [Google Scholar] [CrossRef]
- Thiagarajan, P.S.; Sinyuk, M.; Turaga, S.M.; Mulkearns-Hubert, E.E.; Hale, J.S.; Rao, V.; Demelash, A.; Saygin, C.; China, A.; Alban, T.J.; et al. Cx26 drives self-renewal in triple-negative breast cancer via interaction with NANOG and focal adhesion kinase. Nat. Commun. 2018, 9, 578. [Google Scholar] [CrossRef]
- Stout, C.E.; Costantin, J.L.; Naus, C.C.; Charles, A.C. Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. J. Biol. Chem. 2002, 277, 10482–10488. [Google Scholar] [CrossRef]
- Anselmi, F.; Hernandez, V.H.; Crispino, G.; Seydel, A.; Ortolano, S.; Roper, S.D.; Kessaris, N.; Richardson, W.; Rickheit, G.; Filippov, M.A.; et al. ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc. Natl. Acad. Sci. USA 2008, 105, 18770–18775. [Google Scholar] [CrossRef]
- Kang, J.; Kang, N.; Lovatt, D.; Torres, A.; Zhao, Z.; Lin, J.; Nedergaard, M. Connexin 43 hemichannels are permeable to ATP. J. Neurosci. 2008, 28, 4702–4711. [Google Scholar] [CrossRef] [PubMed]
- Sáez, J.C.; Schalper, K.A.; Retamal, M.A.; Orellana, J.A.; Shoji, K.F.; Bennett, M.V. Cell membrane permeabilization via connexin hemichannels in living and dying cells. Exp. Cell Res. 2010, 316, 2377–2389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.T.; Yu, Y.Q.; Yang, H.; Huo, Y.F.; Huang, Y.; Tian, X.X.; Fang, W.G. Extracellular ATP promotes angiogenesis and adhesion of TNBC cells to endothelial cells via upregulation of CTGF. Cancer Sci. 2022, 113, 2457–2471. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Qian, Y.; Cao, Y.; Shriwas, P.; Zhang, H.; Chen, X. Extracellular ATP, as an energy and phosphorylating molecule, induces different types of drug resistances in cancer cells through ATP internalization and intracellular ATP level increase. Oncotarget 2017, 8, 87860–87877. [Google Scholar] [CrossRef] [PubMed]
- Kepp, O.; Loos, F.; Liu, P.; Kroemer, G. Extracellular nucleosides and nucleotides as immunomodulators. Immunol. Rev. 2017, 280, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.B.; Braunstein, T.H.; Nielsen, M.S.; MacAulay, N. Distinct permeation profiles of the connexin 30 and 43 hemichannels. FEBS Lett. 2014, 588, 1446–1457. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Ma, M.; Zhou, W.; Wang, Y.; Yang, L. pH-dependent channel gating in connexin26 hemichannels involves conformational changes in N-terminus. Biochim. Biophys. Acta 2012, 1818, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Retamal, M.A. Connexin and Pannexin hemichannels are regulated by redox potential. Front. Physiol. 2014, 5, 80. [Google Scholar] [CrossRef]
- Alstrøm, J.S.; Hansen, D.B.; Nielsen, M.S.; MacAulay, N. Isoform-specific phosphorylation-dependent regulation of connexin hemichannels. J. Neurophysiol. 2015, 114, 3014–3022. [Google Scholar] [CrossRef]
- Kuang, J.Y.; Guo, Y.F.; Chen, Y.; Wang, J.; Duan, J.J.; He, X.L.; Li, L.; Yu, S.C.; Bian, X.W. Connexin 43 C-terminus directly inhibits the hyperphosphorylation of Akt/ERK through protein-protein interactions in glioblastoma. Cancer Sci. 2018, 109, 2611–2622. [Google Scholar] [CrossRef]
- González-Sánchez, A.; Jaraíz-Rodríguez, M.; Domínguez-Prieto, M.; Herrero-González, S.; Medina, J.M.; Tabernero, A. Connexin43 recruits PTEN and Csk to inhibit c-Src activity in glioma cells and astrocytes. Oncotarget 2016, 7, 49819–49833. [Google Scholar] [CrossRef]
- Kotini, M.; Barriga, E.H.; Leslie, J.; Gentzel, M.; Rauschenberger, V.; Schambony, A.; Mayor, R. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat. Commun. 2018, 9, 3846. [Google Scholar] [CrossRef]
- Dang, X.; Doble, B.W.; Kardami, E. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell. Biochem. 2003, 242, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Epifantseva, I.; Xiao, S.; Baum, R.E.; Kléber, A.G.; Hong, T.; Shaw, R.M. An Alternatively Translated Connexin 43 Isoform, GJA1-11k, Localizes to the Nucleus and Can Inhibit Cell Cycle Progression. Biomolecules 2020, 10, 473. [Google Scholar] [CrossRef] [PubMed]
- Bugazia, D.; Al-Najjar, E.; Esmail, A.; Abdelrahim, S.; Abboud, K.; Abdelrahim, A.; Umoru, G.; Rayyan, H.A.; Abudayyeh, A.; Al Moustafa, A.E.; et al. Pancreatic ductal adenocarcinoma: The latest on diagnosis, molecular profiling, and systemic treatments. Front. Oncol. 2024, 14, 1386699. [Google Scholar] [CrossRef]
- Tao, J.; Yang, G.; Zhou, W.; Qiu, J.; Chen, G.; Luo, W.; Zhao, F.; You, L.; Zheng, L.; Zhang, T.; et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J. Hematol. Oncol. 2021, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, Q.; Liu, X.; Wang, F.; Yang, Y.; Tian, X. Hypoxic pancreatic cancer derived exosomal miR-30b-5p promotes tumor angiogenesis by inhibiting GJA1 expression. Int. J. Biol. Sci. 2022, 18, 1220–1237. [Google Scholar] [CrossRef]
- Georgikou, C.; Yin, L.; Gladkich, J.; Xiao, X.; Sticht, C.; Torre, C.; Gretz, N.; Gross, W.; Schäfer, M.; Karakhanova, S.; et al. Inhibition of miR30a-3p by sulforaphane enhances gap junction intercellular communication in pancreatic cancer. Cancer Lett. 2020, 469, 238–245. [Google Scholar] [CrossRef]
- Zheng, H.; Ding, B.; Xue, K.; Yu, J.; Lou, W. Construction of a lncRNA/pseudogene-hsa-miR-30d-5p-GJA1 regulatory network related to metastasis of pancreatic cancer. Genomics 2021, 113, 1742–1753. [Google Scholar] [CrossRef]
- Mau, B.; Johnson, B.; Hansel, D.E.; McConkey, D.J. The Many Faces of Muscle-Invasive Bladder Cancer: Histopathological and Molecular Characterization. Semin. Radiat. Oncol. 2023, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, L.; Mu, Z.; Liu, S.; Qu, H.; Xie, Q.; Hu, B. MicroRNA-1298-5p inhibits cell proliferation and the invasiveness of bladder cancer cells via down-regulation of connexin 43. Biochem. Cell Biol. 2020, 98, 227–237. [Google Scholar] [CrossRef]
- Chi, Q.; Wang, Z.Y.; Li, H.Y.; Song, D.B.; Xu, H.; Ma, G.; Wang, Z.M.; Li, X.M. Tumor-suppressor microRNA-139-5p restrains bladder cancer cell line ECV-304 properties via targeting Connexin 43. Chin. Med. J. 2019, 132, 2354–2361. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Fu, Y.; Shao, Z.M.; He, Q.Z.; Jiang, B.Q.; Wu, Y.; Zhuang, Z.G. Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 2091–2104. [Google Scholar]
- Lin, Z.J.; Ming, J.; Yang, L.; Du, J.Z.; Wang, N.; Luo, H.J. Mechanism of Regulatory Effect of MicroRNA-206 on Connexin 43 in Distant Metastasis of Breast Cancer. Chin. Med. J. 2016, 129, 424–434. [Google Scholar] [CrossRef]
- Ming, J.; Zhou, Y.; Du, J.; Fan, S.; Pan, B.; Wang, Y.; Fan, L.; Jiang, J. miR-381 suppresses C/EBPα-dependent Cx43 expression in breast cancer cells. Biosci. Rep. 2015, 35, e00266. [Google Scholar] [CrossRef]
- Ming, J.; Zhou, Y.; Du, J.; Fan, S.; Pan, B.; Wang, Y.; Fan, L.; Jiang, J. Identification of miR-200a as a novel suppressor of connexin 43 in breast cancer cells. Biosci. Rep. 2015, 35, e00251. [Google Scholar] [CrossRef]
- Maqbool, R.; Rashid, R.; Ismail, R.; Niaz, S.; Chowdri, N.A.; Hussain, M.U. The carboxy-terminal domain of connexin 43 (CT-Cx43) modulates the expression of p53 by altering miR-125b expression in low-grade human breast cancers. Cell. Oncol. 2015, 38, 443–451. [Google Scholar] [CrossRef]
- Li, D.; Li, L.; Chen, X.; Zhou, C.; Hao, B.; Cao, Y. Dysregulation of lncRNA-CCRR contributes to brain metastasis of breast cancer by intercellular coupling via regulating connexin 43 expression. J. Cell. Mol. Med. 2021, 25, 4826–4834. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Wang, G.; Wang, B.; Ding, Y.; Zhao, J.; Liu, H.; Cui, S. miR-206 as a prognostic and sensitivity biomarker for platinum chemotherapy in epithelial ovarian cancer. Cancer Cell Int. 2020, 20, 534. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Tian, J.; Zu, Y.; Wang, F.; Yang, Y.; Ma, S.; Cao, J.; Huang, Q.; Ha, C. The role of Connexin26 regulated by MiR-2114-3p in the pathogenesis of ovarian cancer. Biochem. Biophys. Res. Commun. 2023, 640, 105–116. [Google Scholar] [CrossRef]
- Bandarchi, B.; Jabbari, C.A.; Vedadi, A.; Navab, R. Molecular biology of normal melanocytes and melanoma cells. J. Clin. Pathol. 2013, 66, 644–648. [Google Scholar] [CrossRef]
- Scatolini, M.; Patel, A.; Grosso, E.; Mello-Grand, M.; Ostano, P.; Coppo, R.; Vitiello, M.; Venesio, T.; Zaccagna, A.; Pisacane, A.; et al. GJB5 association with BRAF mutation and survival in cutaneous malignant melanoma. Br. J. Dermatol. 2022, 186, 117–128. [Google Scholar] [CrossRef]
- Wang, J.L.; Li, H.; Zhang, J.B.; Zhang, C.H.; Hou, X.Q. Suppression of connexin 43 expression by miR-106a promotes melanoma cell proliferation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 965–971. [Google Scholar] [CrossRef]
- Tittarelli, A.; Navarrete, M.; Lizana, M.; Hofmann-Vega, F.; Salazar-Onfray, F. Hypoxic Melanoma Cells Deliver microRNAs to Dendritic Cells and Cytotoxic T Lymphocytes through Connexin-43 Channels. Int. J. Mol. Sci. 2020, 21, 7567. [Google Scholar] [CrossRef]
- Salvato, I.; Marchini, A. Immunotherapeutic Strategies for the Treatment of Glioblastoma: Current Challenges and Future Perspectives. Cancers 2024, 16, 1276. [Google Scholar] [CrossRef]
- Jin, Z.; Xu, S.; Yu, H.; Yang, B.; Zhao, H.; Zhao, G. miR-125b inhibits Connexin43 and promotes glioma growth. Cell. Mol. Neurobiol. 2013, 33, 1143–1148. [Google Scholar] [CrossRef]
- Hao, J.; Zhang, C.; Zhang, A.; Wang, K.; Jia, Z.; Wang, G.; Han, L.; Kang, C.; Pu, P. miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol. Rep. 2012, 27, 1504–1510. [Google Scholar] [CrossRef]
- Thuringer, D.; Boucher, J.; Jego, G.; Pernet, N.; Cronier, L.; Hammann, A.; Solary, E.; Garrido, C. Transfer of functional microRNAs between glioblastoma and microvascular endothelial cells through gap junctions. Oncotarget 2016, 7, 73925–73934. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, X.; Guo, Y.; Peng, F.; Zheng, N.; He, B.; Ge, H.; Tao, L.; Wang, Q. Pattern of cell-to-cell transfer of microRNA by gap junction and its effect on the proliferation of glioma cells. Cancer Sci. 2019, 110, 1947–1958. [Google Scholar] [CrossRef]
- Hong, X.; Sin, W.C.; Harris, A.L.; Naus, C.C. Gap junctions modulate glioma invasion by direct transfer of microRNA. Oncotarget 2015, 6, 15566–15577. [Google Scholar] [CrossRef]
- Suzhi, Z.; Liang, T.; Yuexia, P.; Lucy, L.; Xiaoting, H.; Yuan, Z.; Qin, W. Gap Junctions Enhance the Antiproliferative Effect of MicroRNA-124-3p in Glioblastoma Cells. J. Cell. Physiol. 2015, 230, 2476–2488. [Google Scholar] [CrossRef]
- Liang, J.; Xie, J.X.; He, J.; Li, Y.; Wei, D.; Zhou, R.; Wei, G.; Liu, X.; Chen, Q.; Li, D. Inhibiting lncRNA NEAT1 Increases Glioblastoma Response to TMZ by Reducing Connexin 43 Expression. Cancer Rep. 2024, 7, e70031. [Google Scholar] [CrossRef]
- Thuringer, D.; Jego, G.; Berthenet, K.; Hammann, A.; Solary, E.; Garrido, C. Gap junction-mediated transfer of miR-145-5p from microvascular endothelial cells to colon cancer cells inhibits angiogenesis. Oncotarget 2016, 7, 28160–28168. [Google Scholar] [CrossRef]
- Alajez, N.M.; Lenarduzzi, M.; Ito, E.; Hui, A.B.; Shi, W.; Bruce, J.; Yue, S.; Huang, S.H.; Xu, W.; Waldron, J.; et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011, 71, 2381–2391. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, Y.; Liu, H.; Zhao, X.; Shen, X. miR-301-3p directly regulates. J. Int. Med. Res. 2021, 49, 3000605211033185. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, J.H.; Song, B.; Xiong, E.Q.; Chen, Z.W.; Zhou, Z.S.; Su, Y.P. Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol. Ther. 2012, 13, 890–898. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Zheng, L. METTL3 promotes choriocarcinoma progression by activating the miR-935/GJA1 pathway in an m6A-dependent manner. Am. J. Reprod. Immunol. 2023, 90, e13791. [Google Scholar] [CrossRef] [PubMed]
- Gindin, Y.; Jiang, Y.; Francis, P.; Walker, R.L.; Abaan, O.D.; Zhu, Y.J.; Meltzer, P.S. miR-23a impairs bone differentiation in osteosarcoma via down-regulation of GJA1. Front. Genet. 2015, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Wang, C.; Zhai, X.; Li, J.; Ju, J.; Zhu, Y.; Zheng, S.; Ren, N.; Huang, B.; Jiang, X.; et al. lncRNA LEF1-AS1 Acts as a Novel Biomarker and Promotes Hypopharyngeal Squamous Cell Carcinoma Progression and Metastasis by Targeting the miR-221-5p/GJA1 Axis. Dis. Markers 2022, 2022, 3881310. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef]
- Svoboda, M.; Slyskova, J.; Schneiderova, M.; Makovicky, P.; Bielik, L.; Levy, M.; Lipska, L.; Hemmelova, B.; Kala, Z.; Protivankova, M.; et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014, 35, 1510–1515. [Google Scholar] [CrossRef]
- Wei, J.; Gao, W.; Zhu, C.J.; Liu, Y.Q.; Mei, Z.; Cheng, T.; Shu, Y.Q. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin. J. Cancer 2011, 30, 407–414. [Google Scholar] [CrossRef]
- Yang, J.; Meng, X.; Pan, J.; Jiang, N.; Zhou, C.; Wu, Z.; Gong, Z. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 2018, 15, 35–43. [Google Scholar] [CrossRef]
- Uppaluri, K.R.; Challa, H.J.; Gaur, A.; Jain, R.; Krishna Vardhani, K.; Geddam, A.; Natya, K.; Aswini, K.; Palasamudram, K.; Sri Manjari, K. Unlocking the potential of non-coding RNAs in cancer research and therapy. Transl. Oncol. 2023, 35, 101730. [Google Scholar] [CrossRef]
- Alzhrani, R.; Alsaab, H.O.; Petrovici, A.; Bhise, K.; Vanamala, K.; Sau, S.; Krinock, M.J.; Iyer, A.K. Improving the therapeutic efficiency of noncoding RNAs in cancers using targeted drug delivery systems. Drug Discov. Today 2020, 25, 718–730. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11, 340–354. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Poyet, C.; Buser, L.; Roudnicky, F.; Detmar, M.; Hermanns, T.; Mannhard, D.; Höhn, A.; Rüschoff, J.; Zhong, Q.; Sulser, T.; et al. Connexin 43 expression predicts poor progression-free survival in patients with non-muscle invasive urothelial bladder cancer. J. Clin. Pathol. 2015, 68, 819–824. [Google Scholar] [CrossRef]
- Naoi, Y.; Miyoshi, Y.; Taguchi, T.; Kim, S.J.; Arai, T.; Tamaki, Y.; Noguchi, S. Connexin26 expression is associated with lymphatic vessel invasion and poor prognosis in human breast cancer. Breast. Cancer Res. Treat. 2007, 106, 11–17. [Google Scholar] [CrossRef]
- Knösel, T.; Emde, A.; Schlüns, K.; Chen, Y.; Jürchott, K.; Krause, M.; Dietel, M.; Petersen, I. Immunoprofiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia 2005, 7, 741–747. [Google Scholar] [CrossRef]
- Willebrords, J.; Maes, M.; Crespo Yanguas, S.; Vinken, M. Inhibitors of connexin and pannexin channels as potential therapeutics. Pharmacol. Ther. 2017, 180, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.M.; Abt, M.; Doud, E.H.; Oblak, A.L.; Yeh, E.S. Mapping the Anti-Cancer Activity of α-Connexin Carboxyl-Terminal (aCT1) Peptide in Resistant HER2+ Breast Cancer. Cancers 2024, 16, 423. [Google Scholar] [CrossRef] [PubMed]
- Martins-Marques, T.; Girão, H. Expression and Function of Connexins in Extracellular Vesicles. Methods Mol. Biol. 2024, 2801, 17–28. [Google Scholar] [CrossRef]
- Acuña, R.A.; Varas-Godoy, M.; Berthoud, V.M.; Alfaro, I.E.; Retamal, M.A. Connexin-46 Contained in Extracellular Vesicles Enhance Malignancy Features in Breast Cancer Cells. Biomolecules 2020, 10, 676. [Google Scholar] [CrossRef]
- Gemel, J.; Kilkus, J.; Dawson, G.; Beyer, E.C. Connecting Exosomes and Connexins. Cancers 2019, 11, 476. [Google Scholar] [CrossRef]
| ncRNA | Cancer Type | Connexin-Associated Mechanism | Functional Effect | Clinicopathological Association | Reference |
|---|---|---|---|---|---|
| miR-30b-5p | Pancreatic Cancer | Binds directly to Cx43 mRNA | Increases angiogenesis | ND | [65] |
| miR-30a-3p | Pancreatic Cancer | Binds directly to Cx43 mRNA | Reduces tumor size | Highly expressed in cancer tissues and a negative correlation with disease staging and Cx43 expression | [66] |
| miR-30d-5p | Pancreatic Cancer | Inversely correlated with Cx43 expression | ND | Better prognosis | [67] |
| miR-1298-5p | Bladder Cancer | Binds directly to Cx43 mRNA | Inhibits invasion, proliferation, and invasion | Reduced expression in bladder cancer tissues | [69] |
| miR-139-5p | Bladder Cancer | Binds directly to Cx43 mRNA | Reduces proliferation, migration, and invasion | Reduced expression in bladder cancer tissues | [70] |
| miR-206 | Breast Cancer | Binds directly to Cx43 mRNA | Decreases cell viability, proliferation, migration, and invasion | Reduced expression in BC tissues | [72,73] |
| Ovarian Cancer | Binds directly to Cx43 mRNA | Increases invasion, migration, and cisplatin resistance | Poor prognosis and highly expressed in EOC tissues | [78] | |
| miR-381 | Breast Cancer | Suppresses C/EBPα-dependent Cx43 expression | Reduces migration and invasion | Reduced expression in BC tissues | [74] |
| miR-200a | Breast Cancer | Binds directly to Cx43 mRNA | Reduces migration | Downregulated in metastatic BC tissues | [75] |
| miR-125b | Breast Cancer | Inversely correlated with CT-Cx43 expression | Increases proliferation | Increased expression in BC tissues | [76] |
| lncRNA-CCRR | Breast Cancer | Directly correlated with Cx43 expression | Promotes communication between BC cells and astrocytes | Highly expressed in metastatic BC cancer tissues | [77] |
| miR-2114-3p | Ovarian Cancer | Binds directly to Cx26 mRNA, inhibiting the PI3k pathway | Inhibits tumor growth and invasion and induces S phase arrest of EOC cells | Reduced expression in EOC tissues and associated with better prognosis | [79] |
| miR-335-5p | Melanoma | Inversely correlated with GJB5 expression | ND | GJB5 underexpression is associated with overall worse survival | [81] |
| miR-106a | Melanoma | Binds directly to Cx43 mRNA | Increases proliferation and colony formation capacity | ND | [82] |
| miR-192-5p | Melanoma | Under hypoxic conditions, miR-192-5p is transferred to cytotoxic T cells via Cx43 | Inhibits the antitumor activity of T cells | ND | [83] |
| miR-19b | Glioblastoma | Transfer of miR-19b Cx43-dependent from GBM cells to astrocytes | Increases invasion | ND | [44] |
| miR-125b | Glioblastoma | Binds directly to Cx43 mRNA | Inhibits apoptosis and promotes colony formation in vitro and tumor growth in vivo | ND | [85] |
| miR-221/222 | Glioblastoma | Binds directly to Cx43 mRNA | Silencing of miR-221/222 reduces proliferation and migration capacities | ND | [86] |
| miR-5096 | Glioblastoma | Transfer of miR-5096 Cx43-dependent from GBM cells to astrocytes | Silencing of miR-5096 reduces migration capacity | ND | [89] |
| miR-124-3p | Glioblastoma | Transfer of miR-124-3p Cx-dependent in GBM cells (Cx not specified) | Reduces proliferation, colony formation, and tumor growth in vivo | ND | [90] |
| NEAT1 | Glioblastoma | Acts as a competitive endogenous RNA for miR-454-3p, increasing Cx43 levels | Increases resistance to TMZ | Highly expressed in recurrent gliomas compared to primary gliomas | [91] |
| miR-145-5p | Colorectal | Transfer of miR-145-5p Cx43-dependent through HMEC and CRC cells | Inhibits angiogenic process in vitro | ND | [92] |
| miR-218 | Nasopharyngeal Cancer | Binds directly to Cx43 mRNA and regulates the SLIT/ROBO pathway | Reduces cell viability, apoptosis in vitro, and tumor growth in vivo | Downregulated in NPC tissues and cell lines | [93] |
| miR-301-3p | Gastric Cancer | Binds directly to Cx43 mRNA | Increases migration, invasion, and proliferation | Increased expression in cell lines and GC tissues and correlated with poor differentiation, advanced TNM stage, vascular invasion, and lymph node metastasis | [94] |
| miR-20a | Prostate Cancer | Binds directly to Cx43 mRNA | Silencing of miR-20a reduces proliferation in vitro and tumor growth in vivo | Overexpressed in tumor tissue | [95] |
| miR-935 | Choriocarcinoma | Binds directly to Cx43 mRNA | Increases cell proliferation, migration, invasion, tube formation, and tumorigenesis in vitro and in vivo | Increased expression in CCA cell lines and tumor tissues | [96] |
| miR-23a | Osteosarcoma | Binds directly to Cx43 mRNA | Delays osteoblast differentiation | ND | [97] |
| LEF1-AS1 | Head and Neck Squamous Cell Carcinoma | Competitive endogenous RNA (ceRNA) by sponging miR-221-5p, leading to the overexpression of Cx43 | Overexpression promotes proliferation, colony formation, resistance to apoptosis, migration, EMT, and invasion | Overexpressed in tumor tissues and associated with worse survival | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Moreno, P.; Muñoz, J.P.; Retamal, M.A. Molecular Interplay Between Non-Coding RNAs and Connexins and Its Possible Role in Cancer. Int. J. Mol. Sci. 2025, 26, 2538. https://doi.org/10.3390/ijms26062538
Pérez-Moreno P, Muñoz JP, Retamal MA. Molecular Interplay Between Non-Coding RNAs and Connexins and Its Possible Role in Cancer. International Journal of Molecular Sciences. 2025; 26(6):2538. https://doi.org/10.3390/ijms26062538
Chicago/Turabian StylePérez-Moreno, Pablo, Juan P. Muñoz, and Mauricio A. Retamal. 2025. "Molecular Interplay Between Non-Coding RNAs and Connexins and Its Possible Role in Cancer" International Journal of Molecular Sciences 26, no. 6: 2538. https://doi.org/10.3390/ijms26062538
APA StylePérez-Moreno, P., Muñoz, J. P., & Retamal, M. A. (2025). Molecular Interplay Between Non-Coding RNAs and Connexins and Its Possible Role in Cancer. International Journal of Molecular Sciences, 26(6), 2538. https://doi.org/10.3390/ijms26062538









