Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis)
Abstract
1. Introduction
2. Results
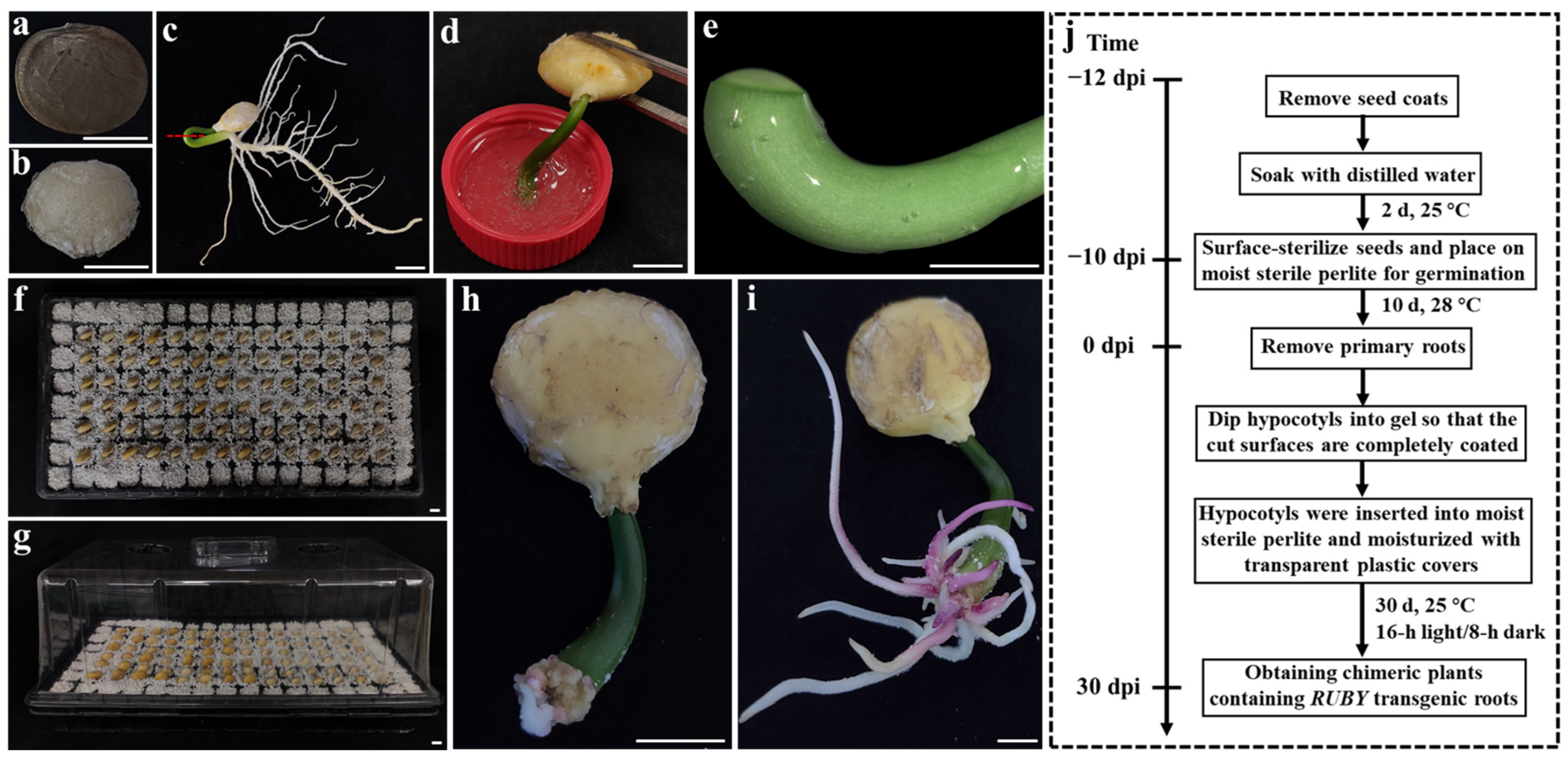
2.1. Establishment of an in Planta A. rhizogenes Gel-Mediated Transformation System for P. volubilis
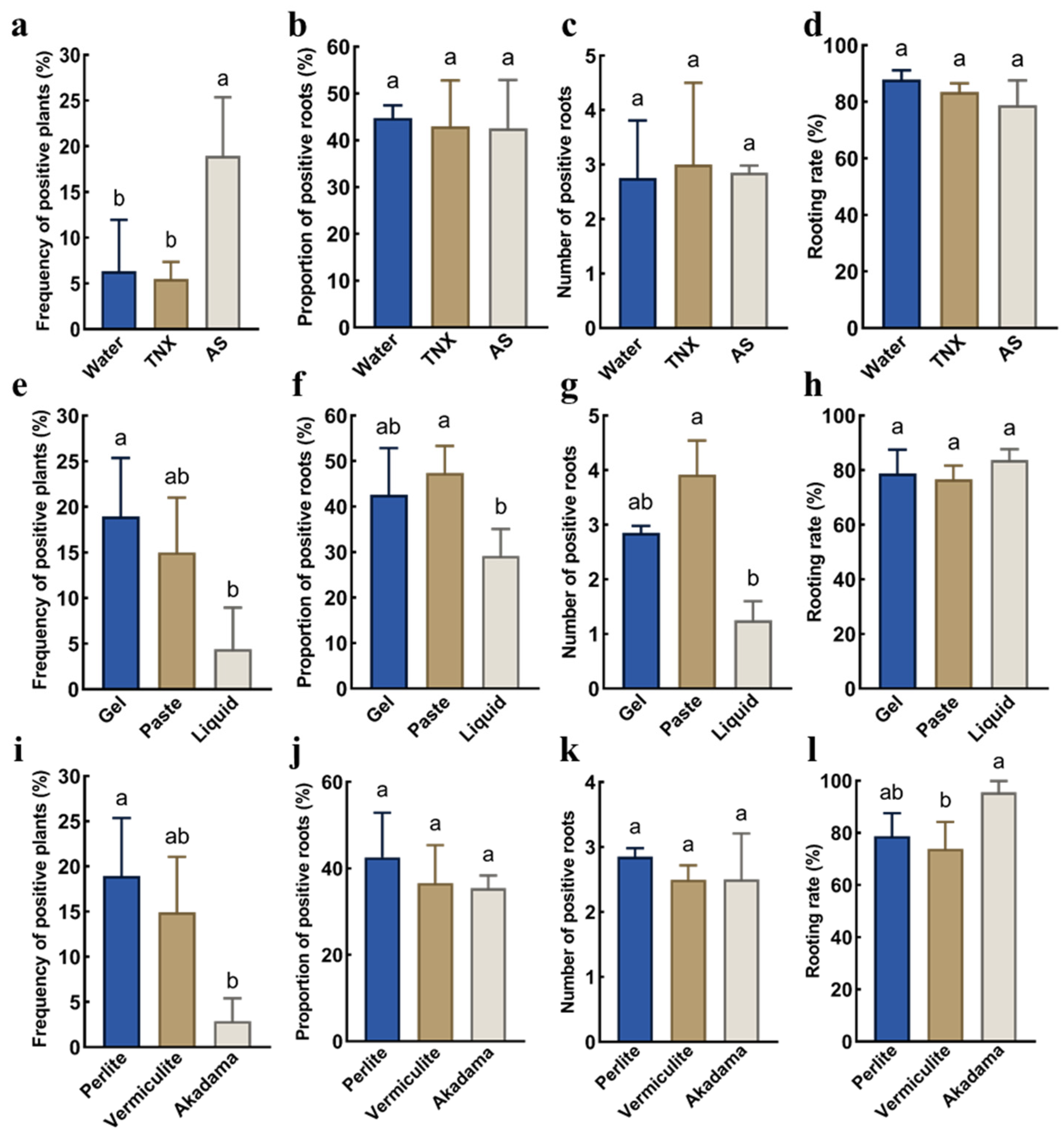
2.2. Effects of Exogenous Additives, Infection Methods, and Potting Substrate Types on Transgenic Hairy Roots of P. volubilis
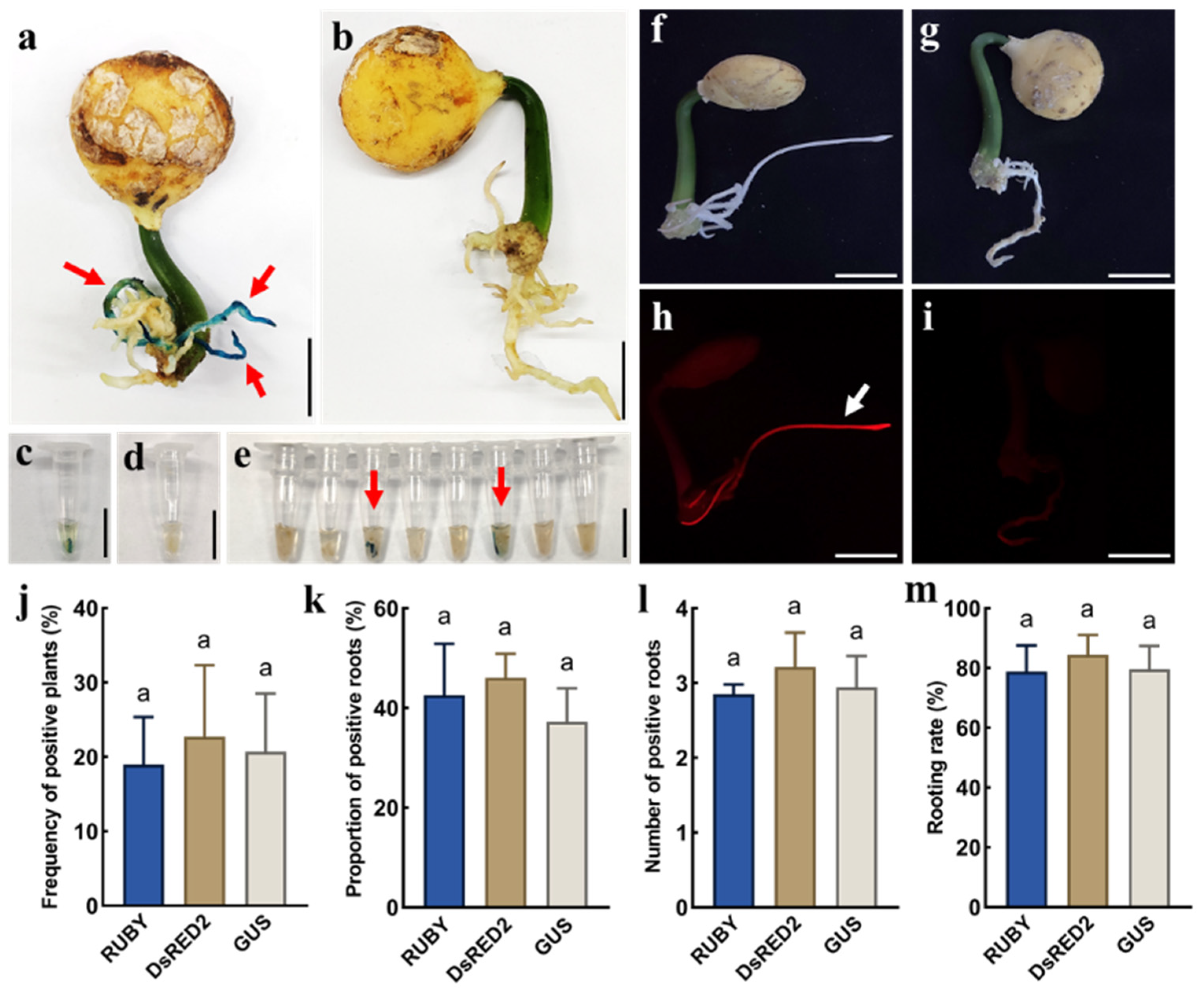
2.3. Transformation of GUS and DsRed2 Reporter Genes via an in Planta Agrobacterium Gel Transformation System
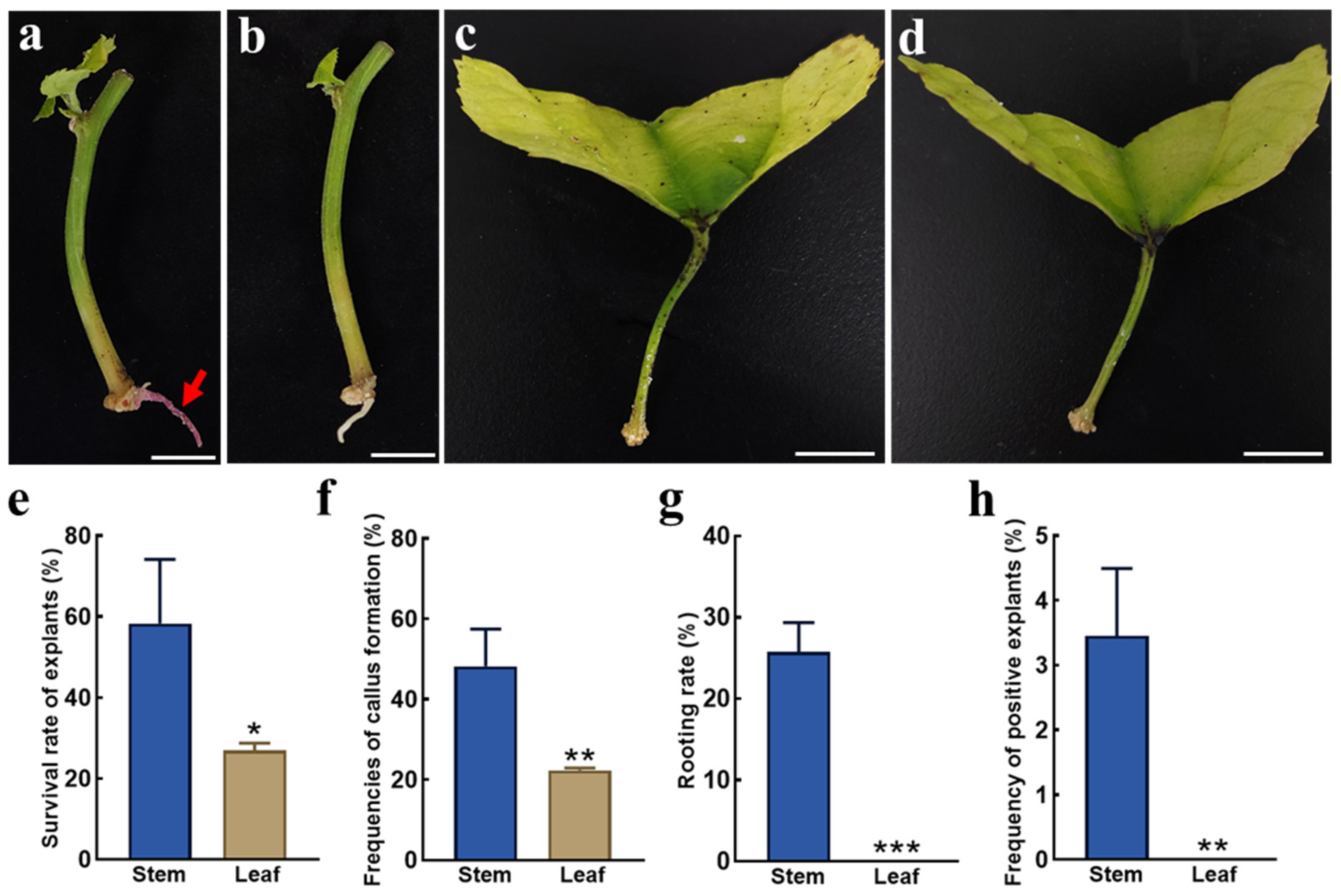
2.4. Applicability of the in Planta Agrobacterium Gel Transformation System in Different Explants of P. volubilis and Other Woody Plants
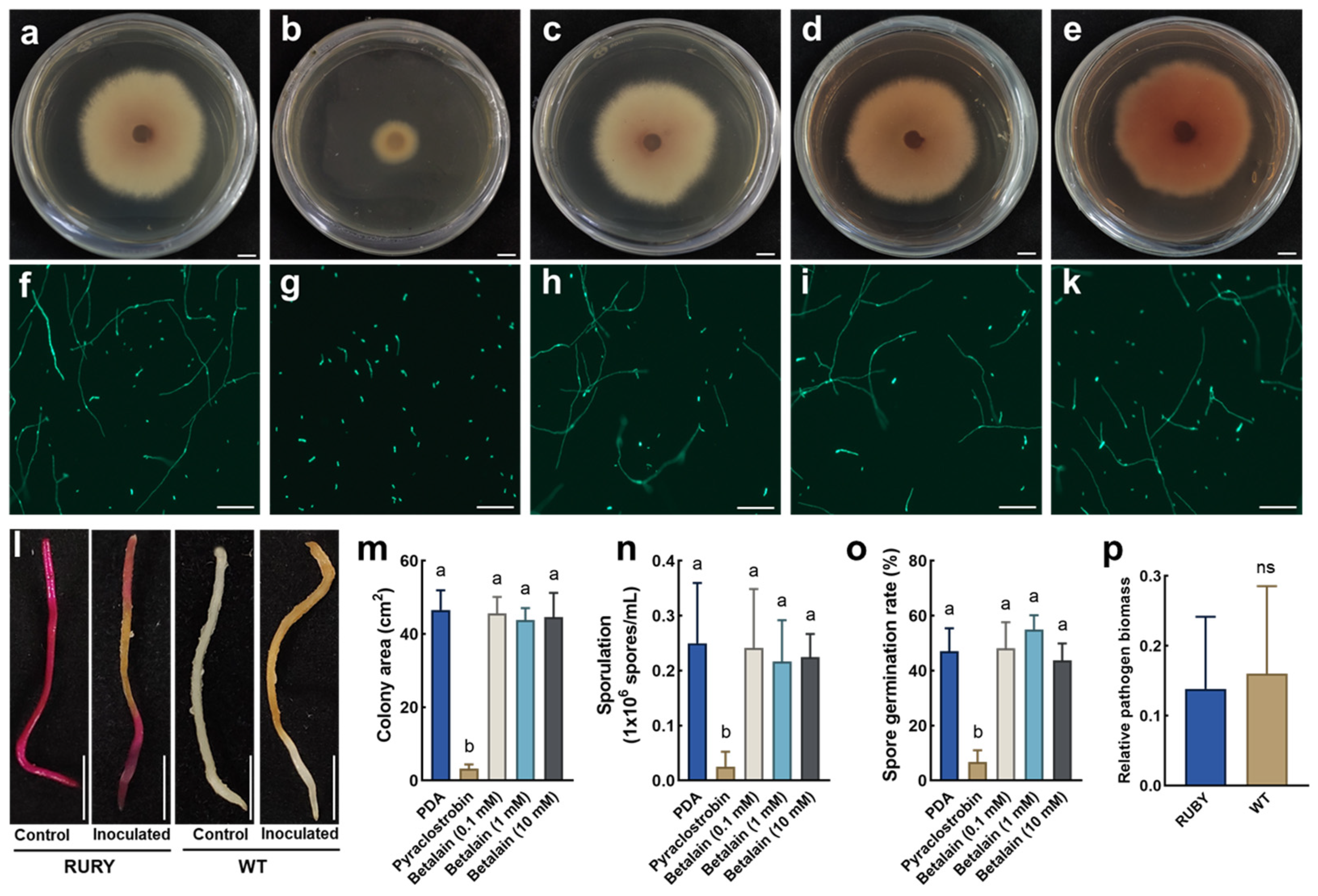
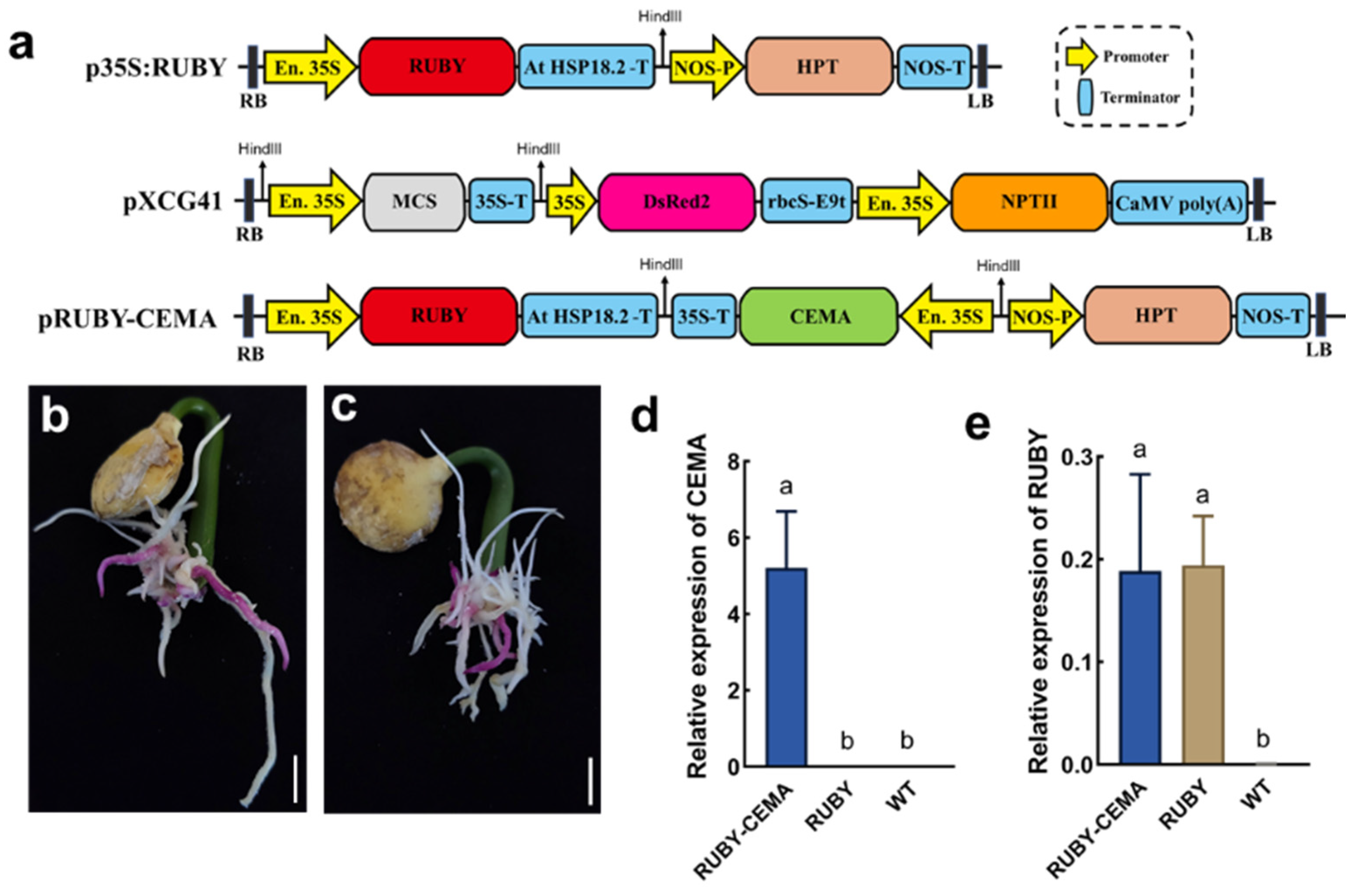
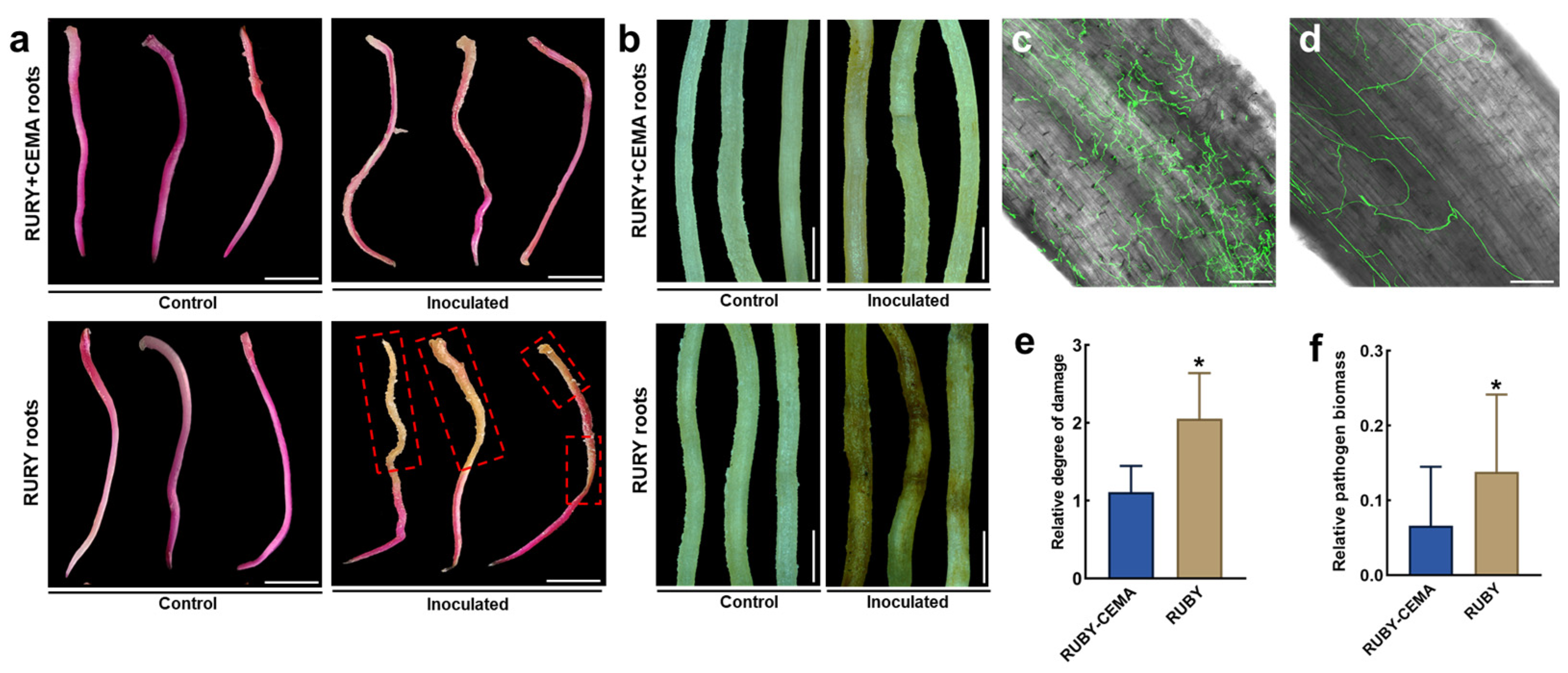
2.5. Analysis of Disease Resistance Gene Function in Roots Using an in Planta Agrobacterium Gel Transformation System
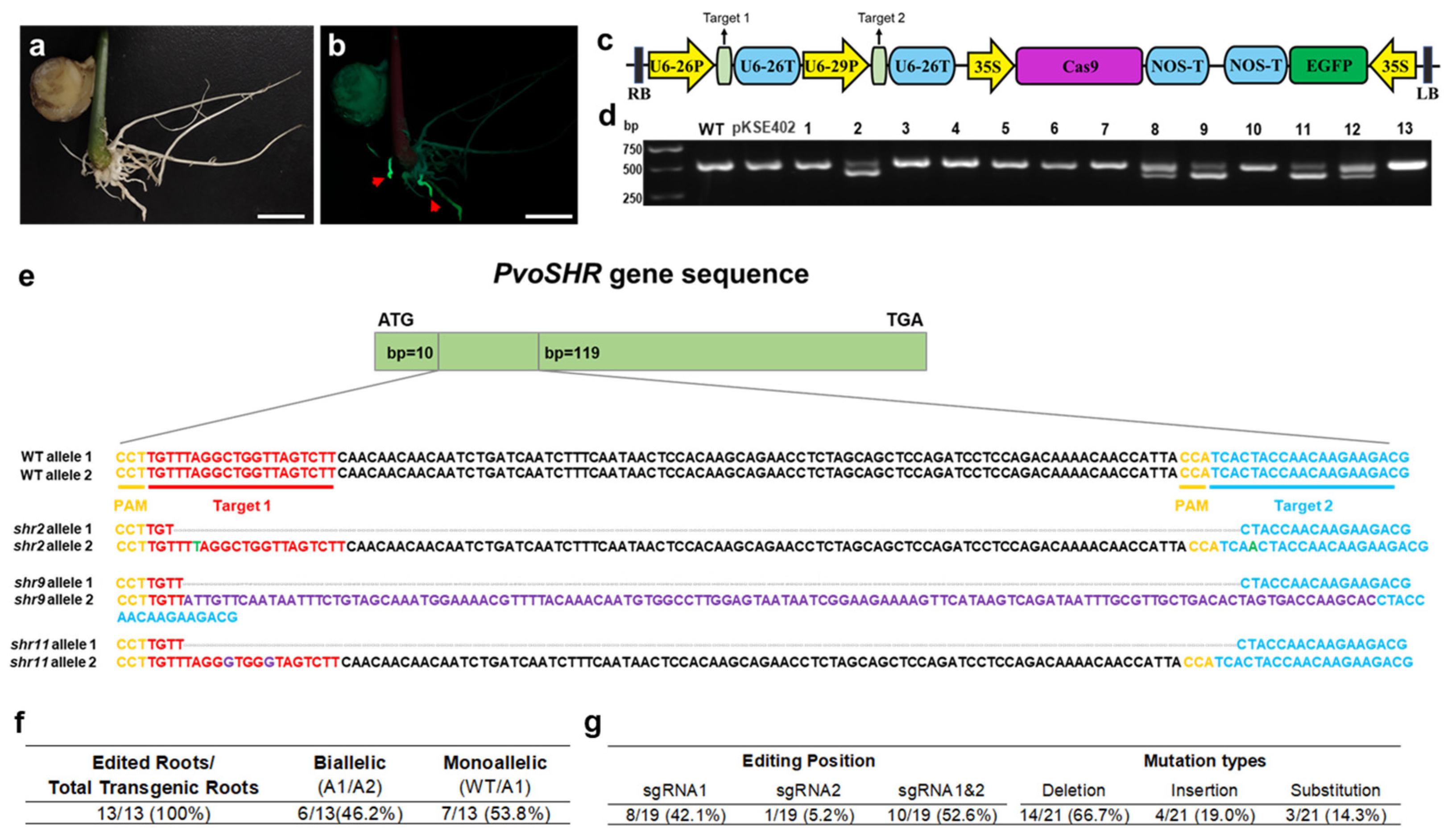
2.6. Gene Editing of P. volubilis Using the in Planta Agrobacterium Gel Transformation System
3. Discussion
3.1. In Planta Agrobacterium-Mediated Transformation of P. volubilis Root System Provides Technical Support for Gene Function Research and Gene Editing
3.2. Gel Is an Ideal Carrier for A. rhizogenes and Helps Induce Hairy Roots
3.3. Feasibility of RUBY as a Visual Screening Reporter for Transgenic Hairy Roots
4. Materials and Methods
4.1. Preparation of Plant Materials
4.2. Vector Construction
4.3. A. rhizogenes-Mediated Hairy Root Transformation System in P. volubilis
4.4. Molecular Identification and Expression Analysis of RUBY Hairy Roots
4.5. Determination of Betalain Content
4.6. Effects of Exogenous Additives on Hairy Root Efficiency in P. volubilis
4.7. Hairy Root Induction Methods for Stem Segments and Petioles of P. volubilis
4.8. Hairy Root Induction in P. corniculata, Malus Domestica, Morus Alba, and Jatropha Curcas
4.9. Measurement of Colony Area, Spore Production, and Spore Germination Rate of F. oxysporum
4.10. Inoculation of P. volubilis Hairy Roots with FoPvo1-GFP
4.11. Observation of F. oxysporum Colonization by Laser Confocal Microscopy
4.12. Antibacterial Activity Assay of Root Extracts on Agar Plates
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kodahl, N.; Sørensen, M. Sacha Inchi (Plukenetia volubilis L.) Is an Underutilized Crop with a Great Potential. Agronomy 2021, 11, 1066. [Google Scholar] [CrossRef]
- Follegatti-Romero, L.A.; Piantino, C.R.; Grimaldi, R.; Cabral, F.A. Supercritical CO2 extraction of omega-3 rich oil from Sacha inchi (Plukenetia volubilis L.) seeds. J. Supercrit. Fluids 2009, 49, 323–329. [Google Scholar] [CrossRef]
- Wang, S.N.; Zhu, F.; Kakuda, Y. Sacha inchi (Plukenetia volubilis L.): Nutritional composition, biological activity, and uses. Food Chem. 2018, 265, 316–328. [Google Scholar] [CrossRef]
- Gonzales, G.F.; Gonzales, C. A randomized, double-blind placebo-controlled study on acceptability, safety and efficacy of oral administration of sacha inchi oil (Plukenetia volubilis L.) in adult human subjects. Food Chem. Toxicol. 2014, 65, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Aspajo, G.; Belkhelfa, H.; Haddioui-Hbabi, L.; Bourdy, G.; Deharo, E. Sacha Inchi Oil (Plukenetia volubilis L.), effect on adherence of Staphylococus aureus to human skin explant and keratinocytes in vitro. J. Ethnopharmacol. 2015, 171, 330–334. [Google Scholar] [CrossRef]
- Hu, X.-D.; Pan, B.-Z.; Fu, Q.; Niu, L.; Chen, M.-S.; Xu, Z.-F. De novo transcriptome assembly of the eight major organs of Sacha Inchi (Plukenetia volubilis) and the identification of genes involved in alpha-linolenic acid metabolism. BMC Genom. 2018, 19, 380. [Google Scholar] [CrossRef]
- Liu, G.; Wu, Z.; Shang, X.; Peng, Y.; Gao, L. Overexpression of PvFAD3 gene from Plukenetia volubilis promotes the biosynthesis of α-Linolenic acid in transgenic tobacco seeds. Genes 2022, 13, 450. [Google Scholar] [CrossRef]
- Fu, Q.; Niu, L.; Chen, M.; Tao, Y.; Wang, X.; He, H.; Pan, B.; Xu, Z.-F. De novo transcriptome assembly and comparative analysis between male and benzyladenine-induced female inflorescence buds of Plukenetia volubilis. J. Plant Physiol. 2018, 221, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wu, Z.; Peng, Y.; Shang, X.; Gao, L. Integrated Transcriptome and Proteome Analysis Provides Insight into the Ribosome Inactivating Proteins in Plukenetia volubilis Seeds. Int. J. Mol. Sci. 2022, 23, 9562. [Google Scholar] [CrossRef]
- Ono, N.N.; Tian, L. The multiplicity of hairy root cultures: Prolific possibilities. Plant Sci. 2011, 180, 439–446. [Google Scholar] [CrossRef]
- Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants 2021, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Irigoyen, S.; Ramasamy, M.; Pant, S.; Niraula, P.; Bedre, R.; Gurung, M.; Rossi, D.; Laughlin, C.; Gorman, Z.; Achor, D.; et al. Plant hairy roots enable high throughput identification of antimicrobials against Candidatus Liberibacter spp. Nat. Commun. 2020, 11, 5802. [Google Scholar] [CrossRef]
- Li, J.; Zeng, T.; Xu, Z.; Li, J.; Hu, H.; Yu, Q.; Zhou, L.; Zheng, R.; Luo, J.; Wang, C. Ribozyme-mediated CRISPR/Cas9 gene editing in pyrethrum (Tanacetum cinerariifolium) hairy roots using a RNA polymerase II-dependent promoter. Plant Methods 2022, 18, 32. [Google Scholar] [CrossRef]
- Yu, G.; Zou, J.; Wang, J.; Zhu, R.; Qi, Z.; Jiang, H.; Hu, Z.; Yang, M.; Zhao, Y.; Wu, X.; et al. A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins. Crop J. 2022, 10, 332–341. [Google Scholar] [CrossRef]
- Xiao, X.; Ma, F.; Chen, C.; Guo, W. High efficient transformation of auxin reporter gene into trifoliate orange via Agrobacterium rhizogenes-mediated co-transformation. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 118, 137–146. [Google Scholar] [CrossRef]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut–dip–budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2023, 4, 100345. [Google Scholar] [CrossRef]
- Liu, L.; Qu, J.; Wang, C.; Liu, M.; Zhang, C.; Zhang, X.; Guo, C.; Wu, C.; Yang, G.; Huang, J.; et al. An efficient genetic transformation system mediated by Rhizobium rhizogenes in fruit trees based on the transgenic hairy root to shoot conversion. Plant Biotechnol. J. 2024, 22, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Saifi, S.K.; Passricha, N.; Tuteja, R.; Kharb, P.; Tuteja, N. In planta transformation: A smart way of crop improvement. In Advancement in Crop Improvement Techniques; Tuteja, N., Tuteja, R., Passricha, N., Saifi, S.K., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 351–362. [Google Scholar]
- Yevtushenko, D.P.; Romero, R.; Forward, B.S.; Hancock, R.E.; Kay, W.W.; Misra, S. Pathogen-induced expression of a cecropin A-melittin antimicrobial peptide gene confers antifungal resistance in transgenic tobacco. J. Exp. Bot. 2005, 56, 1685–1695. [Google Scholar] [CrossRef]
- Li, X.; Niu, G.; Fan, Y.; Liu, W.; Wu, Q.; Yu, C.; Wang, J.; Xiao, Y.; Hou, L.; Jin, D.; et al. Synthetic dual hormone-responsive promoters enable engineering of plants with broad-spectrum resistance. Plant Commun. 2023, 4, 100596. [Google Scholar] [CrossRef]
- Triozzi, P.M.; Schmidt, H.W.; Dervinis, C.; Kirst, M.; Conde, D. Simple, efficient and open-source CRISPR/Cas9 strategy for multi-site genome editing in Populus tremula × alba. Tree Physiol. 2021, 41, 2216–2227. [Google Scholar] [CrossRef]
- Murphy, E. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″) (9). Mol. Gen. Genet. 1985, 200, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Sheikholeslam, S.N.; Weeks, D.P. Acetosyringone promotes high efficiency transformation of Arabidopsis thaliana explants by Agrobacterium tumefaciens. Plant Mol. Biol. 1987, 8, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-w.; Kumaishi, K.; Motohashi, R.; Enoki, H.; Chacuttayapong, W.; Takamizo, T.; Saika, H.; Endo, M.; Yamada, T.; Hirose, A.; et al. Oxicam-type nonsteroidal anti-inflammatory drugs enhance Agrobacterium-mediated transient transformation in plants. Plant Biotechnol. 2022, 39, 323–327. [Google Scholar] [CrossRef]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Xie, L.; Gong, X.; Yang, K.; Huang, Y.; Zhang, S.; Shen, L.; Sun, Y.; Wu, D.; Ye, C.; Zhu, Q.-H. Technology-enabled great leap in deciphering plant genomes. Nat. Plants 2024, 10, 551–566. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, J.; Zhao, J.; Niu, S. Genetic transformation in conifers: Current status and future prospects. For. Res. 2024, 4, e010. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, C.; Xiao, D.; Liang, Y.; Wang, Y. Advances and perspectives of transgenic technology and biotechnological application in forest trees. Front. Plant Sci. 2021, 12, 786328. [Google Scholar] [CrossRef] [PubMed]
- Alagarsamy, K.; Shamala, L.F.; Wei, S. Protocol: High-efficiency in-planta Agrobacterium-mediated transgenic hairy root induction of Camellia sinensis var. sinensis. Plant Methods 2018, 14, 17. [Google Scholar] [CrossRef]
- Gomes, C.; Dupas, A.; Pagano, A.; Grima-Pettenati, J.; Paiva, J.A.P. Hairy Root Transformation: A Useful Tool to Explore Gene Function and Expression in Salix spp. Recalcitrant to Transformation. Front. Plant Sci. 2019, 10, 1427. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, G.; Dupas, A.; Medina, L.; Blandels, N.; Clemente, H.S.; Ladouce, N.; Badawi, M.; Hernandez-Raquet, G.; Mounet, F.; et al. Implementing the CRISPR/Cas9 Technology in Eucalyptus Hairy Roots Using Wood-Related Genes. Int. J. Mol. Sci. 2020, 21, 3408. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lai, E.; Zhao, L.; Cai, Y.; Ogutu, C.; Cherono, S.; Han, Y.; Zheng, B. Development of a fast and efficient root transgenic system for functional genomics and genetic engineering in peach. Sci. Rep. 2020, 10, 2836. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, D.; Fu, J.; Zhang, Z.; Qin, Y.; Hu, G.; Zhao, J. Agrobacterium rhizogenes-mediated hairy root transformation as an efficient system for gene function analysis in Litchi chinensis. Plant Methods 2021, 17, 103. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Chen, J.; Zhang, L.; Zhang, L. The First Report on Transgenic Hairy Root Induction from the Stem of Tung Tree (Vernicia fordii). Plants 2022, 11, 1315. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Zhang, Y.; Chai, X.; Fu, Q.; Geng, X.; Ni, J.; Xu, Z.-F. Establishment of an Agrobacterium rhizogenes–mediated genetic transformation system of hairy roots in sacha inchi (Plukenetia volubilis L.). Plant Physiol. J. 2023, 59, 373–382. [Google Scholar]
- Yu, J.; Deng, S.; Huang, H.; Mo, J.; Xu, Z.-F.; Wang, Y. Exploring the Potential Applications of the Noninvasive Reporter Gene RUBY in Plant Genetic Transformation. Forests 2023, 14, 637. [Google Scholar] [CrossRef]
- Meteier, E.; La Camera, S.; Goddard, M.L.; Laloue, H.; Mestre, P.; Chong, J. Overexpression of the VvSWEET4 Transporter in Grapevine Hairy Roots Increases Sugar Transport and Contents and Enhances Resistance to Pythium irregulare, a Soilborne Pathogen. Front. Plant Sci. 2019, 10, 884. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, L.; Wang, P.; Liao, Y.; Duan, L.; Lin, K.; Chen, X.; Li, L.; Xu, J.; Hu, H.; et al. Transcriptome-Based Construction of the Gibberellin Metabolism and Signaling Pathways in Eucalyptus grandis × E. urophylla, and Functional Characterization of GA20ox and GA2ox in Regulating Plant Development and Abiotic Stress Adaptations. Int. J. Mol. Sci. 2023, 24, 7051. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, Y.; Lin, K.; Wu, W.; Duan, L.; Wang, P.; Xiao, X.; Zhang, T.; Chen, X.; Wang, J.; et al. Cytokinin promotes anthocyanin biosynthesis via regulating sugar accumulation and MYB113 expression in Eucalyptus. Tree Physiol. 2024, 44, tpad154. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, Y.; Zhang, T.; Zeng, Z.; Wang, J.; Duan, L.; Chen, X.; Lin, K.; Liang, X.; Han, Z.; et al. Reactive oxygen species act as the key signaling molecules mediating light-induced anthocyanin biosynthesis in Eucalyptus. Plant Physiol. Biochem. 2024, 212, 108715. [Google Scholar] [CrossRef]
- Liao, Y.; Zeng, Z.; Lin, K.; Jiang, W.; Wang, J.; Duan, L.; Liang, X.; Huang, Y.; Han, Z.; Hu, H.; et al. Gibberellin promotes xylem expansion and cell lignification by regulating sugar accumulation and the expression of JcMYB43 and JcMYB63 in the woody plant Jatropha curcas. Int. J. Biol. Macromol. 2025, 294, 139434. [Google Scholar] [CrossRef]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.-T.; Benfey, P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef]
- Kim, S.-I.; Veena; Gelvin, S.B. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007, 51, 779–791. [Google Scholar] [CrossRef]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the Plant Genome. In Annual Review of Genetics; Bonini, N.M., Ed.; Annual Reviews: Palo Alto, CA, USA, 2017; Volume 51, pp. 195–217. [Google Scholar]
- Salomon, S.; Puchta, H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998, 17, 6086–6095. [Google Scholar] [CrossRef]
- Chilton, M.-D.M.; Que, Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: New insights on the mechanism of T-DNA integration. Plant Physiol. 2003, 133, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xing, H.-L.; Wang, Z.-P.; Zhang, H.-Y.; Yang, F.; Wang, X.-C.; Chen, Q.-J. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol. Biol. 2018, 96, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Eggenberger, A.L.; Banakar, R.; McCaw, M.E.; Zhu, H.; Main, M.; Kang, M.; Gelvin, S.B.; Wang, K. CRISPR/Cas9-mediated targeted T-DNA integration in rice. Plant Mol. Biol. 2019, 99, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, T.; Prange, A.; Schäfer, P.; Iwen, T.; Grützner, R.; Marillonnet, S.; Lepage, A.; Javelle, M.; Paul, W.; Tissier, A. Efficient scar-free knock-ins of several kilobases in plants by engineered CRISPR-Cas endonucleases. Molecular Plant 2024, 17, 824–837. [Google Scholar] [CrossRef]
- Burdock, G.A. Safety assessment of hydroxypropyl methylcellulose as a food ingredient. Food Chem. Toxicol. 2007, 45, 2341–2351. [Google Scholar] [CrossRef]
- Ghadermazi, R.; Hamdipour, S.; Sadeghi, K.; Ghadermazi, R.; Khosrowshahi Asl, A. Effect of various additives on the properties of the films and coatings derived from hydroxypropyl methylcellulose—A review. Food Sci. Nutr. 2019, 7, 3363–3377. [Google Scholar] [CrossRef]
- Fan, Y.L.; Zhang, X.H.; Zhong, L.J.; Wang, X.Y.; Jin, L.S.; Lyu, S.H. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 2020, 20, 208. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, Z.; Ma, J.; Chen, H.; He, J.; Liu, X.; Zhang, Y.; Liu, H.; Ye, D.; Tang, C. Efficient vegetative propagation and genetic transformation of Russian dandelion (Taraxacum kok-saghyz Rodin) from leaf explants. Ind. Crops Prod. 2024, 209, 118072. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Wang, W.; Wang, Y.; Chen, X.; Wu, H.; Gao, Z.; Xu, H.; Liu, T.; Li, Y.; et al. Efficient genetic transformation and gene editing of Chinese cabbage using Agrobacterium rhizogenes. Plant Physiol. 2024, 197, kiae543. [Google Scholar] [CrossRef] [PubMed]
- Jogam, P.; Anumula, V.; Sandhya, D.; Manokari, M.; Venkatapuram, A.K.; Achary, V.M.M.; Shekhawat, M.S.; Peddaboina, V.; Allini, V.R. Monitoring genetic transformation with RUBY visible reporter in Nicotiana tabaccum L. Plant Cell Tissue Organ Cult. (PCTOC) 2024, 157, 23. [Google Scholar] [CrossRef]
- He, Y.; Zhang, T.; Sun, H.; Zhan, H.; Zhao, Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020, 7, 152. [Google Scholar] [CrossRef]
- Pan, W.; Cheng, Z.; Han, Z.; Yang, H.; Zhang, W.; Zhang, H. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing of watermelon assisted by genes encoding developmental regulators. J. Zhejiang Univ.-Sci. B 2022, 23, 339–344. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002, 129, 706–716. [Google Scholar] [CrossRef]
- Engler, C.; Gruetzner, R.; Kandzia, R.; Marillonnet, S. Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS ONE 2009, 4, e5553. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Cheng, Z.; Song, X.; Liu, X.; Yan, S.; Song, W.; Wang, Z.; Han, L.; Zhao, J.; Yan, L.; Zhou, Z.; et al. SPATULA and ALCATRAZ confer female sterility and fruit cavity via mediating pistil development in cucumber. Plant Physiol. 2022, 189, 1553–1569. [Google Scholar] [CrossRef]
- Fu, Q.; Li, C.; Tang, M.; Tao, Y.-B.; Pan, B.-Z.; Zhang, L.; Niu, L.; He, H.; Wang, X.; Xu, Z.-F. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 2015, 9, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Tao, Y.-B.; Chen, M.-S.; Fu, Q.; Li, C.; Dong, Y.; Wang, X.; He, H.; Xu, Z.-F. Selection of Reliable Reference Genes for Gene Expression Studies of a Promising Oilseed Crop, Plukenetia volubilis, by Real-Time Quantitative PCR. Int. J. Mol. Sci. 2015, 16, 12513–12530. [Google Scholar] [CrossRef] [PubMed]
- Herbach, K.M.; Maier, C.; Stintzing, F.C.; Carle, R. Effects of processing and storage on juice colour and betacyanin stability of purple pitaya (Hylocereus polyrhizus) juice. Eur. Food Res. Technol. 2007, 224, 649–658. [Google Scholar] [CrossRef]
- Chai, X.; Yang, Z.; Fu, Q.; Pan, B.Z.; Tang, M.; Li, C.; Xu, Z.-F. First Report of Root and Basal Stem Rot in Sacha Inchi (Plukenetia volubilis) Caused by Fusarium oxysporum in China. Plant Dis. 2018, 102, 242. [Google Scholar] [CrossRef]
- Lorang, J.; Tuori, R.; Martinez, J.; Sawyer, T.; Redman, R.; Rollins, J.; Wolpert, T.; Johnson, K.; Rodriguez, R.; Dickman, M.; et al. Green Fluorescent Protein Is Lighting Up Fungal Biology. Appl. Environ. Microbiol. 2001, 67, 1987–1994. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Ishimoto, A.; Sakai, Y.; Sato, M.; Nishihama, R.; Abe, K.; Sano, Y.; Furuichi, T.; Tsuji, H.; Kohchi, T.; et al. Improved clearing method contributes to deep imaging of plant organs. Commun. Biol. 2022, 5, 12. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, K.; Lu, L.-X.; Pan, B.-Z.; Chai, X.; Fu, Q.-T.; Geng, X.-C.; Mo, Y.; Fei, Y.-C.; Xu, J.-J.; Li, M.; et al. Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis). Int. J. Mol. Sci. 2025, 26, 2496. https://doi.org/10.3390/ijms26062496
Lin K, Lu L-X, Pan B-Z, Chai X, Fu Q-T, Geng X-C, Mo Y, Fei Y-C, Xu J-J, Li M, et al. Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis). International Journal of Molecular Sciences. 2025; 26(6):2496. https://doi.org/10.3390/ijms26062496
Chicago/Turabian StyleLin, Kai, Li-Xin Lu, Bang-Zhen Pan, Xia Chai, Qian-Tang Fu, Xian-Chen Geng, Yi Mo, Yu-Chong Fei, Jia-Jing Xu, Meng Li, and et al. 2025. "Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis)" International Journal of Molecular Sciences 26, no. 6: 2496. https://doi.org/10.3390/ijms26062496
APA StyleLin, K., Lu, L.-X., Pan, B.-Z., Chai, X., Fu, Q.-T., Geng, X.-C., Mo, Y., Fei, Y.-C., Xu, J.-J., Li, M., Ni, J., & Xu, Z.-F. (2025). Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis). International Journal of Molecular Sciences, 26(6), 2496. https://doi.org/10.3390/ijms26062496







