CD44 Methylation Levels in Androgen-Deprived Prostate Cancer: A Putative Epigenetic Modulator of Tumor Progression
Abstract
1. Introduction
2. Results
2.1. Clinicopathologic Characteristics of PCa Cases
2.2. Intratumoral Microvessel Density (MVD) Related to Hormone Treatment
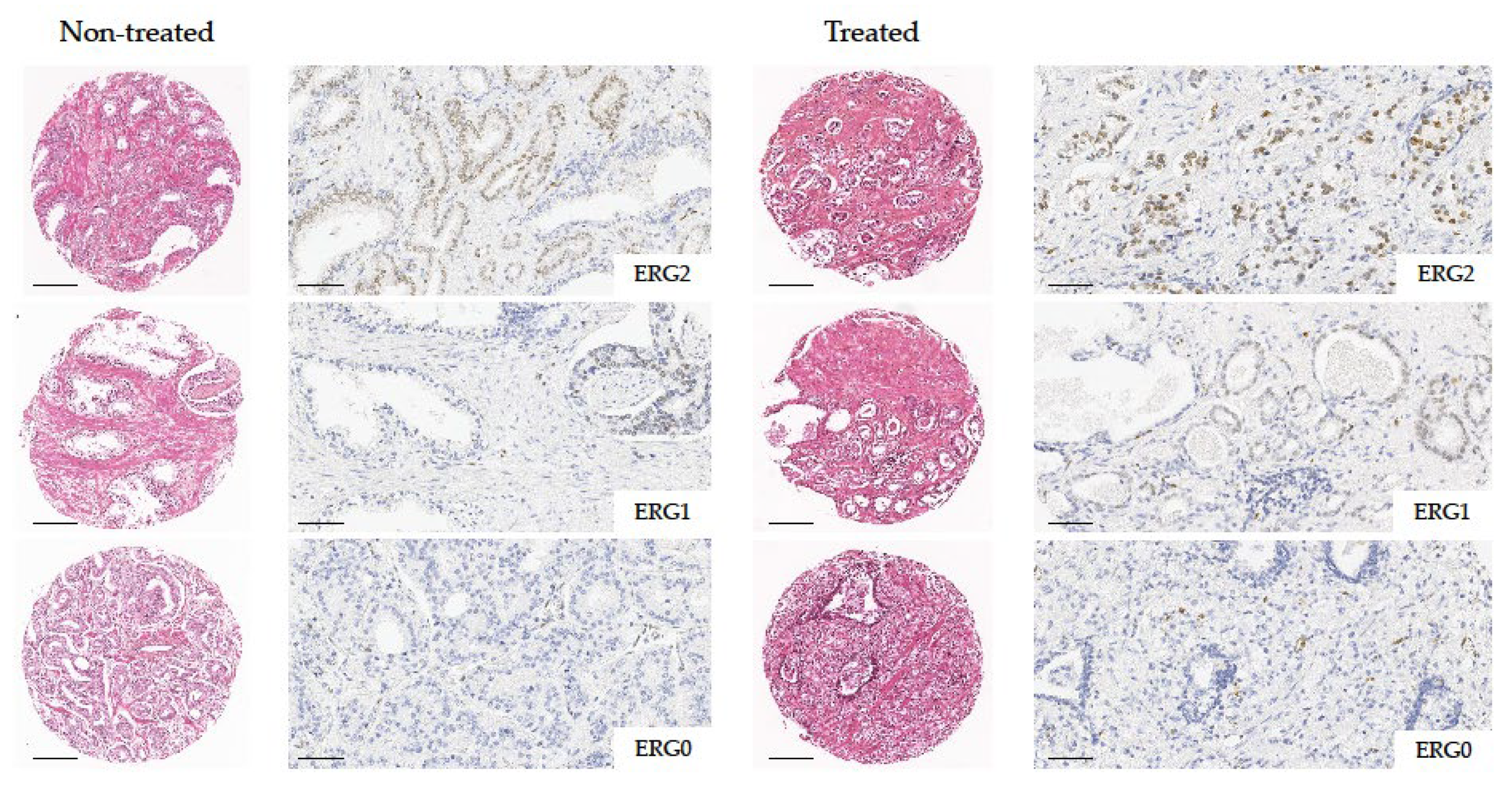
2.3. ERG Immunohistochemical Expression Related to Hormone Treatment
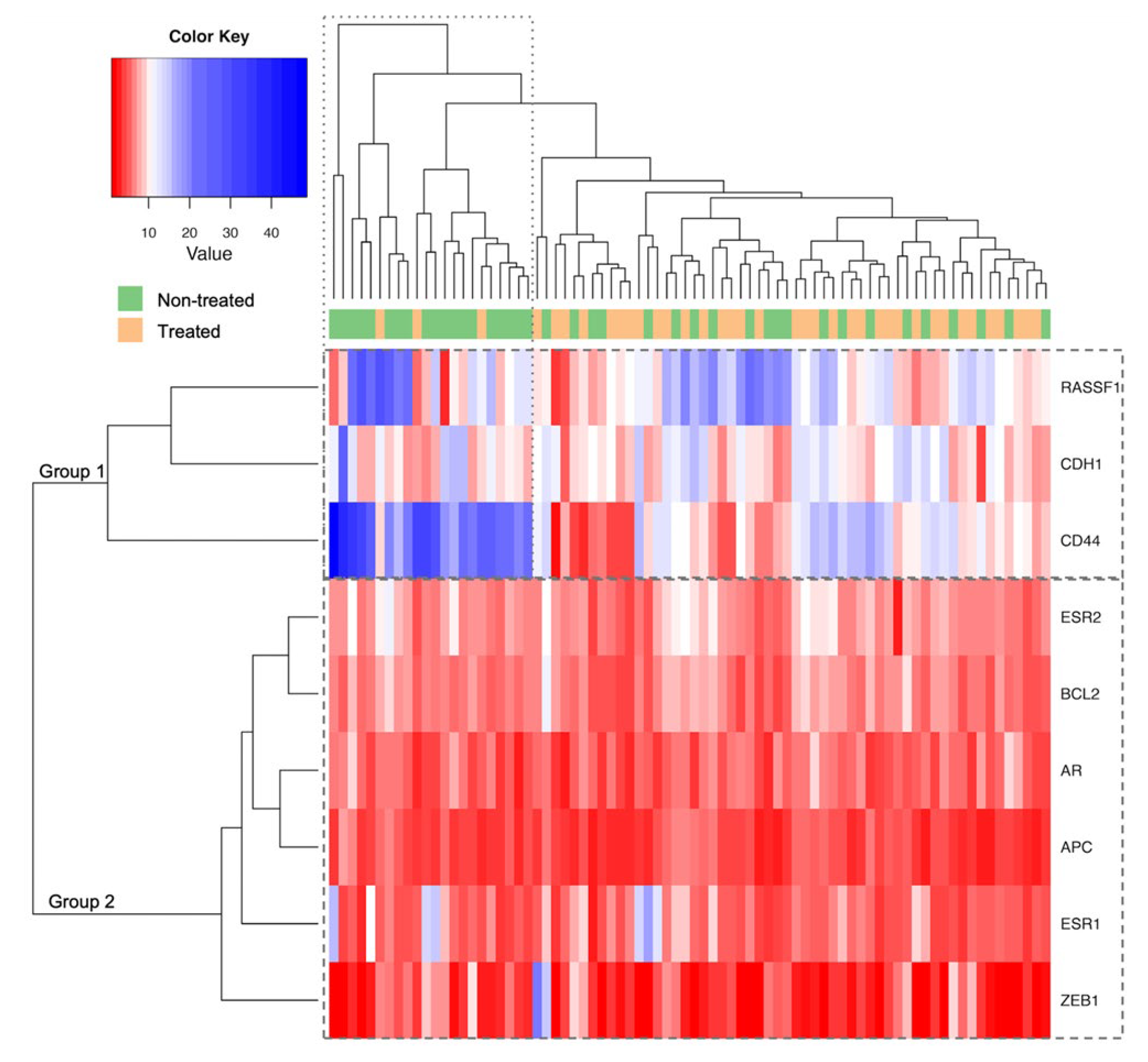
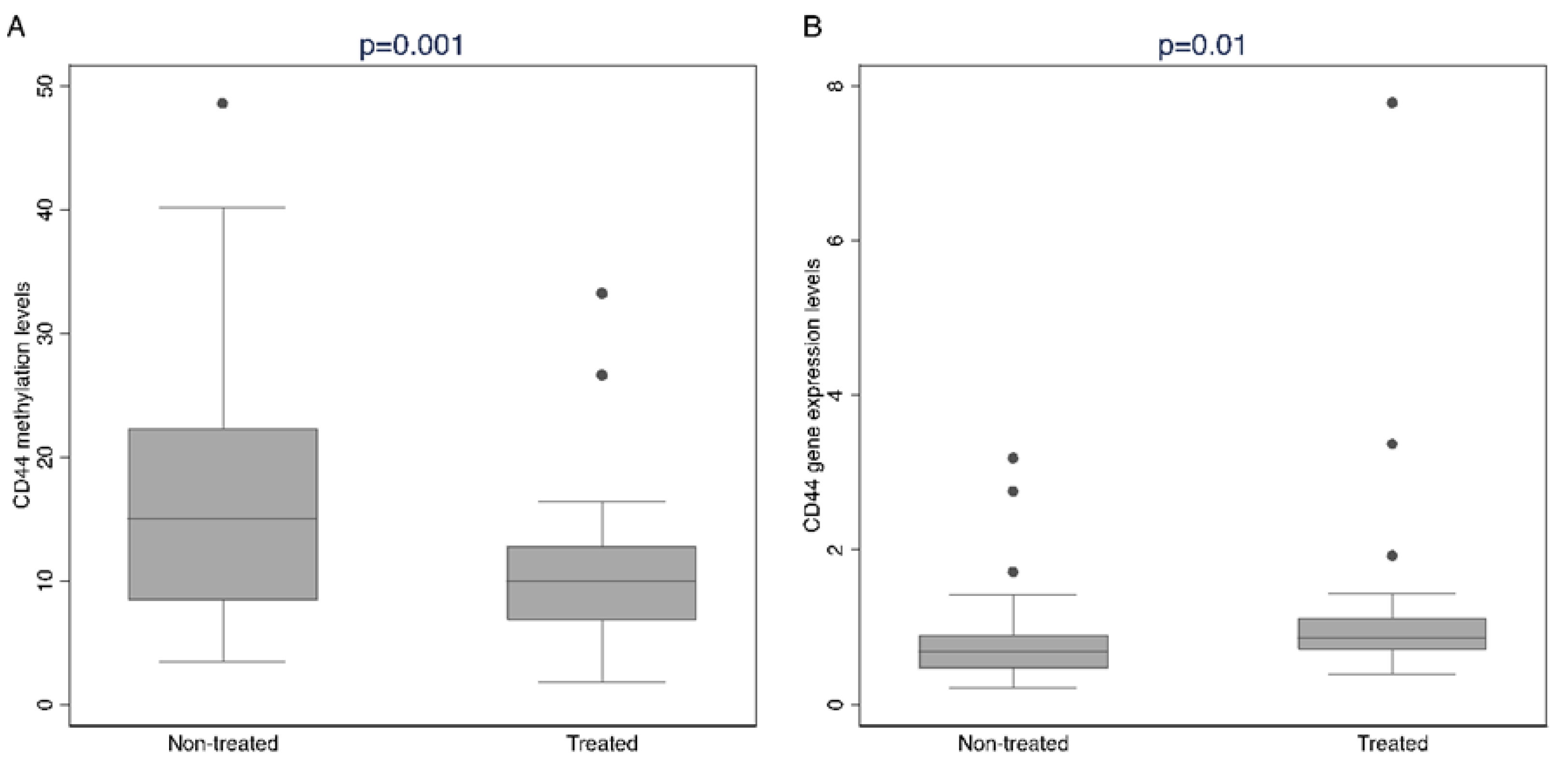
2.4. Methylation Analysis Related to Hormone Treatment
2.5. CD44 Methylation Levels According to MVD and ERG Expression
3. Discussion
4. Materials and Methods
4.1. Tissue Specimens
4.2. Traditional Whole Sections and Tissue Microarray Construction
4.3. Immunohistochemical Analysis
4.4. Methylation Analysis
4.5. CD44 Gene Expression Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B. The role of the androgen receptor in the development of prostatic hyperplasia and prostate cancer. Mol. Cell. Biochem. 2003, 253, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on prostate cancer. Part II—2020 Update: Treatment of relapsing and metastatic prostate cancer. Eur. Urol. 2021, 79, 263–282. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, X.; Liang, X.; Jiang, G. Molecular and cellular mechanisms of castration resistant prostate cancer. Oncol. Lett. 2018, 15, 6063–6076. [Google Scholar] [CrossRef]
- Li, P.; Yang, R.; Gao, W.Q. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Mol. Cancer 2014, 13, 55. [Google Scholar] [CrossRef]
- Zhang, W.; Meng, Y.; Liu, N.; Wen, X.F.; Yang, T. Insights into chemoresistance of prostate cancer. Int. J. Biol. Sci. 2015, 11, 1160–1170. [Google Scholar] [CrossRef]
- Ojo, D.; Lin, X.; Wong, N.; Gu, Y.; Tang, D. Prostate cancer stem-like cells contribute to the development of castration-resistant prostate cancer. Cancers 2015, 7, 2290–2308. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Xiong, X.; Zhang, S.; Tan, D.; Yang, L.; Wei, Q. Different predictive values of microvessel density for biochemical recurrence among different PCa populations: A systematic review and meta-analysis. Cancer Med. 2023, 12, 2166–2178. [Google Scholar] [CrossRef]
- Adamo, P.; Ladomery, M.R. The oncogene ERG: A key factor in prostate cancer. Oncogene 2016, 35, 403–414. [Google Scholar] [CrossRef]
- Rafikova, G.; Gilyazova, I.; Enikeeva, K.; Pavlov, V.; Kzhyshkowska, J. Prostate cancer: Genetics, epigenetics and the need for immunological biomarkers. Int. J. Mol. Sci. 2023, 24, 12797. [Google Scholar] [CrossRef]
- Phé, V.; Cussenot, O.; Rouprêt, M. Methylated genes as potential biomarkers in prostate cancer. BJU Int. 2010, 105, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Albany, C.; Alva, A.S.; Aparicio, A.M.; Singal, R.; Yellapragada, S.; Sonpavde, G.; Hahn, N.M. Epigenetics in prostate cancer. Prostate Cancer 2011, 2011, 580318. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Massie, C.E.; Mills, I.G.; Lynch, A.G. The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 2017, 166, 1–15. [Google Scholar] [CrossRef]
- Ragavi, R.; Muthukumaran, P.; Nandagopal, S.; Ahirwar, D.K.; Tomo, S.; Misra, S.; Guerriero, G.; Shukla, K.K. Epigenetics regulation of prostate cancer: Biomarker and therapeutic potential. Urol. Oncol. 2023, 41, 340–353. [Google Scholar] [CrossRef]
- Daniunaite, K.; Bakavicius, A.; Zukauskaite, K.; Rauluseviciute, I.; Lazutka, J.R.; Ulys, A.; Jankevicius, F.; Jarmalaite, S. Promoter methylation of PRKCB, ADAMTS12, and NAALAD2 is specific to prostate cancer and predicts biochemical disease recurrence. Int. J. Mol. Sci. 2021, 22, 6091. [Google Scholar] [CrossRef]
- Friedlander, T.W.; Roy, R.; Tomlins, S.A.; Ngo, V.T.; Kobayashi, Y.; Azameera, A.; Rubin, M.A.; Pienta, K.J.; Chinnaiyan, A.; Ittmann, M.M.; et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012, 72, 616–625. [Google Scholar] [CrossRef]
- Angulo, J.C.; Andrés, G.; Ashour, N.; Sánchez-Chapado, M.; López, J.I.; Ropero, S. Development of castration resistant prostate cancer can be predicted by a DNA hypermethylation profile. J. Urol. 2016, 195, 619–626. [Google Scholar] [CrossRef]
- Dillinger, T.; Sheibani-Tezerji, R.; Pulverer, W.; Stelzer, I.; Hassler, M.R.; Scheibelreiter, J.; Pérez Malla, C.U.; Kuroll, M.; Domazet, S.; Redl, E.; et al. Identification of tumor tissue-derived DNA methylation biomarkers for the detection and therapy response evaluation of metastatic castration resistant prostate cancer in liquid biopsies. Mol. Cancer 2022, 21, 7. [Google Scholar] [CrossRef]
- Li, L.C.; Carrol, P.R.; Dahiya, R. Epigenetic changes in prostate cancer: Implication for diagnosis and treatment. J. Natl. Cancer Inst. 2005, 97, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Sato, H.; Kanesaka, M.; Imamura, Y.; Sakamoto, S.; Ichikawa, T.; Kaneda, A. Epigenetic modifications in prostate cancer. Int. J. Urol. 2021, 28, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Kito, H.; Suzuki, H.; Ichikawa, T.; Sekita, N.; Kamiya, N.; Akakura, K.; Igarashi, T.; Nakayama, T.; Watanabe, M.; Harigaya, K.; et al. Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer. Prostate 2001, 49, 110–115. [Google Scholar] [CrossRef]
- de la Taille, A.; Martínez-Piñeiro, L.; Cabri, P.; Houchard, A.; Schalken, J.; Triptocare LT Study Group. Factors predicting progression to castrate resistant prostate cancer in patients with advanced prostate cancer receiving long-term androgen-deprivation therapy. BJU Int. 2017, 119, 74–81. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Dashzeveg, N.; Cao, Y.; Jia, Y.; Liu, X.; Shen, Y.; Liu, H. Extracellular Domains I and II of cell-surface glycoprotein CD44 mediate its trans-homophilic dimerization and tumor cluster aggregation. J. Biol. Chem. 2020, 295, 2640–2649. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The role of CD44 in epithelial-mesenchymal transition and cancer development. Oncol. Targets Ther. 2015, 8, 3783–3792. [Google Scholar]
- Zöller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A multifunctional mediator of cancer progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef]
- Hurt, E.M.; Kawasaki, B.T.; Klarmann, G.J.; Thomas, S.B.; Farrar, W.L. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer 2008, 98, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Korski, K.; Malicka-Durczak, A.; Bręborowicz, J. Expression of stem cell marker CD44 in prostate cancer biopsies predicts cancer grade in radical prostatectomy specimens. Pol. J. Pathol. 2014, 65, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, B.E.; Leong, K.G.; Yue, P.; Li, L.; Jhunjhunwala, S.; Chen, D.; Seo, K.; Modrusan, Z.; Gao, W.Q.; et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: Implications for androgen deprivation therapy. Cancer Res. 2012, 72, 527–536. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Verma, S.; Kumar, S.; Gupta, S. Role of prostate cancer stem-like cells in the development of antiandrogen resistance. Cancer Drug Resist. 2022, 5, 459–471. [Google Scholar] [CrossRef]
- Verma, P.; Shukla, N.; Kumari, S.; Ansari, M.S.; Gautam, N.K.; Patel, G.K. Cancer stem cell in prostate cancer progression, metastasis and therapy resistance. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188887. [Google Scholar] [CrossRef]
- Niu, Y.; Chang, T.M.; Yeh, S.; Ma, W.L.; Wang, Y.Z.; Chang, C. Differential androgen receptor signals in different cells explain why androgen-deprivation therapy of prostate cancer fails. Oncogene 2010, 29, 3593–3604. [Google Scholar] [CrossRef]
- Frame, F.M.; Maitland, N.J. Epigenetic control of gene expression in the normal and malignant human prostate: A rapid response which promotes therapeutic resistance. Int. J. Mol. Sci. 2019, 20, 2437. [Google Scholar] [CrossRef]
- Skvortsov, S.; Skvortsova, I.I.; Tang, D.G.; Dubrovska, A. Prostate cancer stem cells: Current understanding. Stem Cells 2018, 36, 1457–1474. [Google Scholar] [CrossRef]
- Zoma, M.; Curti, L.; Shinde, D.; Albino, D.; Mitra, A.; Sgrignani, J.; Mapelli, S.N.; Sandrini, G.; Civenni, G.; Merulla, J.; et al. EZH2-induced lysine K362 methylation enhances TMPRSS2-ERG oncogenic activity in prostate cancer. Nat. Commun. 2021, 12, 4147. [Google Scholar] [CrossRef]
- Tolkach, Y.; Zarbl, R.; Bauer, S.; Ritter, M.; Ellinger, J.; Hauser, S.; Hüser, L.; Klauck, S.M.; Altevogt, P.; Sültmann, H.; et al. DNA promoter methylation and ERG regulate the expression of CD24 in prostate cancer. Am. J. Pathol. 2021, 191, 618–630. [Google Scholar] [CrossRef]
- Zhao, S.G.; Chen, W.S.; Li, H.; Foye, A.; Zhang, M.; Sjöström, M.; Aggarwal, R.; Playdle, D.; Liao, A.; Alumkal, J.J.; et al. The DNA methylation landscape of advanced prostate cancer. Nat. Genet. 2020, 52, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Belluti, S.; Imbriano, C.; Casarini, L. Nuclear estrogen receptors in prostate cancer: From genes to function. Cancers 2023, 15, 4653. [Google Scholar] [CrossRef] [PubMed]
- Bernasocchi, T.; El Tekle, G.; Bolis, M.; Mutti, A.; Vallerga, A.; Brandt, L.P.; Spriano, F.; Svinkina, T.; Zoma, M.; Ceserani, V.; et al. Dual functions of SPOP and ERG dictate androgen therapy responses in prostate cancer. Nat. Commun. 2021, 12, 734. [Google Scholar] [CrossRef] [PubMed]
- Tzelepi, V.; Logotheti, S.; Efstathiou, E.; Troncoso, P.; Aparicio, A.; Sakellakis, M.; Hoang, A.; Perimenis, P.; Melachrinou, M.; Logothetis, C.; et al. Epigenetics and prostate cancer: Defining the timing of DNA methyltransferase deregulation during prostate cancer progression. Pathology 2020, 52, 218–227. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017; pp. 715–726. [Google Scholar]
- Netto, G.J.; Amin, M.B.; Kench, J.G. Tumours of the prostate: Introduction. In WHO Classification of Tumours Editorial Board. Urinary and Male Genital Tumours, 5th ed.; IARC Press: Lyon, France, 2022; Volume 8, pp. 194–195. [Google Scholar]
- Evans, A.J. Treatment effects in prostate cancer. Mod. Pathol. 2018, 31, S110–S121. [Google Scholar] [CrossRef]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor angiogenesis and metastasis—Correlation in invasive breast carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef]
- Ceccarelli, V.; Valentini, V.; Ronchetti, S.; Cannarile, L.; Billi, M.; Riccardi, C.; Ottini, L.; Talesa, V.N.; Grignani, F.; Vecchini, A. Eicosapentaenoic acid induces DNA demethylation in carcinoma cells through a TET1-dependent mechanism. FASEB J. 2018, 32, 5990–6001. [Google Scholar] [CrossRef]
| Characteristic a | All Cases (n = 90; 100.0%) | Non-Treated Cases (n = 46; 51.1%) | Treated Cases (n = 44; 48.9%) | p-Value b |
|---|---|---|---|---|
| Age at diagnosis | ||||
| Mean ± SD (range) | 65.3 ± 5.4 (49–73) | 65.3 ± 5.6 (49–73) | 65.3 ± 5.3 (53–73) | 0.89 |
| Gleason grade group | ||||
| 1–2 | 27 (58.7) | 27 (58.7) | - | |
| 3–5 | 19 (41.3) | 19 (41.3) | - | |
| Tumor extent (pT) | ||||
| pT2 | 47 (52.2) | 26 (56.5) | 21 (47.7) | 0.53 |
| pT3 | 43 (47.8) | 20 (43.5) | 23 (52.3) | |
| Lymph node involvement (pN) | ||||
| pN0 | 68 (90.7) | 33 (94.3) | 35 (87.5) | 0.44 |
| pN1 | 7 (9.3) | 2 (5.7) | 5 (12.5) | |
| Surgical margins | ||||
| Negative | 80 (88.9) | 38 (82.6) | 42 (95.5) | 0.09 |
| Positive | 10 (11.1) | 8 (17.4) | 2 (4.5) | |
| Serum CgA, ng/mL | ||||
| Mean ± SD (range) | 85.6 ± 55.2 (27.7–430) | 88.2 ± 43.0 (32.6–218) | 83.1 ± 65.8 (27.7–430) | 0.29 |
| Status | ||||
| No evidence of disease (NED) | 67 (77.9) | 39 (86.7) | 28 (68.3) | 0.067 |
| Disease progression | 19 (22.1) | 6 (13.3) | 13 (31.7) | |
| Gene | All Cases Median (n) | Non-Treated Cases Median (n) | Treated Cases Median (n) | p-Value a |
|---|---|---|---|---|
| AR | 5.00 (82) | 5.00 (41) | 5.00 (41) | 0.59 |
| ESR1 | 4.75 (81) | 4.63 (40) | 4.75 (41) | 0.82 |
| ESR2 | 6.40 (82) | 6.60 (41) | 6.30 (41) | 0.34 |
| APC | 3.70 (80) | 3.70 (39) | 3.70 (41) | 0.22 |
| BCL2 | 6.00 (82) | 6.00 (41) | 5.70 (41) | 0.57 |
| CD44 | 11.70 (82) | 15.20 (41) | 10.00 (41) | 0.001 |
| CDH1 | 10.00 (81) | 9.05 (40) | 10.70 (41) | 0.20 |
| RASSF1 | 11.13 (80) | 12.00 (39) | 10.25 (41) | 0.24 |
| ZEB1 | 3.00 (76) | 3.00 (38) | 2.65 (38) | 0.39 |
| Gene | ERG0 (n = 61) | ERG1 (n = 11) | ERG2 (n = 18) | p-Value a |
|---|---|---|---|---|
| AR | 5.0 (55) | 5.3 (9) | 4.9 (18) | 0.53 |
| ESR1 | 5.0 (54) | 4.5 (9) | 4.3 (18) | 0.014 |
| ESR2 | 6.4 (55) | 6.6 (9) | 6.0 (18) | 0.69 |
| APC | 3.7 (53) | 3.7 (9) | 3.4 (18) | 0.80 |
| BCL2 | 6.0 (55) | 6.0 (9) | 5.5 (18) | 0.57 |
| CD44 | 10.0 (55) | 19.0 (9) | 13.3 (18) | 0.004 |
| CDH1 | 9.8 (55) | 11.3 (8) | 9.8 (18) | 0.72 |
| RASSF1 | 11.5 (53) | 12.3 (9) | 10.0 (18) | 0.50 |
| ZEB1 | 3.0 (50) | 2.7 (9) | 2.7 (17) | 0.83 |
| Non-Treated Cases (n = 46) | Treated Cases (n = 44) | |||
|---|---|---|---|---|
| % a | p-Value b | % a | p-Value b | |
| ERG0 | 13.3 | 0.11 | 9.0 | 0.029 |
| ERG1 | 21.8 | 15.4 | ||
| ERG2 | 15.5 | 10.7 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valentini, V.; Santi, R.; Silvestri, V.; Saieva, C.; Roviello, G.; Amorosi, A.; Compérat, E.; Ottini, L.; Nesi, G. CD44 Methylation Levels in Androgen-Deprived Prostate Cancer: A Putative Epigenetic Modulator of Tumor Progression. Int. J. Mol. Sci. 2025, 26, 2516. https://doi.org/10.3390/ijms26062516
Valentini V, Santi R, Silvestri V, Saieva C, Roviello G, Amorosi A, Compérat E, Ottini L, Nesi G. CD44 Methylation Levels in Androgen-Deprived Prostate Cancer: A Putative Epigenetic Modulator of Tumor Progression. International Journal of Molecular Sciences. 2025; 26(6):2516. https://doi.org/10.3390/ijms26062516
Chicago/Turabian StyleValentini, Virginia, Raffaella Santi, Valentina Silvestri, Calogero Saieva, Giandomenico Roviello, Andrea Amorosi, Eva Compérat, Laura Ottini, and Gabriella Nesi. 2025. "CD44 Methylation Levels in Androgen-Deprived Prostate Cancer: A Putative Epigenetic Modulator of Tumor Progression" International Journal of Molecular Sciences 26, no. 6: 2516. https://doi.org/10.3390/ijms26062516
APA StyleValentini, V., Santi, R., Silvestri, V., Saieva, C., Roviello, G., Amorosi, A., Compérat, E., Ottini, L., & Nesi, G. (2025). CD44 Methylation Levels in Androgen-Deprived Prostate Cancer: A Putative Epigenetic Modulator of Tumor Progression. International Journal of Molecular Sciences, 26(6), 2516. https://doi.org/10.3390/ijms26062516








