Genome-Wide Identification of GLK Family Genes in Phoebe bournei and Their Transcriptional Analysis Under Abiotic Stresses
Abstract
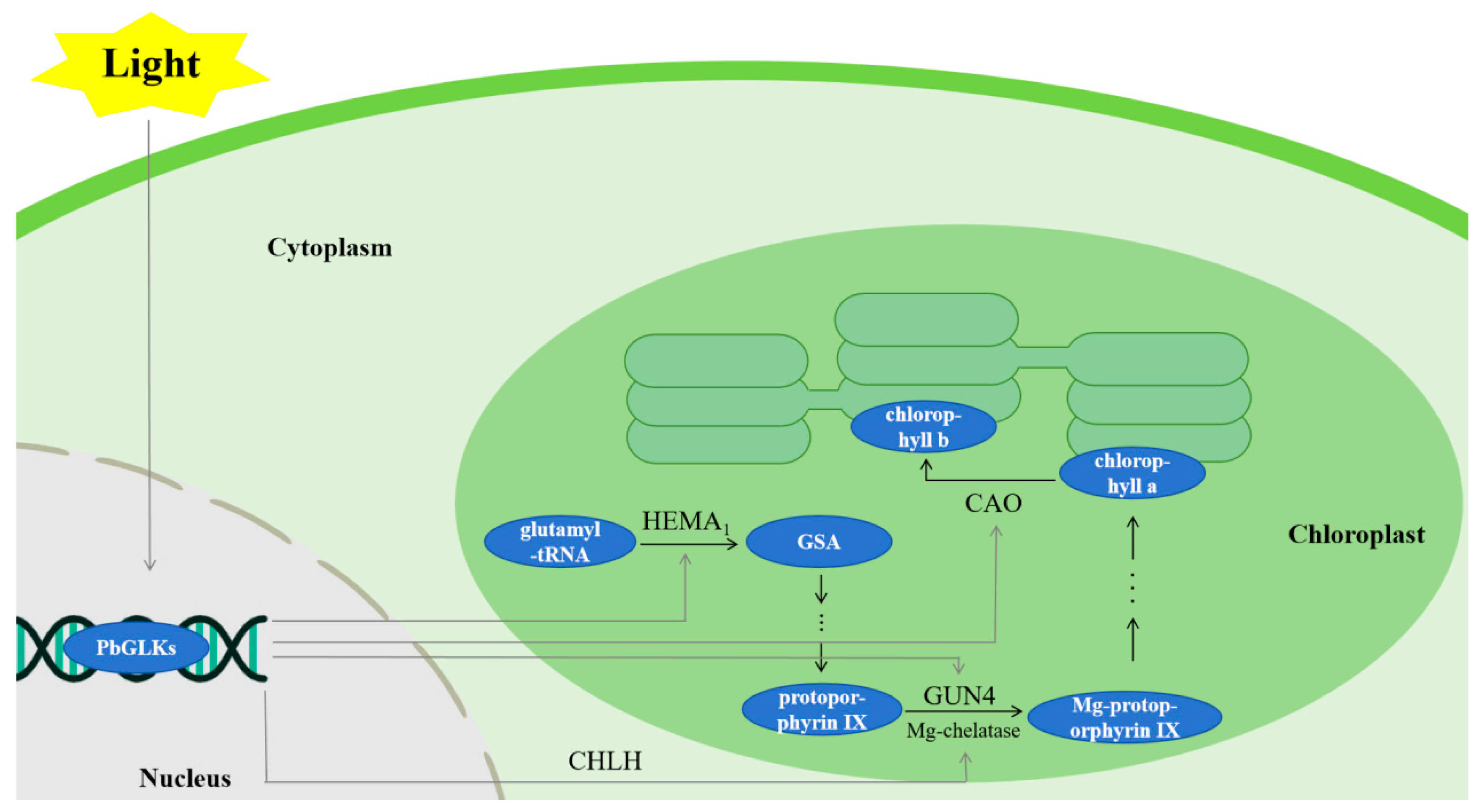
1. Introduction
2. Results
2.1. Identification of PbGLK Genes in P. bournei
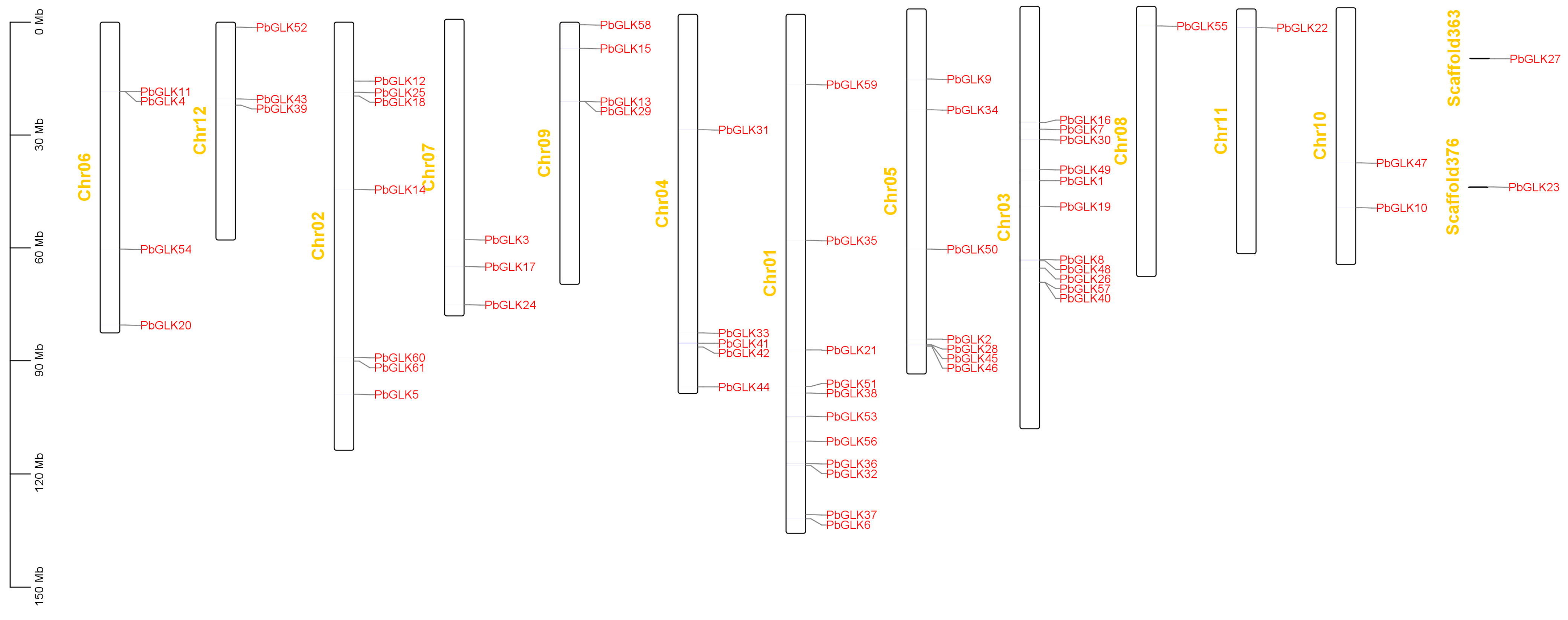
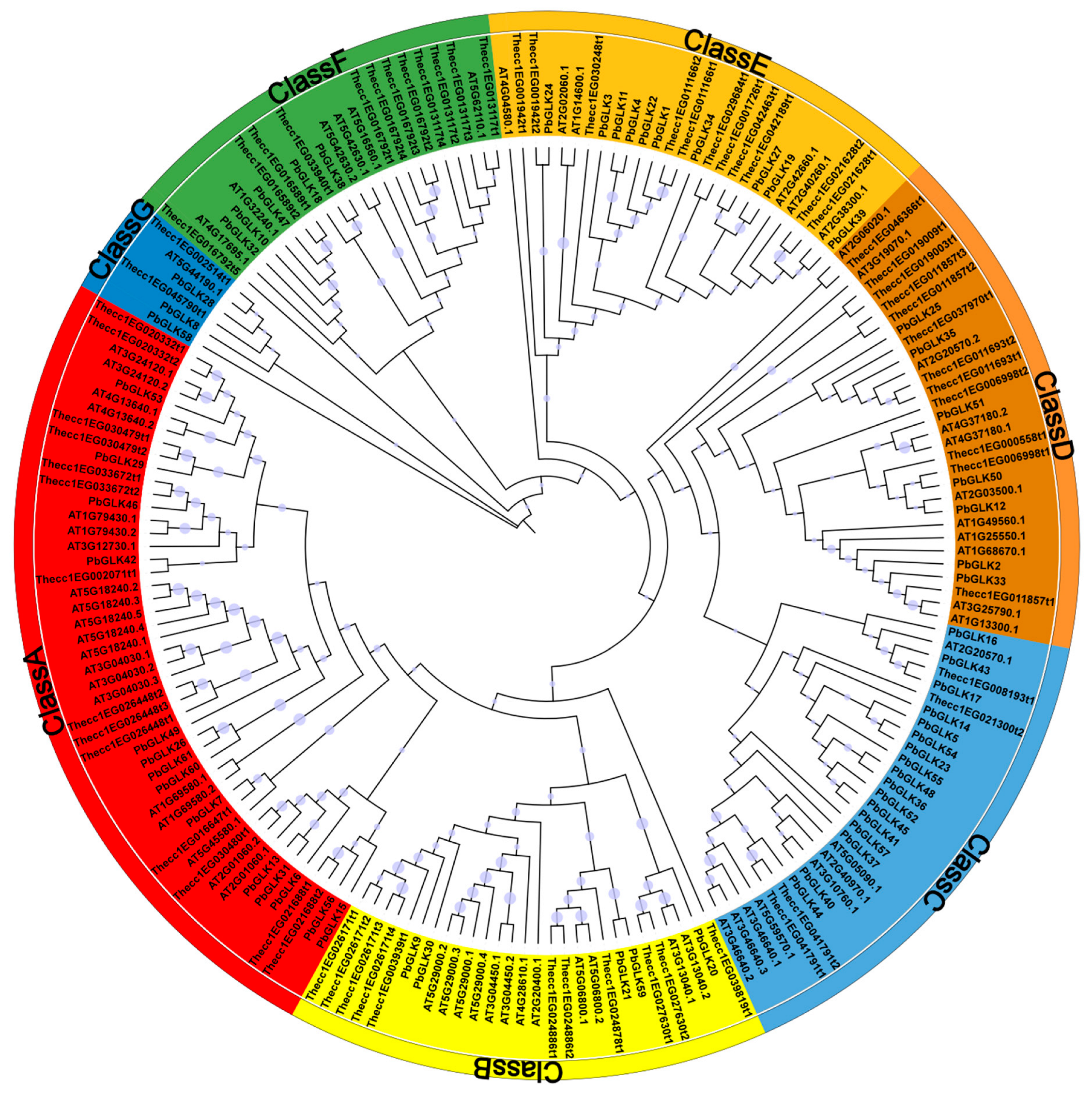
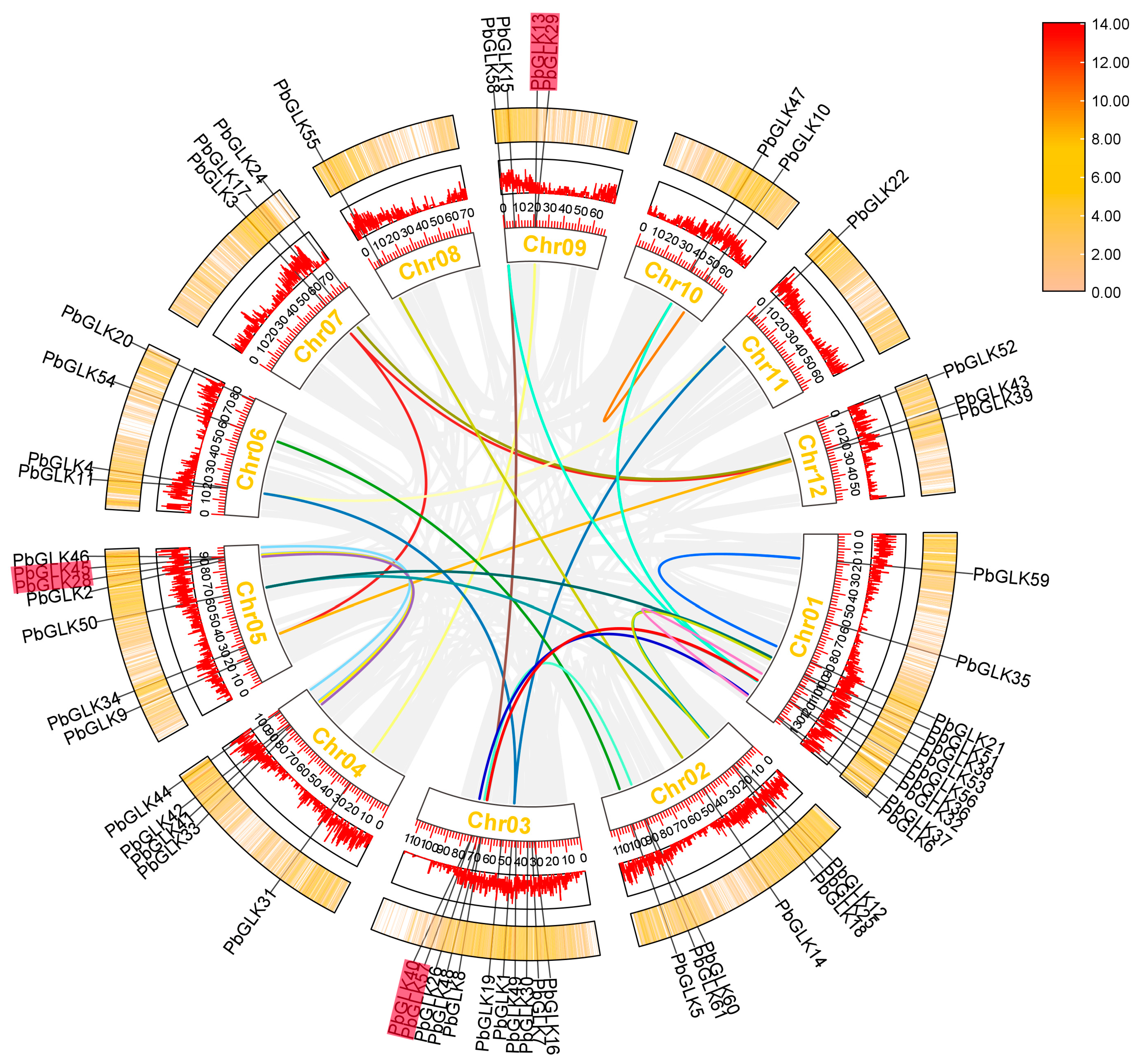
2.2. Phylogenetic Analysis and PbGLK’s Chromosomal Locations
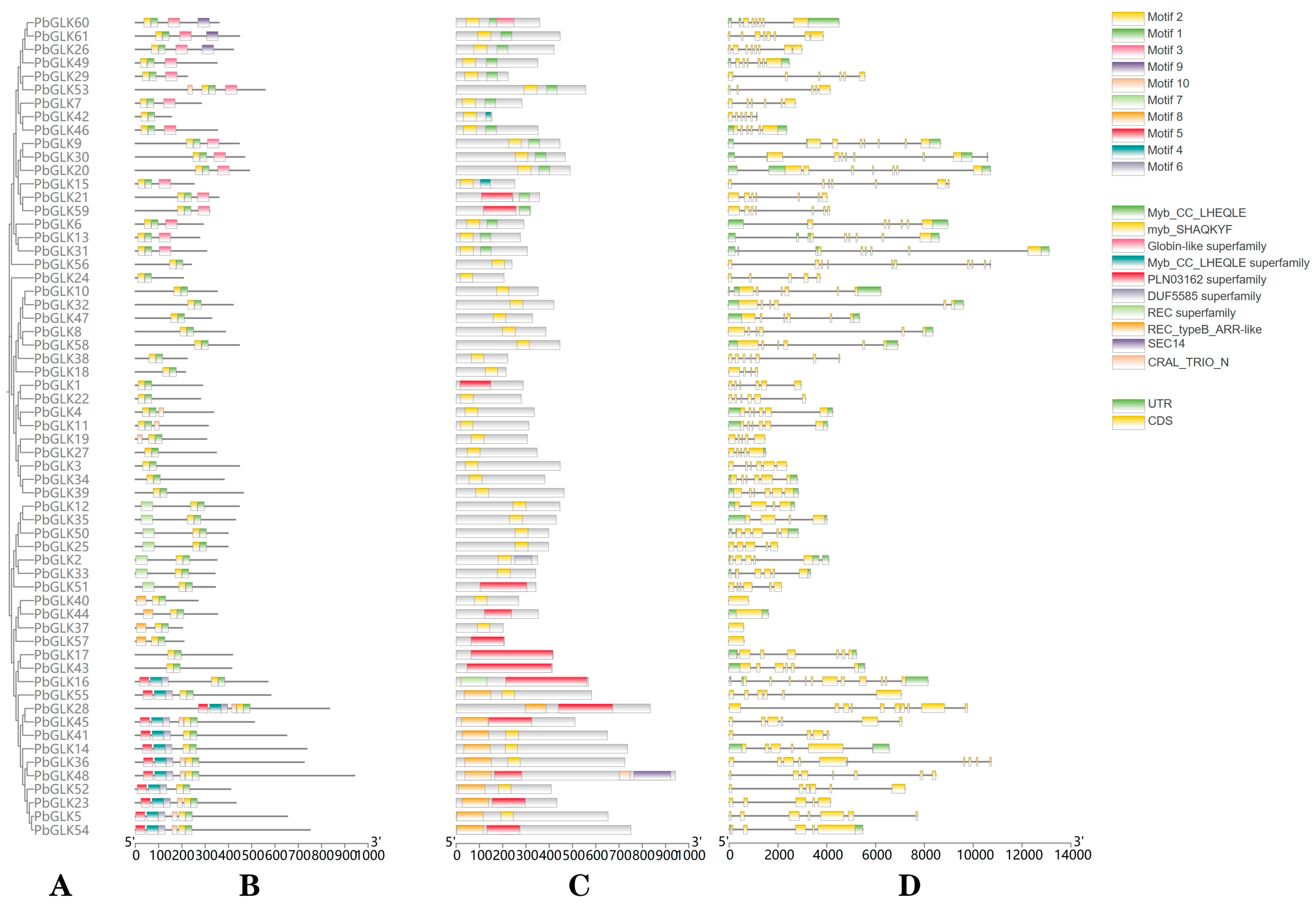
2.3. PbGLK Gene Structure, Conserved Motif, and Domain Analysis
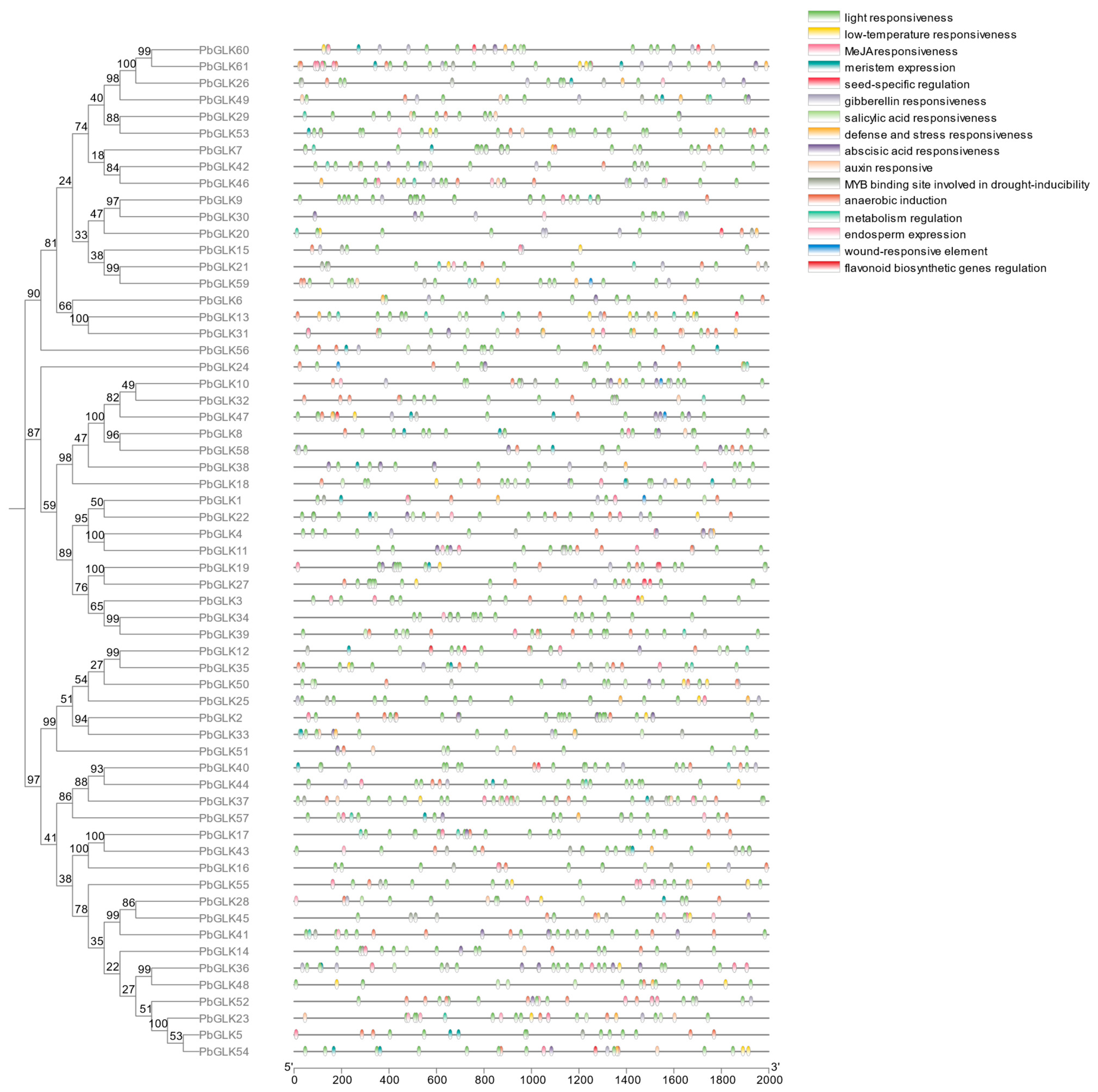
2.4. Cis-Acting Elements: Analysis of the PbGLK Gene Family
2.5. Intraspecific Collinearity Analysis of PbGLK Genes
2.6. Synteny Analysis Among PbGLK Genes
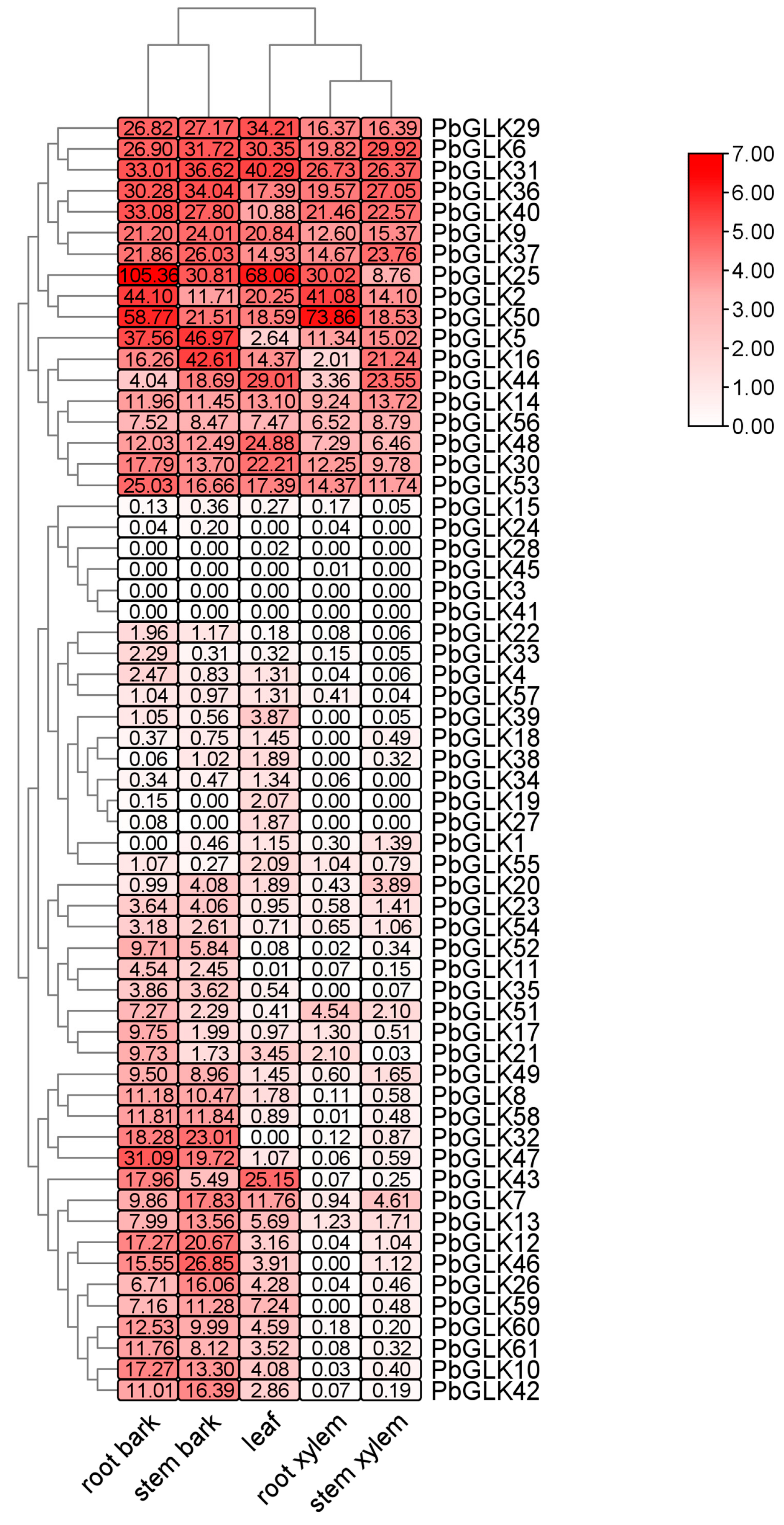
2.7. Expression Analysis of PbGLKs in P. bournei Tissues
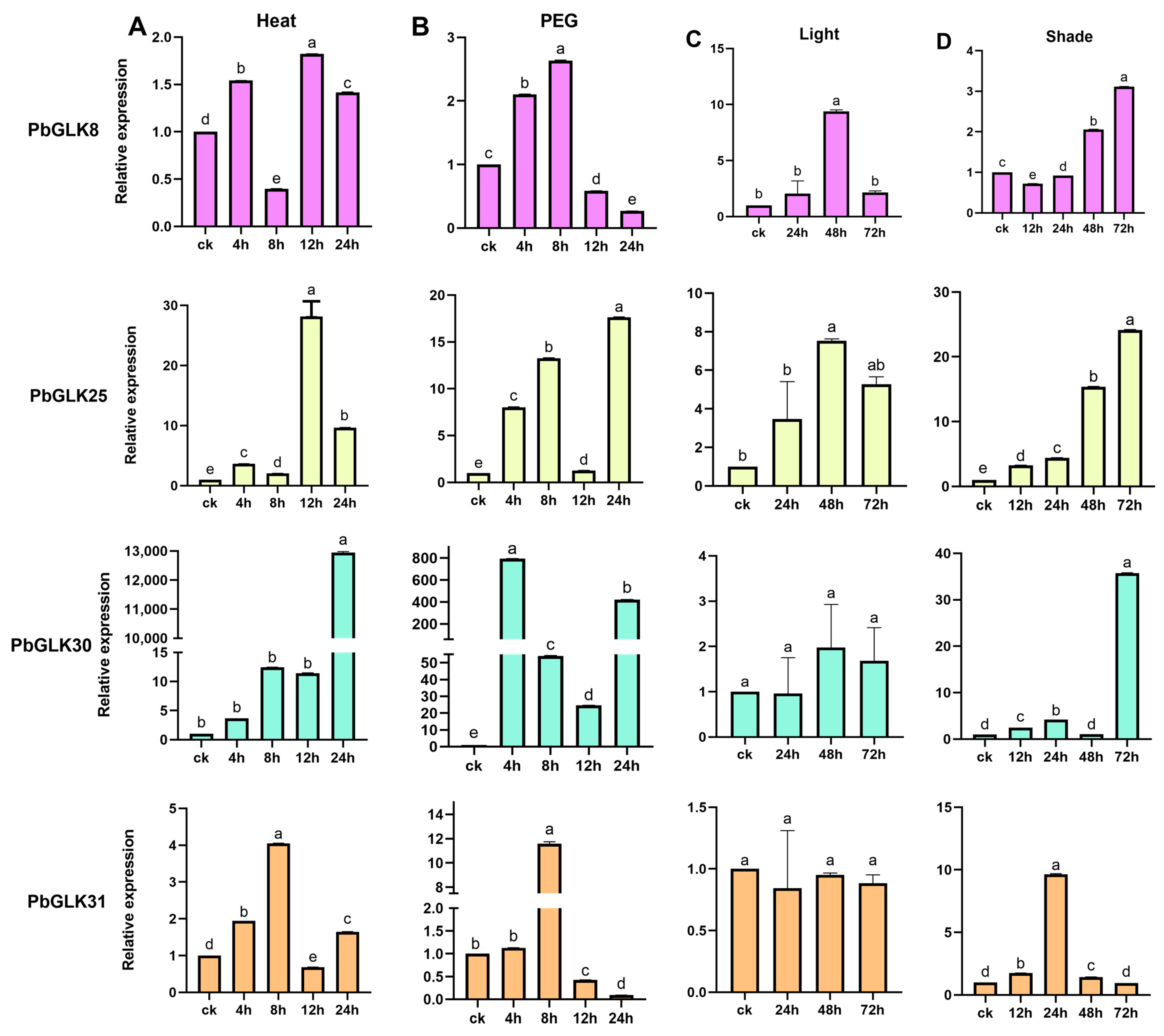
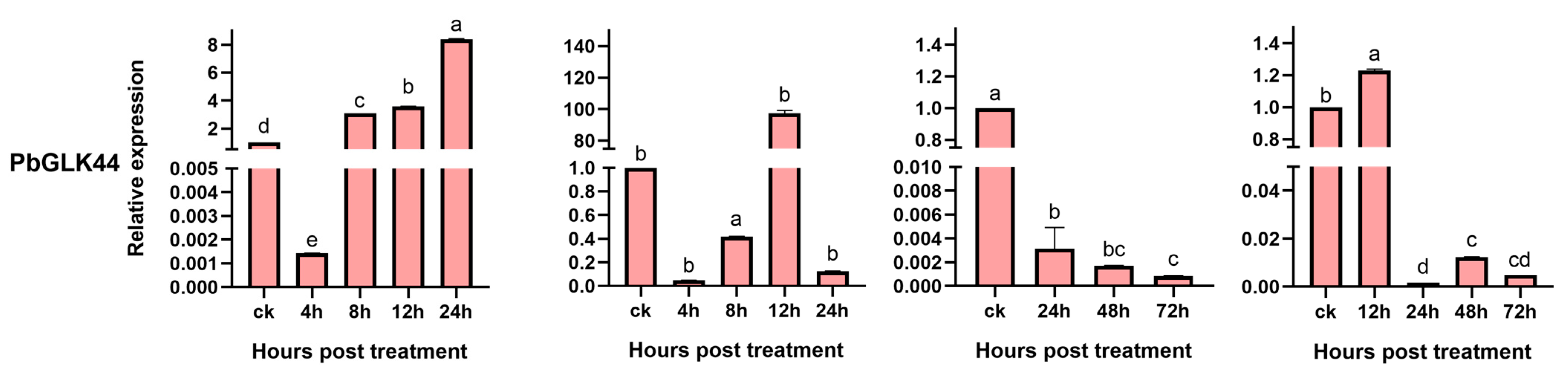
2.8. Expression of PbGLK Genes Under Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Genome Data and Plant Material Source
4.2. Identification and Physical and Chemical Property Analysis
4.3. Chromosomal Distribution and Gene Duplication of PbGLK Genes
4.4. Collinearity Analysis of PbGLK Genes
4.5. Phylogenetic Analysis
4.6. Analysis of Conserved Motifs and Gene Structures
4.7. Promoter Cis-Element Analysis of PbGLK Genes
4.8. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, C.F.; Liu, X.Y.; Tang, Y.Q.; Fu, Y.Q.; Zhang, J.M.; Yang, L.T.; Li, P.H.; Zhu, Z.L.; Dong, P. A comprehensive review of TGA transcription factors in plant growth, stress responses, and beyond. Int. J. Biol. Macromol. 2024, 258, 128880. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.Y.; Kagale, S.; Ferrie, A.M.R. Multifaceted roles of transcription factors during plant embryogenesis. Front. Plant Sci. 2024, 14, 1322728. [Google Scholar] [CrossRef]
- Bernal-Gallardo, J.J.; Zuñiga-Mayo, V.M.; Marsch-Martinez, N.; de Folter, S. Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis. Plants 2023, 12, 596. [Google Scholar] [CrossRef] [PubMed]
- Drewell, R.A.; Klonaros, D.; Dresch, J.M. Transcription factor expression landscape in Drosophila embryonic cell lines. BMC Genom. 2024, 25, 307. [Google Scholar] [CrossRef]
- Ma, Z.M.; Hu, L.J. WRKY Transcription Factor Responses and Tolerance to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2024, 25, 6845. [Google Scholar] [CrossRef]
- Yan, T.Y.; Shu, X.C.; Ning, C.L.; Li, Y.H.; Wang, Z.; Wang, T.; Zhuang, W.B. Functions and Regulatory Mechanisms of bHLH Transcription Factors during the Responses to Biotic and Abiotic Stresses in Woody Plants. Plants 2024, 13, 2315. [Google Scholar] [CrossRef]
- Radani, Y.; Li, R.X.; Korboe, H.M.; Ma, H.Y.; Yang, L.M. Transcriptional and Post-Translational Regulation of Plant bHLH Transcription Factors during the Response to Environmental Stresses. Plants 2023, 12, 2113. [Google Scholar] [CrossRef]
- Dhatterwal, P.; Sharma, N.; Prasad, M. Decoding the functionality of plant transcription factors. J. Exp. Bot. 2024, 75, 4745–4759. [Google Scholar] [CrossRef]
- Bakery, A.; Vraggalas, S.; Shalha, B.; Chauchan, H.; Benhamed, M.; Fragkostefanakis, S. Heat stress transcription factors as the central molecular rheostat to optimize plant survival and recovery from heat stress. New Phytol. 2024, 244, 51–64. [Google Scholar] [CrossRef]
- Claeys, H.; Inzé, D. The Agony of Choice: How Plants Balance Growth and Survival under Water-Limiting Conditions. Plant Physiol. 2013, 162, 1768–1779. [Google Scholar] [CrossRef]
- Shu, L.; Li, L.H.; Jiang, Y.Q.; Yan, J.L. Advances in membrane-tethered NAC transcription factors in plants. Plant Sci. 2024, 342, 112034. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Chen, Q.Q.; Hu, J.; Li, C.C.; Luo, L.Y.; Zeng, L. Genome-Wide Identification and Characterization of GARP Transcription Factor Gene Family Members Reveal Their Diverse Functions in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 947072. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.J.; You, S.M.; Kong, W.Y.; Tang, Q.Y.; Bai, W.T.; Cai, Y.; Zheng, H.; Wang, C.L.; Jiang, L.; Wang, C.M.; et al. A GARP transcription factor anther dehiscence defected 1 (OsADD1) regulates rice anther dehiscence. Plant Mol. Biol. 2019, 101, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.P.; Wu, P.J.; Zhang, T.Y.; Song, H.L.; Zhang, Y.F.; Chen, J.F.; Yue, C.P.; Huang, J.Y.; Sun, T.; Zhou, T. Genome-Scale Investigation of GARP Family Genes Reveals Their Pivotal Roles in Nutrient Stress Resistance in Allotetraploid Rapeseed. Int. J. Mol. Sci. 2022, 23, 14484. [Google Scholar] [CrossRef]
- Nagatoshi, Y.; Mitsuda, N.; Hayashi, M.; Inoue, S.; Okuma, E.; Kubo, A.; Murata, Y.; Seo, M.; Saji, H.; Kinoshita, T.; et al. GOLDEN 2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. USA 2016, 113, 4218–4223. [Google Scholar] [CrossRef]
- Kim, N.; Jeong, J.; Kim, J.; Oh, J.; Choi, G. Shade represses photosynthetic genes by disrupting the DNA binding of GOLDEN2-LIKE1. Plant Physiol. 2023, 192, 680. [Google Scholar] [CrossRef]
- Tang, R.; Zhou, X.; Weng, S.S.; Wang, F.; Li, R.; Xie, Q.L.; Li, Z.H.; Xie, S.Q.; Cao, A.P.; Zhuo, L.; et al. Genome-Wide Analysis of GLK Gene Family in Four Cotton Species Provides Insights into Their Involvement in Cotton Abiotic Stress Response. Agriculture 2024, 14, 2086. [Google Scholar] [CrossRef]
- Alam, I.; Manghwar, H.; Zhang, H.Y.; Yu, Q.X.; Ge, L.F. Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Front. Plant Sci. 2022, 13, 1052659. [Google Scholar] [CrossRef]
- Zhou, X.R.; Yu, W.; Gong, F.S.; Xu, H.W.; Lyu, J.; Zhou, X.F. Golden 2-like Transcription Factors Regulate Photosynthesis under UV-B Stress by Regulating the Calvin Cycle. Plants 2024, 13, 1856. [Google Scholar] [CrossRef]
- Muino, J.M.; Grossmann, C.; Tatjana, K.; Kerstin, K. Natural genetic variation in GLK1-mediated photosynthetic acclimation in response to light. BMC Plant Biol. 2024, 24, 87. [Google Scholar] [CrossRef]
- Wang, H.T.; Xu, F.F. Identification and expression analysis of the GLK gene family in tea plant (Camellia sinensis) and a functional study of CsGLK54 under low-temperature stress. Sci. Rep. 2024, 14, 12465. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.X.; Liang, S.; Hu, L.F.; Yu, L.; Liang, P.X.; Hao, Z.D.; Peng, Y.; Yang, J.; Shi, J.S.; Chen, J.H. Overexpression of Liriodendron Hybrid LhGLK1 in Arabidopsis Leads to Excessive Chlorophyll Synthesis and Improved Growth. Int. J. Mol. Sci. 2024, 25, 6968. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.Y.; Ren, S.B.; Shen, W.; Li, J.J.; Li, Y.X.; Li, C.S.; Li, Y.M.H.; Zong, Z.X.; Xie, W.B.; Grierson, D.; et al. Limited conservation in cross-species comparison of GLK transcription factor binding suggested wide-spread cistrome divergence. Nat. Commun. 2022, 13, 7632. [Google Scholar] [CrossRef]
- Zhang, D.W.; Tan, W.R.; Yang, F.; Han, Q.; Deng, X.G.; Guo, H.Q.; Liu, B.H.; Yin, Y.H.; Lin, H.H. A BIN2-GLK1 Signaling Module Integrates Brassinosteroid and Light Signaling to Repress Chloroplast Development in the Dark. Dev. Cell 2021, 56, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, R.; Zeng, X.Y.; Lee, S.H.; Ye, L.H.; Tian, S.L.; Zhang, Y.J.; Busch, W.; Zhou, W.B.; Zhu, X.G.; et al. GLK transcription factors accompany ELONGATED HYPOCOTYL5 to orchestrate light-induced seedling development in Arabidopsis. Plant Physiol. 2024, 194, 2400–2421. [Google Scholar] [CrossRef]
- Choi, H.; Gwak, D.; Kim, S.; Yi, T.G.Y.; Ha, S.H. Molecular Action of GOLDEN2-LIKE Transcription Factor Family with Diverse Interacting Promoters and Proteins. Physiol. Plant. 2024, 176, e14164. [Google Scholar] [CrossRef]
- Hernández-Verdeja, T.; Lundgren, M.R. GOLDEN2-LIKE transcription factors: A golden ticket to improve crops? Plants People Planet 2024, 6, 79–93. [Google Scholar] [CrossRef]
- Lambret-Frotte, J.; Smith, G.; Langdale, J.A. GOLDEN2-like is sufficient but not necessary for chloroplast biogenesis in mesophyll cells of C4 grasses. Plant J. 2024, 117, 416–431. [Google Scholar] [CrossRef]
- Taketa, S.; Hattori, M.; Takami, T.; Himi, E.; Sakamoto, W. Mutations in a Golden2-Like Gene Cause Reduced Seed Weight in Barley albino lemma 1 Mutants. Plant Cell Physiol. 2021, 62, 447–457. [Google Scholar] [CrossRef]
- Wu, R.H.; Guo, L.; Wang, R.Y.; Zhang, Q.; Yao, H.J. Genome-Wide Identification and Characterization of G2-Like Transcription Factor Genes in Moso Bamboo (Phyllostachys edulis). Molecules 2022, 27, 5491. [Google Scholar] [CrossRef]
- Hills, A.C.; Khan, S.; López-Juez, E. Chloroplast Biogenesis-Associated Nuclear Genes: Control by Plastid Signals Evolved Prior to Their Regulation as Part of Photomorphogenesis. Front. Plant Sci. 2015, 6, 1078. [Google Scholar] [CrossRef]
- Tachibana, R.; Abe, S.; Marugami, M.; Yamagami, A.; Akema, R.; Ohashi, T.; Nishida, K.; Nosaki, S.; Miyakawa, T.; Tanokura, M.; et al. BPG4 regulates chloroplast development and homeostasis by suppressing GLK transcription factors and involving light and brassinosteroid signaling. Nat. Commun. 2024, 15, 370. [Google Scholar] [CrossRef] [PubMed]
- Susila, H.; Nasim, Z.; Gawarecka, K.; Jung, J.Y.; Jin, S.; Youn, G.; Ahn, J.H. Chloroplasts prevent precocious flowering through a GOLDEN2-LIKE-B-BOX DOMAIN PROTEIN module. Plant Commun. 2023, 4, 100515. [Google Scholar] [CrossRef] [PubMed]
- Li, M.P.; Lee, K.P.; Liu, T.; Dogra, V.; Duan, J.L.; Li, M.S.; Xing, W.M.; Kim, C.H. Antagonistic modules regulate photosynthesis-associated nuclear genes via GOLDEN2-LIKE transcription factors. Plant Physiol. 2022, 188, 2308–2324. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Jiang, S.H.; Wen, B.B.; Yang, C.; Xiao, W.; Fu, X.L.; Li, D.M.; Chen, X.D.; Gao, D.S.; et al. Transcriptomic and Functional Analyses Reveal That PpGLK1 Regulates Chloroplast Development in Peach (Prunus persica). Front. Plant Sci. 2018, 9, 34. [Google Scholar] [CrossRef]
- Han, Y.T.; Li, F.F.; Wu, Y.; Wang, D.; Luo, G.B.; Wang, X.N.; Wang, X.; Kuang, H.H.; Larkin, R.M. PSEUDO-ETIOLATION IN LIGHT proteins reduce greening by binding GLK transcription factors. Plant Physiol. 2024, 194, 1722–1744. [Google Scholar] [CrossRef]
- Frangedakis, E.; Yelina, N.E.; Billakurthi, K.; Hua, L.; Schreier, T.; Dickinson, P.J.; Tomaselli, M.; Haseloff, J.; Hibberd, J.M. MYB-related transcription factors control chloroplast biogenesis. Cell 2024, 187, 4859–4876. [Google Scholar] [CrossRef]
- Zubo, Y.O.; Blakley, I.C.; Franco-Zorrilla, J.M.; Yamburenko, M.V.; Solano, R.; Kieber, J.J.; Loraine, A.E.; Schaller, G.E. Coordination of Chloroplast Development through the Action of the GNC and GLK Transcription Factor Families. Plant Physiol. 2018, 178, 130–147. [Google Scholar] [CrossRef]
- Veciana, N.; Martin, G.; Leivar, P.; Monte, E. BBX16 mediates the repression of seedling photomorphogenesis downstream of the GUN1/GLK1 module during retrograde signalling. New Phytol. 2022, 234, 93–106. [Google Scholar] [CrossRef]
- Yu, J.J.; Yin, K.; Liu, Y.; Li, Y.H.; Zhang, J.H.; Han, X.; Tong, Z.K. Co-expression network analysis reveals PbTGA4 and PbAPRR2 as core transcription factors of drought response in an important timber species Phoebe bournei. Front. Plant Sci. 2024, 14, 1297235. [Google Scholar] [CrossRef]
- Li, Y.R.; Chen, Y.F.; Li, P.Y.; Huang, H.F.; Xue, K.X.; Cai, S.Y.; Liao, X.L.; Jin, S.F.; Zheng, D.X. Microplastics in soil affect the growth and physiological characteristics of Chinese fir and Phoebe bournei seedlings. Environ. Pollut. 2024, 358, 124503. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.L.; Sun, S.X.; Li, Y.M.; Jia, L.; Ye, S.L.; Yu, Y.X.; Dossa, K.; Luan, Y.P. Leaf-transcriptome profiles of phoebe bournei provide insights into temporal drought stress responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Wu, X.H.; Grossnickle, S.C.; Chen, L.H.; Yu, X.X.; El-Kassaby, Y.A.; Feng, J.L. Formula Fertilization Promotes Phoebe bournei Robust Seedling Cultivation. Forests 2020, 11, 781. [Google Scholar] [CrossRef]
- An, J.; Wei, X.L.; Huo, H.H. Transcriptome analysis reveals the accelerated expression of genes related to photosynthesis and chlorophyll biosynthesis contribution to shade-tolerant in Phoebe bournei. BMC Plant Biol. 2022, 22, 268. [Google Scholar] [CrossRef]
- Yin, Z.Y.; Liao, W.H.; Li, J.S.; Pan, J.X.; Yang, S.J.; Chen, S.P.; Cao, S.J. Genome-Wide Identification of GATA Family Genes in Phoebe bournei and Their Transcriptional Analysis under Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 10342. [Google Scholar] [CrossRef]
- Zhang, Y.; Wan, S.Q.; Xing, B.C.; Peng, C.Y.; Zhu, J.J.; Shao, Q.S.; Lv, A.M. An HD-Zip transcription factor ArHDZ22 regulates plant height and decreases salt tolerance in Anoectochilus roxburghii. Ind. Crops Prod. 2025, 223, 120251. [Google Scholar] [CrossRef]
- Girin, T.; Lejay, L.; Wirth, J.; Widiez, T.; Palenchar, P.M.; Nazoa, P.; Touraine, B.; Gojon, A.; Lepetit, M. Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant Cell Environ. 2007, 30, 1366–1380. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; van Wijnen, A.J.; Rice, D.; Raffi, M.Y.; Azizi, P.; Osman, M.; Taheri, S.; Abu Bakar, M.F.; Isa, M.N.M.; et al. Contribution of transposable elements in the plant’s genome. Gene 2018, 665, 155–166. [Google Scholar] [CrossRef]
- Deng, K.X.; Zhang, Q.Z.; Hong, Y.X.; Yan, J.B.; Hu, X.H. A deep learning-based web server for identifying base-resolution cis-regulatory elements within plant core promoters. Plant Commun. 2023, 4, 100455. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Zhou, T.; Wu, Y.; Yuan, M.; Zhang, X.; Liu, Y. Genome-wide characterization and expression analysis of the bHLH gene family in response to abiotic stresses in Zingiber officinale Roscoe. BMC Genom. 2025, 26, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, K.; Veluchamy, A.; Fatima, A.; Garcia-Ramirez, G.X.; Reichheld, J.-P.; Artyukh, O.; Frohlich, K.; Polussa, A.; Parween, S.; Nagarajan, A.P.; et al. Microbe-induced coordination of plant iron-sulfur metabolism enhances high-light-stress tolerance of Arabidopsis. Plant Commun. 2024, 5, 101012. [Google Scholar] [CrossRef]
- Han, R.; Ma, L.; Terzaghi, W.; Guo, Y.; Li, J.G. Molecular mechanisms underlying coordinated responses of plants to shade and environmental stresses. Plant J. 2024, 117, 1893–1913. [Google Scholar] [CrossRef] [PubMed]
- Alem, A.L.; Ariel, F.D.; Cho, Y.; Hong, J.C.; Gonzalez, D.H.; Viola, I.L. TCP15 interacts with GOLDEN2-LIKE 1 to control cotyledon opening in Arabidopsis. Plant J. 2022, 110, 748–763. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Wei, S.B.; Gao, Y.; Pei, H.C.; Geng, R.D.; Lu, Z.F.; Wang, P.; Zhou, W.B. Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure. Plant Physiol. 2024, 194, 774–786. [Google Scholar] [CrossRef]
- Wijnants, S.; Riedelberger, M.; Penninger, P.; Kuchler, K.; Van Dijck, P. Sugar Phosphorylation Controls Carbon Source Utilization and Virulence of Candida albicans. Front. Microbiol. 2020, 11, 1274. [Google Scholar] [CrossRef]
- Wang, P.; Fouracre, J.; Kelly, S.; Karki, S.; Gowik, U.; Aubry, S.; Shaw, M.K.; Westhoff, P.; Slamet-Loedin, I.H.; Quick, W.P.; et al. Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 2013, 237, 481–495. [Google Scholar] [CrossRef]
- Liu, Z.; Xiong, T.; Zhao, Y.W.; Qiu, B.F.; Chen, H.; Kang, X.Y.; Yang, J. Genome-wide characterization and analysis of Golden 2-Like transcription factors related to leaf chlorophyll synthesis in diploid and triploid Eucalyptus urophylla. Front. Plant Sci. 2022, 13, 952877. [Google Scholar] [CrossRef]
- Chuang, H.C.; Chang, C.C.; Teng, C.F.; Hsueh, C.H.; Chiu, L.L.; Hsu, P.M.; Lee, M.C.; Hsu, C.P.; Chen, Y.R.; Liu, Y.C.; et al. MAP4K3/GLK Promotes Lung Cancer Metastasis by Phosphorylating and Activating IQGAP1. Cancer Res. 2019, 79, 4978–4993. [Google Scholar] [CrossRef]
- Powell, A.L.T.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernández-Muñoz, R.; Vicente, A.; et al. Uniform ripening Encodes a Golden 2-like Transcription Factor Regulating Tomato Fruit Chloroplast Development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef] [PubMed]
- Fraimovitch, E.; Hagai, T. Promoter evolution of mammalian gene duplicates. Bmc Biol. 2023, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Murmu, J.; Wilton, M.; Allard, G.; Pandeya, R.; Desveaux, D.; Singh, J.; Subramaniam, R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol. Plant Pathol. 2014, 15, 174–184. [Google Scholar] [PubMed]
- Liu, Q.; Huang, H.D.; Chen, Y.Q.; Yue, Z.C.; Wang, Z.P.; Qu, T.T.; Xu, D.Y.; Lu, S.Y.; Hu, H.H. Two Arabidopsis MYB-SHAQKYF transcription repressors regulate leaf wax biosynthesis via transcriptional suppression on DEWAX. New Phytol. 2022, 236, 2115–2130. [Google Scholar] [CrossRef]
- Xiong, B.; Chen, H.; Ma, Q.; Yao, J.; Wang, J.; Wu, W.; Liao, L.; Wang, X.; Zhang, M.; He, S.; et al. Genome-Wide Analysis of the GLK Gene Family and Its Expression at Different Leaf Ages in the Citrus Cultivar Kanpei. Plants 2024, 13, 936. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Zhao, P.; Zhang, A.H.; Wang, J.Z.; Ha, M. Genome-Wide Analysis of HSP70s in Hexaploid Wheat: Tandem Duplication, Heat Response, and Regulation. Cells 2022, 11, 818. [Google Scholar] [CrossRef]
- Wani, S.H. Editorial: Mechanisms of abiotic stress responses and tolerance in plants: Physiological, biochemical and molecular interventions, volume II. Front. Plant Sci. 2023, 14, 1272255. [Google Scholar] [CrossRef]
- Foyer, C.H.; Hanke, G. ROS production and signalling in chloroplasts: Cornerstones and evolving concepts. Plant J. 2022, 111, 642–661. [Google Scholar] [CrossRef]
- Pottosin, I.; Shabala, S. Transport Across Chloroplast Membranes: Optimizing Photosynthesis for Adverse Environmental Conditions. Mol. Plant 2016, 9, 356–370. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.H.; Han, S.; Li Chong, S.; Meng, G.L.; Song, M.Y.; Wang, Y.; Zhou, S.C.; Liu, C.C.; Lou, L.H.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef]
- Zhao, X.W.; Liu, D.K.; Wang, Q.Q.; Ke, S.J.; Li, Y.Y.; Zhang, D.Y.; Zheng, Q.Y.; Zhang, C.L.; Liu, Z.J.; Lan, S.R. Genome-wide identification and expression analysis of the GRAS gene family in Dendrobium chrysotoxum. Front. Plant Sci. 2022, 13, 1058287. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Yan, Q.; Li, J.W.; Feng, L.; Zhang, Y.; Xu, J.; Xia, R.; Zeng, Z.H.; Liu, Y.L. The GRAS gene family and its roles in seed development in litchi (Litchi chinensis Sonn). BMC Plant Biol. 2021, 21, 423. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Manuel, M. A new semi-subterranean diving beetle of the Hydroporus normandi-complex from south-eastern France, with notes on other taxa of the complex (Coleoptera: Dytiscidae). Zootaxa 2013, 3652, 453–474. [Google Scholar] [CrossRef][Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef]
- Day, R.W.; Quinn, G.P. Comparisons of Treatments After an Analysis of Variance in Ecology. Ecol. Monogr. 1989, 59, 433–463. [Google Scholar] [CrossRef]

| Gene Accession | Gene Id | Size/aa 1 | MW 2/Da | Theoretical pI 3 | Instability Index | Aliphatic Index | GRAVY 4 | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| OF25727-RA | PbGLK1 | 287 | 32,398.86 | 8.34 | 51.17 | 80.17 | −0.559 | Mitochondrion |
| OF02278-RA | PbGLK2 | 349 | 39,145.71 | 6.66 | 78.03 | 57.02 | −0.98 | Nucleus |
| OF27064-RA | PbGLK3 | 446 | 50,230.21 | 5.27 | 52.6 | 73.52 | −0.682 | Nucleus |
| OF17914-RA | PbGLK4 | 335 | 38,287.71 | 8.28 | 70.16 | 70.42 | −0.796 | Nucleus |
| OF08332-RA | PbGLK5 | 652 | 70,642.4 | 5.9 | 46.14 | 78.97 | −0.452 | Nucleus |
| OF28298-RA | PbGLK6 | 290 | 31,657.62 | 6.55 | 45.16 | 77.34 | −0.644 | Chloroplast |
| OF09746-RA | PbGLK7 | 282 | 31,675.63 | 8.59 | 33.76 | 72.98 | −0.827 | Nucleus |
| OF23687-RA | PbGLK8 | 385 | 43,157.17 | 6.67 | 58.11 | 57.77 | −0.671 | Nucleus |
| OF01303-RA | PbGLK9 | 445 | 49,256.73 | 5.14 | 65.21 | 65.73 | −0.716 | Nucleus |
| OF20594-RA | PbGLK10 | 351 | 39,354.17 | 8.62 | 50.29 | 69.97 | −0.599 | Nucleus |
| OF17917-RA | PbGLK11 | 312 | 35,624.58 | 6.31 | 65.28 | 66.25 | −0.877 | Nucleus |
| OF04301-RA | PbGLK12 | 445 | 48,861.35 | 6.26 | 60.64 | 59.28 | −0.807 | Nucleus |
| OF03170-RA | PbGLK13 | 275 | 30,294.98 | 9.1 | 51.95 | 73.75 | −0.757 | Nucleus |
| OF08790-RA | PbGLK14 | 736 | 79,576.63 | 6.16 | 43.31 | 79.47 | −0.43 | Nucleus |
| OF16131-RA | PbGLK15 | 251 | 28,606.27 | 8.7 | 54.3 | 72.31 | −0.747 | Nucleus |
| OF09825-RA | PbGLK16 | 568 | 62,996.06 | 5.98 | 52.61 | 72.29 | −0.571 | Nucleus |
| OF24628-RA | PbGLK17 | 416 | 45,705.35 | 5.89 | 48.05 | 72.88 | −0.576 | Nucleus |
| OF04095-RA | PbGLK18 | 214 | 24,016.97 | 8.59 | 33.18 | 72.99 | −0.636 | Nucleus |
| OF24353-RA | PbGLK19 | 305 | 34,766.86 | 8.88 | 63.42 | 62 | −1.178 | Nucleus |
| OF26355-RA | PbGLK20 | 489 | 53,954.06 | 6.33 | 67.79 | 71.25 | −0.722 | Nucleus |
| OF02538-RA | PbGLK21 | 358 | 39,814.67 | 6.19 | 61.43 | 64.78 | −0.637 | Chloroplast |
| OF14101-RA | PbGLK22 | 279 | 32,386.57 | 6.32 | 54.62 | 72.01 | −0.706 | Nucleus |
| OF08001-RA | PbGLK23 | 432 | 47,244.12 | 6.06 | 38.97 | 83.47 | −0.52 | Nucleus |
| OF29722-RA | PbGLK24 | 205 | 23,241.21 | 10.18 | 68.23 | 62.78 | −0.989 | Nucleus |
| OF04128-RA | PbGLK25 | 396 | 44,315.52 | 8.64 | 74.03 | 58.66 | −0.851 | Nucleus |
| OF23584-RA | PbGLK26 | 420 | 46,906.48 | 5.44 | 52.42 | 74.6 | −0.658 | Nucleus |
| OF14499-RA | PbGLK27 | 347 | 39,998.96 | 9.31 | 52.45 | 59.28 | −1.022 | Nucleus |
| OF02188-RA | PbGLK28 | 833 | 90,318.07 | 6.12 | 50.93 | 84.53 | −0.268 | Nucleus |
| OF03171-RA | PbGLK29 | 223 | 24,556.97 | 7.71 | 36.31 | 84.48 | −0.481 | Nucleus |
| OF09627-RA | PbGLK30 | 468 | 51,610.95 | 5.53 | 55.73 | 63.95 | −0.746 | Nucleus |
| OF29382-RA | PbGLK31 | 305 | 33,298.39 | 8.97 | 50.98 | 70.98 | −0.755 | Nucleus |
| OF11944-RA | PbGLK32 | 419 | 46,471.49 | 6.91 | 58.42 | 60.79 | −0.741 | Nucleus |
| OF21061-RA | PbGLK33 | 341 | 37,903.13 | 7.83 | 74.11 | 54.37 | −0.945 | Nucleus |
| OF00450-RA | PbGLK34 | 380 | 43,108.4 | 8.89 | 58.67 | 68.5 | −0.782 | Nucleus |
| OF06602-RA | PbGLK35 | 429 | 47,287.61 | 5.92 | 59.7 | 57.58 | −0.879 | Nucleus |
| OF11915-RA | PbGLK36 | 724 | 78,445.02 | 6.5 | 49.96 | 81.69 | −0.31 | Nucleus |
| OF28330-RA | PbGLK37 | 201 | 21,574.26 | 9.1 | 70.15 | 67.06 | −0.602 | Nucleus |
| OF15632-RA | PbGLK38 | 221 | 24,509.65 | 6.36 | 41.82 | 76.33 | −0.524 | Nucleus |
| OF09129-RA | PbGLK39 | 463 | 51,364.98 | 6.64 | 55.4 | 65.1 | −0.794 | Nucleus |
| OF23451-RA | PbGLK40 | 268 | 29,072.64 | 6.59 | 53.16 | 66.72 | −0.504 | Nucleus |
| OF20950-RA | PbGLK41 | 649 | 72,221.34 | 5.1 | 52.37 | 66.98 | −0.565 | Nucleus |
| OF14662-RA | PbGLK42 | 153 | 17,563.21 | 9.6 | 37.73 | 79.61 | −0.644 | Cytosol |
| OF09041-RA | PbGLK43 | 413 | 45,500.32 | 6.01 | 56.38 | 71.23 | −0.616 | Nucleus |
| OF02019-RA | PbGLK44 | 352 | 38,517.24 | 5.29 | 73.37 | 56.56 | −0.716 | Nucleus |
| OF02187-RA | PbGLK45 | 510 | 56,431.14 | 6.07 | 45.44 | 74.12 | −0.487 | Nucleus |
| OF02164-RA | PbGLK46 | 351 | 38,823.34 | 7.76 | 45.75 | 67.29 | −0.605 | Nucleus |
| OF18065-RA | PbGLK47 | 327 | 36,990.15 | 9.27 | 53.86 | 61.47 | −0.823 | Nucleus |
| OF23673-RA | PbGLK48 | 941 | 103,959.54 | 6.24 | 45.5 | 87.3 | −0.273 | Nucleus |
| OF25917-RA | PbGLK49 | 350 | 38,844.98 | 8.14 | 59.67 | 83.34 | −0.635 | Nucleus |
| OF11234-RA | PbGLK50 | 397 | 43,851.69 | 8.55 | 61.5 | 70.98 | −0.675 | Nucleus |
| OF15566-RA | PbGLK51 | 342 | 38,314.62 | 8.14 | 62.87 | 73.04 | −0.696 | Nucleus |
| OF27986-RA | PbGLK52 | 408 | 46,172.71 | 7.65 | 38.54 | 75.15 | −0.554 | Nucleus |
| OF11599-RA | PbGLK53 | 556 | 62,619.62 | 6.79 | 41.06 | 88.87 | −0.354 | Nucleus |
| OF09936-RA | PbGLK54 | 750 | 81,821.93 | 5.89 | 45.54 | 78.49 | −0.409 | Nucleus |
| OF06026-RA | PbGLK55 | 581 | 63,602.17 | 5.84 | 51.12 | 82.34 | −0.306 | Cytosol |
| OF11698-RA | PbGLK56 | 239 | 27,611.85 | 10.21 | 57.9 | 83.6 | −0.657 | Nucleus |
| OF23452-RA | PbGLK57 | 207 | 22,962.79 | 5.9 | 63.35 | 66.96 | −0.757 | Nucleus |
| OF23315-RA | PbGLK58 | 445 | 49,250.66 | 6.89 | 49.39 | 59.46 | −0.747 | Nucleus |
| OF19961-RA | PbGLK59 | 319 | 35,748.06 | 9.48 | 66.58 | 76.08 | −0.688 | Nucleus |
| OF22052-RA | PbGLK60 | 358 | 39,590.35 | 7.21 | 55.22 | 72.79 | −0.659 | Nucleus |
| OF23001-RA | PbGLK61 | 446 | 50,176.54 | 8.74 | 52.64 | 69.37 | −0.666 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, Y.; Peng, L.; Shi, X.; Zheng, Q.; Fan, D.; Feng, Z.; Liu, X.; Ma, H.; Cao, S.; Chang, W. Genome-Wide Identification of GLK Family Genes in Phoebe bournei and Their Transcriptional Analysis Under Abiotic Stresses. Int. J. Mol. Sci. 2025, 26, 2387. https://doi.org/10.3390/ijms26062387
Lian Y, Peng L, Shi X, Zheng Q, Fan D, Feng Z, Liu X, Ma H, Cao S, Chang W. Genome-Wide Identification of GLK Family Genes in Phoebe bournei and Their Transcriptional Analysis Under Abiotic Stresses. International Journal of Molecular Sciences. 2025; 26(6):2387. https://doi.org/10.3390/ijms26062387
Chicago/Turabian StyleLian, Yiran, Liang Peng, Xinying Shi, Qiumian Zheng, Dunjin Fan, Zhiyi Feng, Xiaomin Liu, Huanhuan Ma, Shijiang Cao, and Weiyin Chang. 2025. "Genome-Wide Identification of GLK Family Genes in Phoebe bournei and Their Transcriptional Analysis Under Abiotic Stresses" International Journal of Molecular Sciences 26, no. 6: 2387. https://doi.org/10.3390/ijms26062387
APA StyleLian, Y., Peng, L., Shi, X., Zheng, Q., Fan, D., Feng, Z., Liu, X., Ma, H., Cao, S., & Chang, W. (2025). Genome-Wide Identification of GLK Family Genes in Phoebe bournei and Their Transcriptional Analysis Under Abiotic Stresses. International Journal of Molecular Sciences, 26(6), 2387. https://doi.org/10.3390/ijms26062387








