Differential Transcriptional Programs Reveal Modular Network Rearrangements Associated with Late-Onset Alzheimer’s Disease
Abstract
1. Introduction
Transcriptional Networks
2. Results
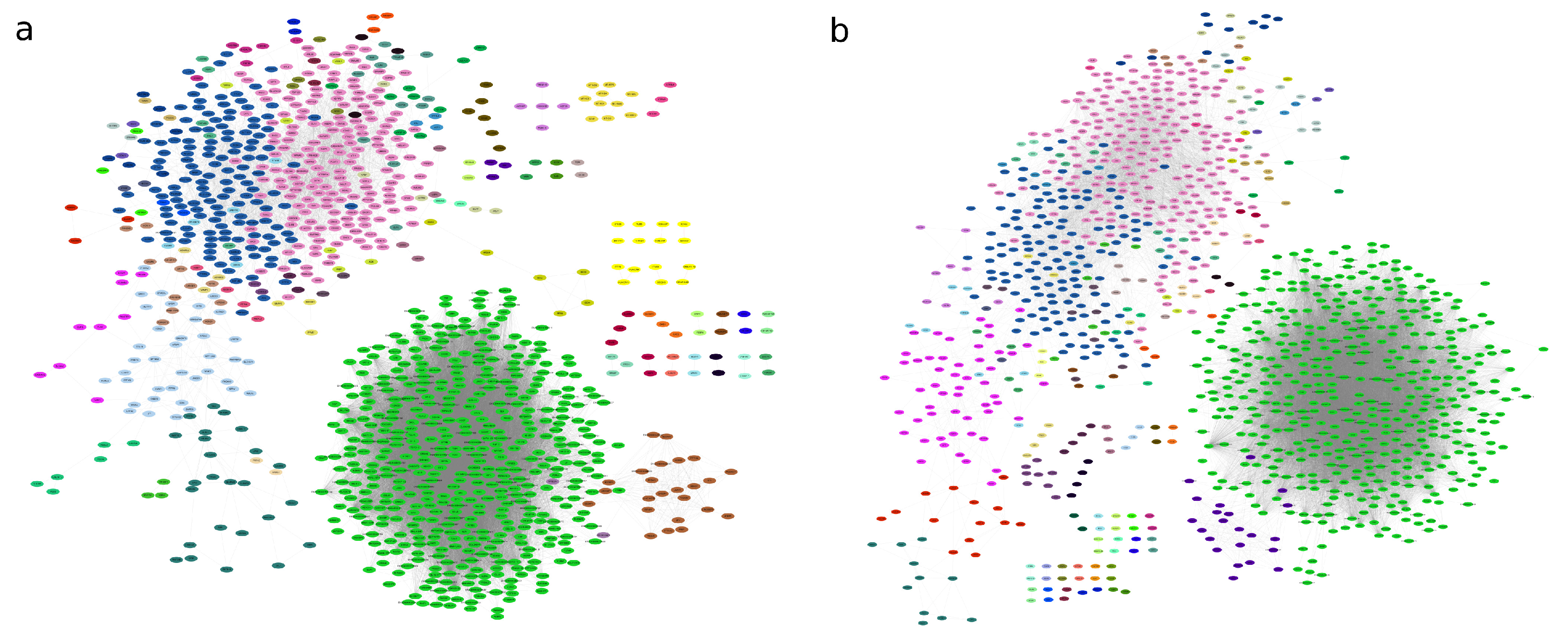
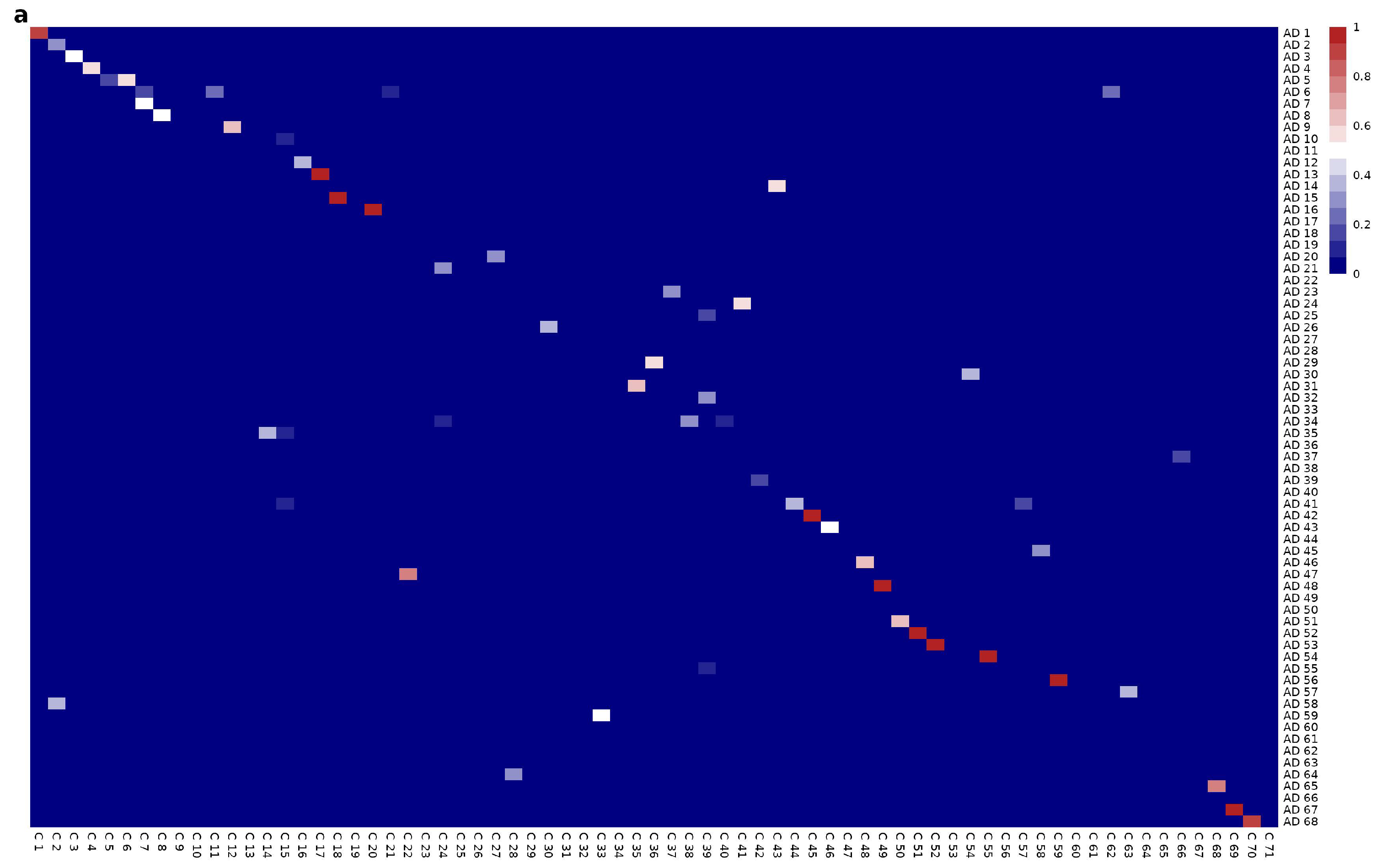
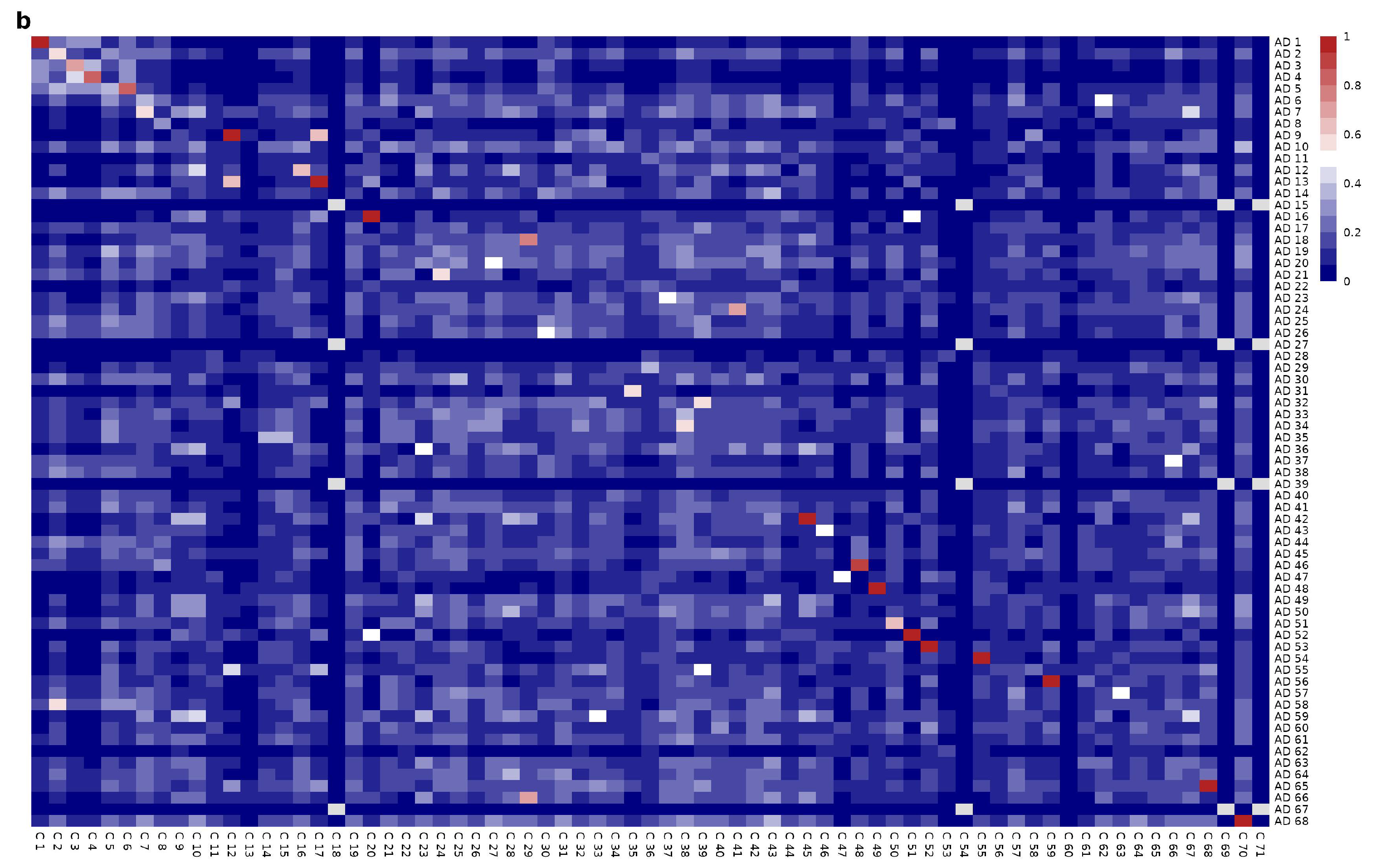
Mesoscopic Network Analysis
3. Discussion
3.1. Gene Co-Expression Network Alterations in Alzheimer’S Disease: Structural and Connectivity Insights
3.2. Functional Insights into Co-Expression Changes in the AD Network: Linking Epigenetic, Cytoskeletal, Immune, and Post-Transcriptional Pathways
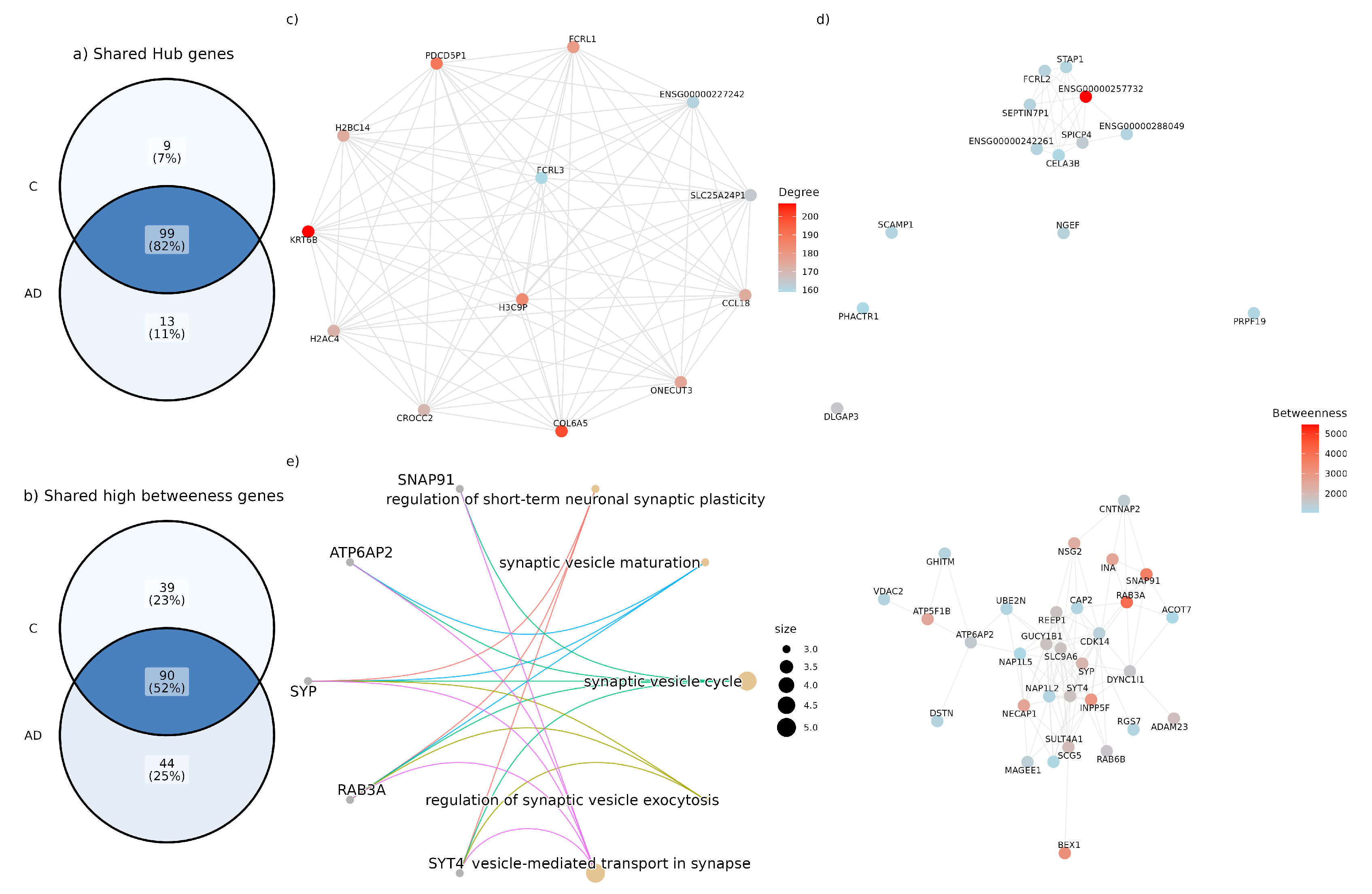
3.3. High Betweenness Genes Only Present in the Disease Network Are Involved in Diverse Biological Pathways
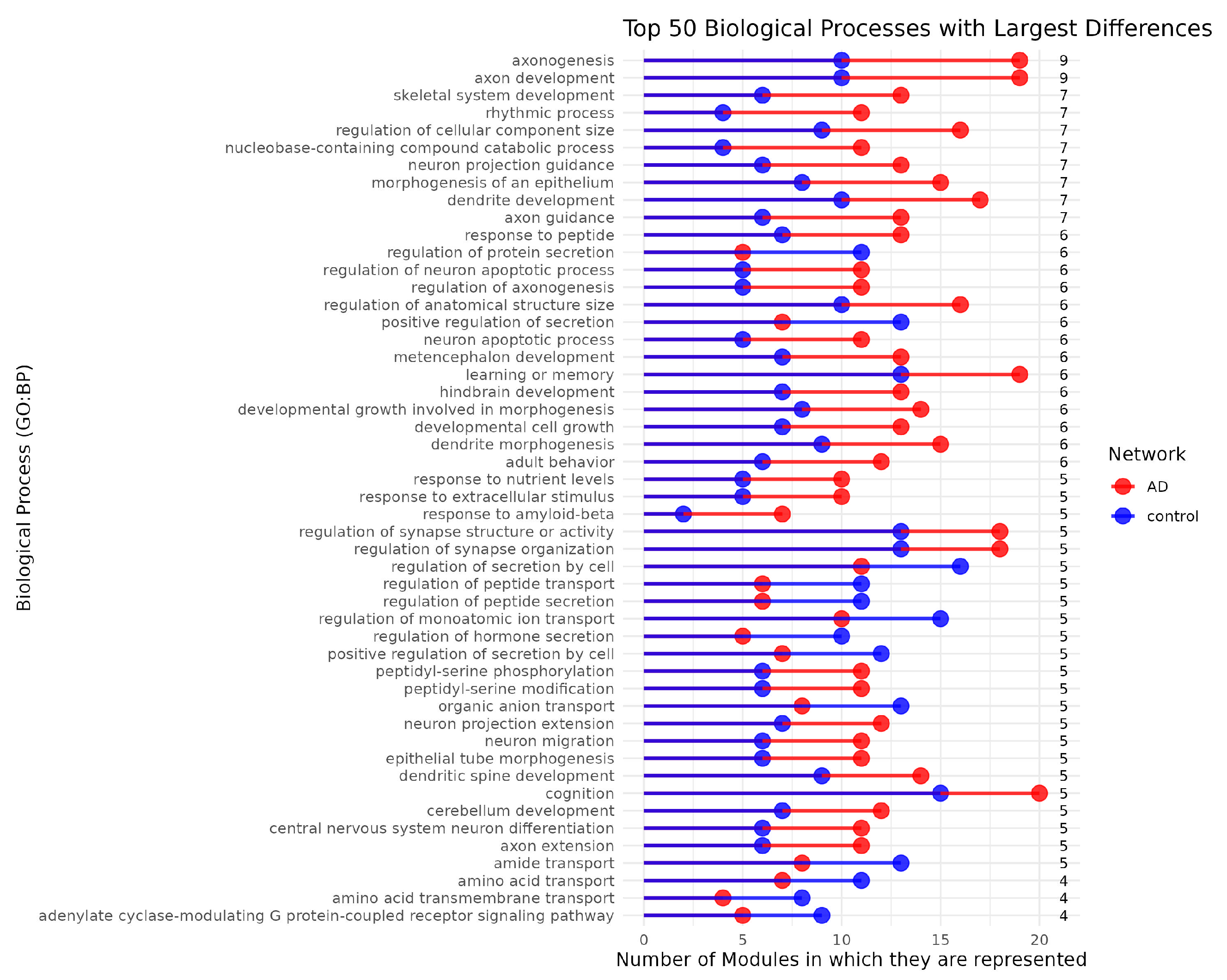
3.4. Network Functional Analysis Show Gene Modular Rearrangement
4. Methods
4.1. Data Acquisition and Classification
4.2. Quality Control
4.3. Inference of Co-Expression Networks
4.4. Topological Analysis and Network Centralities Measure
4.5. Inference of Modular Structure and Functional Analysis
5. Conclusions
Linking Basic and Clinical Research
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aisen, P.S.; Cummings, J.; Jack, C.R.; Morris, J.C.; Sperling, R.; Frölich, L.; Jones, R.W.; Dowsett, S.A.; Matthews, B.R.; Raskin, J.; et al. On the path to 2025: Understanding the Alzheimer’s disease continuum. Alzheimer’s Res. Ther. 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Games, D.; Adams, D.; Alessandrini, R.; Barbour, R.; Berthelette, P.; Blackwell, C.; Carr, T.; Clemens, J.; Donaldson, T.; Gillespie, F. Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature 1995, 373, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Jara, C.; Cerpa, W.; Tapia-Rojas, C.; Quintanilla, R.A. Tau Deletion Prevents Cognitive Impairment and Mitochondrial Dysfunction Age Associated by a Mechanism Dependent on Cyclophilin-D. Front. Neurosci. 2021, 14, 586710. [Google Scholar] [CrossRef]
- Wingo, T.S.; Lah, J.J.; Levey, A.I.; Cutler, D.J. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol. 2012, 69, 59–64. [Google Scholar] [CrossRef]
- Arvanitakis, Z.; Shah, R.C.; Bennett, D.A. Diagnosis and Management of Dementia: A Review. JAMA 2019, 322, 1589–1599. [Google Scholar] [CrossRef]
- Guerreiro, R.; Bras, J. The age factor in Alzheimer’s disease. Genome Med. 2015, 7, 106. [Google Scholar] [CrossRef]
- Reynolds, C.F.; Jeste, D.V.; Sachdev, P.S.; Blazer, D.G. Mental health care for older adults: Recent advances and new directions in clinical practice and research. World Psychiatry 2022, 21, 336–363. [Google Scholar] [CrossRef]
- Kao, Y.C.; Ho, P.C.; Tu, Y.K.; Jou, I.M.; Tsai, K.J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef]
- Tesi, N.; van der Lee, S.J.; Hulsman, M.; Jansen, I.E.; Stringa, N.; van Schoor, N.M.; Scheltens, P.; van der Flier, W.M.; Huisman, M.; Reinders, M.J.T.; et al. Immune response and endocytosis pathways are associated with the resilience against Alzheimer’s disease. Transl. Psychiatry 2020, 10, 332. [Google Scholar] [CrossRef]
- Chung, Y.; Lee, H. Correlation between Alzheimer’s disease and type 2 diabetes using non-negative matrix factorization. Sci. Rep. 2021, 11, 15265. [Google Scholar] [CrossRef] [PubMed]
- Hamzé, R.; Delangre, E.; Tolu, S.; Moreau, M.; Janel, N.; Bailbé, D.; Movassat, J. Type 2 Diabetes Mellitus and Alzheimer’s Disease: Shared Molecular Mechanisms and Potential Common Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 15287. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Alkhayyat, M.; Abou Saleh, M.; Sarmini, M.T.; Singh, A.; Garg, R.; Garg, P.; Mansoor, E.; Padival, R.; Cohen, B.L. Alzheimer Disease Occurs More Frequently in Patients with Inflammatory Bowel Disease: Insight From a Nationwide Study. J. Clin. Gastroenterol. 2023, 57, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.; Sahu, B.; Kaur, H.; Hasler, W.; Prakash, A.; Combs, C.K. Gastrointestinal Changes and Alzheimer’s Disease. Curr. Alzheimer Res. 2022, 19, 335–350. [Google Scholar] [CrossRef]
- Tini, G.; Scagliola, R.; Monacelli, F.; La Malfa, G.; Porto, I.; Brunelli, C.; Rosa, G.M. Alzheimer’s Disease and Cardiovascular Disease: A Particular Association. Cardiol. Res. Pract. 2020, 2020, 2617970. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Gan, Y.H.; Yang, L.; Cheng, W.; Yu, J.T. Depression in Alzheimer’s Disease: Epidemiology, Mechanisms, and Treatment. Biol. Psychiatry 2024, 95, 992–1005. [Google Scholar] [CrossRef]
- Tong, Q.; Chen, L. Associations of Alzheimer’s Disease Neuropathologic Changes with Clinical Presentations of Parkinson’s Disease. J. Alzheimer’s Dis. 2021, 81, 201–207. [Google Scholar] [CrossRef]
- Hampel, H.; Nisticò, R.; Seyfried, N.T.; Levey, A.I.; Modeste, E.; Lemercier, P.; Baldacci, F.; Toschi, N.; Garaci, F.; Perry, G.; et al. Omics sciences for systems biology in Alzheimer’s disease: State-of-the-art of the evidence. Ageing Res. Rev. 2021, 69, 101346. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological Alterations in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404. [Google Scholar] [CrossRef]
- Wilson, R.S.; Boyle, P.A.; Yu, L.; Barnes, L.L.; Sytsma, J.; Buchman, A.S.; Bennett, D.A.; Schneider, J.A. Temporal course and pathologic basis of unawareness of memory loss in dementia. Neurology 2015, 85, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Pàmies-Vilà, R.; Fabregat-Sanjuan, A.; Ros-Alsina, A.; Rigo-Vidal, A.; Pascual-Rubio, V. Analysis of the Dorsolateral Prefrontal Cortex MNI Coordinates. In Proceedings of the XV Ibero-American Congress of Mechanical Engineering; Vizán Idoipe, A., García Prada, J.C., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 132–138. [Google Scholar] [CrossRef]
- Le Reste, P.J.; Haegelen, C.; Gibaud, B.; Moreau, T.; Morandi, X. Connections of the dorsolateral prefrontal cortex with the thalamus: A probabilistic tractography study. Surg. Radiol. Anat. 2016, 38, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Bähner, F.; Demanuele, C.; Schweiger, J.; Gerchen, M.F.; Zamoscik, V.; Ueltzhöffer, K.; Hahn, T.; Meyer, P.; Flor, H.; Durstewitz, D.; et al. Hippocampal–Dorsolateral Prefrontal Coupling as a Species-Conserved Cognitive Mechanism: A Human Translational Imaging Study. Neuropsychopharmacology 2015, 40, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Barbey, A.K.; Koenigs, M.; Grafman, J. Dorsolateral prefrontal contributions to human working memory. Cortex 2013, 49, 1195–1205. [Google Scholar] [CrossRef]
- Hertrich, I.; Dietrich, S.; Blum, C.; Ackermann, H. The Role of the Dorsolateral Prefrontal Cortex for Speech and Language Processing. Front. Hum. Neurosci. 2021, 15, 645209. [Google Scholar] [CrossRef]
- Rilling, J.K.; Sanfey, A.G. Social Interaction. In Encyclopedia of Neuroscience; Squire, L.R., Ed.; Academic Press: Oxford, UK, 2009; pp. 41–48. [Google Scholar] [CrossRef]
- Uddin, L.Q. Cognitive and behavioural flexibility: Neural mechanisms and clinical considerations. Nat. Rev. Neurosci. 2021, 22, 167–179. [Google Scholar] [CrossRef]
- Kaller, C.P.; Rahm, B.; Spreer, J.; Weiller, C.; Unterrainer, J.M. Dissociable contributions of left and right dorsolateral prefrontal cortex in planning. Cereb. Cortex 2011, 21, 307–317. [Google Scholar] [CrossRef]
- Obeso, I.; Herrero, M.T.; Ligneul, R.; Rothwell, J.C.; Jahanshahi, M. A Causal Role for the Right Dorsolateral Prefrontal Cortex in Avoidance of Risky Choices and Making Advantageous Selections. Neuroscience 2021, 458, 166–179. [Google Scholar] [CrossRef]
- Greene, J.D.; Nystrom, L.E.; Engell, A.D.; Darley, J.M.; Cohen, J.D. The neural bases of cognitive conflict and control in moral judgment. Neuron 2004, 44, 389–400. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 15th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Kumar, S.; Zomorrodi, R.; Ghazala, Z.; Goodman, M.S.; Blumberger, D.M.; Cheam, A.; Fischer, C.; Daskalakis, Z.J.; Mulsant, B.H.; Pollock, B.G.; et al. Extent of Dorsolateral Prefrontal Cortex Plasticity and Its Association with Working Memory in Patients With Alzheimer Disease. JAMA Psychiatry 2017, 74, 1266–1274. [Google Scholar] [CrossRef]
- Thomas, S.; Bonchev, D. A survey of current software for network analysis in molecular biology. Hum. Genom. 2010, 4, 353. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.M.; Segal, E.; Koller, D.; Kim, S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Oldham, M.C.; Geschwind, D.H. A Systems Level Analysis of Transcriptional Changes in Alzheimer’s Disease and Normal Aging. J. Neurosci. 2008, 28, 1410. [Google Scholar] [CrossRef]
- Altaf-Ul-Amin, M.; Afendi, F.M.; Kiboi, S.K.; Kanaya, S. Systems Biology in the Context of Big Data and Networks. BioMed Res. Int. 2014, 2014, 428570. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Barabási, A.L. Network science. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2013, 371, 20120375. [Google Scholar] [CrossRef]
- He, B.; Xu, J.; Tian, Y.; Liao, B.; Lang, J.; Lin, H.; Mo, X.; Lu, Q.; Tian, G.; Bing, P. Gene Coexpression Network and Module Analysis across 52 Human Tissues. BioMed Res. Int. 2020, 2020, 6782046. [Google Scholar] [CrossRef]
- Nakamura-García, A.K.; Espinal-Enríquez, J. Pseudogenes in Cancer: State of the Art. Cancers 2023, 15, 4024. [Google Scholar] [CrossRef]
- Gómez-Pascual, A.; Rocamora-Pérez, G.; Ibanez, L.; Botía, J.A. Targeted co-expression networks for the study of traits. Sci. Rep. 2024, 14, 16675. [Google Scholar] [CrossRef]
- Conte, F.; Paci, P. Alzheimer’s disease: Insights from a network medicine perspective. Sci. Rep. 2022, 12, 16846. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Bp, K.; C R, S.; Saikumar, N.V.; Philip, P.; Narayanan, M. Alzheimer’s disease rewires gene coexpression networks coupling different brain regions. npj Syst. Biol. Appl. 2024, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.S.; Madaj, Z.; Cheema, C.T.; Kara, B.; Bennett, D.A.; Schneider, J.A.; Gordon, M.N.; Ginsberg, S.D.; Mufson, E.J.; Counts, S.E. Co-expression network analysis of frontal cortex during the progression of Alzheimer’s disease. Cereb. Cortex 2022, 32, 5108–5120. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lin, J.; Zhang, L. The application of weighted gene co-expression network analysis in identifying key modules and hub genes associated with disease status in Alzheimer’s disease. Ann. Transl. Med. 2019, 7, 800. [Google Scholar] [CrossRef]
- Mathys, H.; Boix, C.A.; Akay, L.A.; Xia, Z.; Davila-Velderrain, J.; Ng, A.P.; Jiang, X.; Abdelhady, G.; Galani, K.; Mantero, J.; et al. Single-cell multiregion dissection of Alzheimer’s disease. Nature 2024, 632, 858–868. [Google Scholar] [CrossRef]
- Saint-André, V. Computational biology approaches for mapping transcriptional regulatory networks. Comput. Struct. Biotechnol. J. 2021, 19, 4884–4895. [Google Scholar] [CrossRef]
- Datta, V.; Siddharthan, R.; Krishna, S. Detection of cooperatively bound transcription factor pairs using ChIP-seq peak intensities and expectation maximization. PLoS ONE 2018, 13, e0199771. [Google Scholar] [CrossRef]
- Giorgi, F.M.; Lopez, G.; Woo, J.H.; Bisikirska, B.; Califano, A.; Bansal, M. Inferring protein modulation from gene expression data using conditional mutual information. PLoS ONE 2014, 9, e109569. [Google Scholar] [CrossRef][Green Version]
- de Anda-Jáuregui, G. Guideline for comparing functional enrichment of biological network modular structures. Appl. Netw. Sci. 2019, 4, 13. [Google Scholar] [CrossRef]
- Hartl, C.L.; Ramaswami, G.; Pembroke, W.G.; Muller, S.; Pintacuda, G.; Saha, A.; Parsana, P.; Battle, A.; Lage, K.; Geschwind, D.H. Co-expression network architecture reveals the brain-wide and multi-regional basis of disease susceptibility. Nat. Neurosci. 2021, 24, 1313. [Google Scholar] [CrossRef]
- Poliseno, L. Pseudogenes: Newly discovered players in human cancer. Sci. Signal. 2012, 5, re5. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Esposito, R.; Aprile, M.; Ciccodicola, A. Non-coding RNA and pseudogenes in neurodegenerative diseases: “The (un)Usual Suspects”. Front. Genet. 2012, 3, 231. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zong, Y.; Zhu, L.; Wang, W.; Han, Y. Chemokines in patients with Alzheimer’s disease: A meta-analysis. Front. Aging Neurosci. 2023, 15, 1047810. [Google Scholar] [CrossRef] [PubMed]
- Konijnenberg, A.; Butterer, A.; Sobott, F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta-Proteins Proteom. 2013, 1834, 1239–1256. [Google Scholar] [CrossRef]
- Chen, Z.; Gui, B.; Zhang, Y.; Xie, G.; Li, W.; Liu, S.; Xu, B.; Wu, C.; He, L.; Yang, J.; et al. Identification of a 35S U4/U6.U5 tri-small nuclear ribonucleoprotein (tri-snRNP) complex intermediate in spliceosome assembly. J. Biol. Chem. 2017, 292, 18113–18128. [Google Scholar] [CrossRef]
- Patel, P.J.; Ren, Y.; Yan, Z. Epigenomic analysis of Alzheimer’s disease brains reveals diminished CTCF binding on genes involved in synaptic organization. Neurobiol. Dis. 2023, 184, 106192. [Google Scholar] [CrossRef]
- Wong, Y.Y.; Xiong, L.Y.; Mori-Fegan, D.K.; Noor, S.; Chenoweth, M.J.; Mirza, S.S.; Masellis, M.; Black, S.E.; Swardfager, W. Alzheimer’s Disease Neuroimaging Initiative. Relationships between polymorphisms in keratin genes and Alzheimer’s disease phenotypes. Alzheimer’s Dement. 2023, 19, e074162. [Google Scholar] [CrossRef]
- Richens, J.L.; Spencer, H.L.; Butler, M.; Cantlay, F.; Vere, K.A.; Bajaj, N.; Morgan, K.; O’Shea, P. Rationalising the role of Keratin 9 as a biomarker for Alzheimer’s disease. Sci. Rep. 2016, 6, 22962. [Google Scholar] [CrossRef]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef]
- Song, Q.; Yu, H.; Cheng, Y.; Han, J.; Li, K.; Zhuang, J.; Lv, Q.; Yang, X.; Yang, H. Bladder cancer-derived exosomal KRT6B promotes invasion and metastasis by inducing EMT and regulating the immune microenvironment. J. Transl. Med. 2022, 20, 308. [Google Scholar] [CrossRef]
- Moir, R.D.; Lathe, R.; Tanzi, R.E. The antimicrobial protection hypothesis of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 1602–1614. [Google Scholar] [CrossRef] [PubMed]
- Vojtechova, I.; Machacek, T.; Kristofikova, Z.; Stuchlik, A.; Petrasek, T. Infectious origin of Alzheimer’s disease: Amyloid beta as a component of brain antimicrobial immunity. PLoS Pathog. 2022, 18, e1010929. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. Int. J. Nanomed. 2019, 14, 5541–5554. [Google Scholar] [CrossRef] [PubMed]
- Roedl, D.; Traidl-Hoffmann, C.; Ring, J.; Behrendt, H.; Braun-Falco, M. Serine protease inhibitor lymphoepithelial Kazal type-related inhibitor tends to be decreased in atopic dermatitis. J. Eur. Acad. Dermatol. Venereol. 2009, 23, 1263–1266. [Google Scholar] [CrossRef]
- Lehmann, S.M.; Leube, R.E.; Schwarz, N. Keratin 6a mutations lead to impaired mitochondrial quality control. Br. J. Dermatol. 2020, 182, 636–647. [Google Scholar] [CrossRef]
- Kallunki, T.; Su, B.; Tsigelny, I.; Sluss, H.K.; Dérijard, B.; Moore, G.; Davis, R.; Karin, M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994, 8, 2996–3007. [Google Scholar] [CrossRef]
- Benvenga, S. Conformational mutations in neuroserpin and familial dementias. Lancet 2002, 360, 1696. [Google Scholar] [CrossRef]
- Davis, R.L.; Shrimpton, A.E.; Carrell, R.W.; Lomas, D.A.; Gerhard, L.; Baumann, B.; Lawrence, D.A.; Yepes, M.; Kim, T.S.; Ghetti, B.; et al. Association between conformational mutations in neuroserpin and onset and severity of dementia. Lancet 2002, 359, 2242–2247. [Google Scholar] [CrossRef]
- Reumann, R.; Vierk, R.; Zhou, L.; Gries, F.; Kraus, V.; Mienert, J.; Romswinkel, E.; Morellini, F.; Ferrer, I.; Nicolini, C.; et al. The serine protease inhibitor neuroserpin is required for normal synaptic plasticity and regulates learning and social behavior. Learn. Mem. 2017, 24, 650–659. [Google Scholar] [CrossRef]
- Yepes, M.; Sandkvist, M.; Wong, M.K.K.; Coleman, T.A.; Smith, E.; Cohan, S.L.; Lawrence, D.A. Neuroserpin reduces cerebral infarct volume and protects neurons from ischemia-induced apoptosis. Blood 2000, 96, 569–576. [Google Scholar] [CrossRef]
- Rodríguez-González, R.; Sobrino, T.; Rodríguez-Yáñez, M.; Millán, M.; Brea, D.; Miranda, E.; Moldes, O.; Pérez, J.; Lomas, D.A.; Leira, R.; et al. Association between neuroserpin and molecular markers of brain damage in patients with acute ischemic stroke. J. Transl. Med. 2011, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, P.K.; Jackson, M.B. The Functional Significance of Synaptotagmin Diversity in Neuroendocrine Secretion. Front. Endocrinol. 2013, 4, 124. [Google Scholar] [CrossRef] [PubMed]
- Weingarten, D.J.; Shrestha, A.; Juda-Nelson, K.; Kissiwaa, S.A.; Spruston, E.; Jackman, S.L. Fast resupply of synaptic vesicles requires synaptotagmin-3. Nature 2022, 611, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chen, S.; Nie, Q.; Xue, Q.; Fan, H.; Wang, Y.; Fan, S.; Zhu, J.; Shen, H.; Li, H.; et al. Synaptotagmin-3 interactions with GluA2 mediate brain damage and impair functional recovery in stroke. Cell Rep. 2023, 42, 112233. [Google Scholar] [CrossRef]
- Chum, P.P.; Bishara, M.A.; Solis, S.R.; Behringer, E.J. Cerebrovascular miRNAs Track Early Development of Alzheimer’s Disease and Target Molecular Markers of Angiogenesis and Blood Flow Regulation. J. Alzheimer’s Dis. 2024, 99, S187–S234. [Google Scholar] [CrossRef]
- Esashi, E.; Bao, M.; Wang, Y.H.; Cao, W.; Liu, Y.J. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur. J. Immunol. 2012, 42, 573–579. [Google Scholar] [CrossRef]
- Floyd, S.R.; Porro, E.B.; Slepnev, V.I.; Ochoa, G.C.; Tsai, L.H.; De Camilli, P. Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2. J. Biol. Chem. 2001, 276, 8104–8110. [Google Scholar] [CrossRef]
- Alwadei, A.H.; Benini, R.; Mahmoud, A.; Alasmari, A.; Kamsteeg, E.J.; Alfadhel, M. Loss-of-function mutation in RUSC2 causes intellectual disability and secondary microcephaly. Dev. Med. Child Neurol. 2016, 58, 1317–1322. [Google Scholar] [CrossRef]
- MacKenzie, K.C.; de Graaf, B.M.; Syrimis, A.; Zhao, Y.; Brosens, E.; Mancini, G.M.S.; Schot, R.; Halley, D.; Wilke, M.; Vøllo, A.; et al. Goldberg–Shprintzen syndrome is determined by the absence, or reduced expression levels, of KIFBP. Hum. Mutat. 2020, 41, 1906–1917. [Google Scholar] [CrossRef]
- Melo, D.; Pallares, L.F.; Ayroles, J.F. Reassessing the modularity of gene co-expression networks using the Stochastic Block Model. PLoS Comput. Biol. 2024, 20, e1012300. [Google Scholar] [CrossRef]
- Yin, W.; Mendoza, L.; Monzon-Sandoval, J.; Urrutia, A.O.; Gutierrez, H. Emergence of co-expression in gene regulatory networks. PLoS ONE 2021, 16, e0247671. [Google Scholar] [CrossRef] [PubMed]
- García-Cortés, D.; Hernández-Lemus, E.; Espinal-Enríquez, J. Luminal A Breast Cancer Co-expression Network: Structural and Functional Alterations. Front. Genet. 2021, 12, 629475. [Google Scholar] [CrossRef] [PubMed]
- Dorostkar, M.M.; Zou, C.; Blazquez-Llorca, L.; Herms, J. Analyzing dendritic spine pathology in Alzheimer’s disease: Problems and opportunities. Acta Neuropathol. 2015, 130, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Daniel Estrella, L.; Trease, A.J.; Sheldon, L.; Roland, N.J.; Fox, H.S.; Stauch, K.L. Tau association with synaptic mitochondria coincides with energetic dysfunction and excitatory synapse loss in the P301S tauopathy mouse model. Neurobiol. Aging 2025, 147, 163–175. [Google Scholar] [CrossRef]
- Lu, X.; Luo, C.; Wu, J.; Deng, Y.; Mu, X.; Zhang, T.; Yang, X.; Liu, Q.; Li, Z.; Tang, S.; et al. Ion channels and transporters regulate nutrient absorption in health and disease. J. Cell. Mol. Med. 2023, 27, 2631–2642. [Google Scholar] [CrossRef]
- Li, M.; Hao, X.; Hu, Z.; Tian, J.; Shi, J.; Ma, D.; Guo, M.; Li, S.; Zuo, C.; Liang, Y.; et al. Microvascular and cellular dysfunctions in Alzheimer’s disease: An integrative analysis perspective. Sci. Rep. 2024, 14, 20944. [Google Scholar] [CrossRef]
- Bennett, D.A.; Buchman, A.S.; Boyle, P.A.; Barnes, L.L.; Wilson, R.S.; Schneider, J.A. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimer’s Dis. 2018, 64, S161–S189. [Google Scholar] [CrossRef]
- De Jager, P.L.; Ma, Y.; McCabe, C.; Xu, J.; Vardarajan, B.N.; Felsky, D.; Klein, H.U.; White, C.C.; Peters, M.A.; Lodgson, B.; et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 2018, 5, 180142. [Google Scholar] [CrossRef]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimer’s Dement. 2012, 8, 1–13. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Nueda, M.J.; Ferrer, A.; Conesa, A. ARSyN: A method for the identification and removal of systematic noise in multifactorial time course microarray experiments. Biostatistics 2012, 13, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.; Liu, C. Approximating discrete probability distributions with dependence trees. IEEE Trans. Inf. Theory 1968, 14, 462–467. [Google Scholar] [CrossRef]
- Steuer, R.; Kurths, J.; Daub, C.O.; Weise, J.; Selbig, J. The mutual information: Detecting and evaluating dependencies between variables. Bioinformatics 2002, 18 (Suppl. S2), S231–S240. [Google Scholar] [CrossRef]
- Meyer, P. Information-Theoretic Variable Selection and Network Inference from Microarray Data. Ph.D. Thesis, Université Libre de Bruxelles, Brussels, Belgium, 2008. [Google Scholar]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Favera, R.D.; Califano, A. ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. Complex Syst. 2005, 1695, 1–9. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Schank, T.; Wagner, D. Approximating Clustering Coefficient and Transitivity. J. Graph Algorithms Appl. 2005, 9, 265–275. [Google Scholar] [CrossRef]
- Wasserman, S.; Faust, K. Social Network Analysis: Methods and Applications; Cambridge University Press: New York, NY, USA, 1994; p. xxxi + 825. [Google Scholar] [CrossRef]
- Barrat, A.; Barthélemy, M.; Pastor-Satorras, R.; Vespignani, A. The architecture of complex weighted networks. Proc. Natl. Acad. Sci. USA 2004, 101, 3747–3752. [Google Scholar] [CrossRef]
- Rosvall, M.; Axelsson, D.; Bergstrom, C.T. The map equation. Eur. Phys. J. Spec. Top. 2009, 178, 13–23. [Google Scholar] [CrossRef]
- Emmons, S.; Mucha, P.J. Map equation with metadata: Varying the role of attributes in community detection. Phys. Rev. E 2019, 100, 022301. [Google Scholar] [CrossRef] [PubMed]
- Bohlin, L.; Edler, D.; Lancichinetti, A.; Rosvall, M. Community Detection and Visualization of Networks with the Map Equation Framework. In Measuring Scholarly Impact: Methods and Practice; Ding, Y., Rousseau, R., Wolfram, D., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 3–34. [Google Scholar] [CrossRef]
- Lancichinetti, A.; Fortunato, S.; Radicchi, F. Benchmark graphs for testing community detection algorithms. Phys. Rev. E 2008, 78, 046110. [Google Scholar] [CrossRef] [PubMed]
- Pilosof, S.; Alcalá-Corona, S.A.; Wang, T.; Kim, T.; Maslov, S.; Whitaker, R.; Pascual, M. The network structure and eco-evolutionary dynamics of CRISPR-induced immune diversification. Nat. Ecol. Evol. 2020, 4, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Bhatia, M.P.S. Analyzing the Structures of Clusters in Multi-layer Biological Networks. In Proceedings of the IEEE 2018 First International Conference on Secure Cyber Computing and Communication (ICSCCC), Jalandhar, India, 15–17 December 2018; pp. 502–507. [Google Scholar] [CrossRef]
- Alcalá-Corona, S.A.; Sandoval-Motta, S.; Espinal-Enríquez, J.; Hernández-Lemus, E. Modularity in Biological Networks. Front. Genet. 2021, 12, 701331. [Google Scholar] [CrossRef]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Verma, P.; Goyal, R. Influence propagation based community detection in complex networks. Mach. Learn. Appl. 2021, 3, 100019. [Google Scholar] [CrossRef]
- Angioni, D.; Cummings, J.; Lansdall, C.; Middleton, L.; Sampaio, C.; Gauthier, S.; Cohen, S.; Petersen, R.; Rentz, D.; Wessels, A.; et al. Clinical Meaningfulness in Alzheimer’s Disease Clinical Trials. A Report from the EU-US CTAD Task Force. J. Prev. Alzheimer’s Dis. 2024, 11, 1219–1227. [Google Scholar] [CrossRef]
- Knuuttila, T.; Loettgers, A. Biological Control Variously Materialized: Modeling, Experimentation and Exploration in Multiple Media. Perspect. Sci. 2021, 29, 468–492. [Google Scholar] [CrossRef]


| Control Subjects, n = 307 | Pathological AD Subjects, n = 486 | |
|---|---|---|
| Age | 85.8 ± 5.20 | 87.8 ± 3.67 |
| Years of education | 16.5 ± 3.59 | 16.1 ± 3.61 |
| Sex | ||
| Males | 125 | 140 |
| Females | 182 | 346 |
| APOE genotype | ||
| 22 | 3 | 0 |
| 23 | 61 | 54 |
| 24 | 4 | 11 |
| 33 | 209 | 274 |
| 34 | 28 | 129 |
| 44 | 0 | 12 |
| Unknown | 2 | 6 |
| Control Network | AD Network | |
|---|---|---|
| Total genes | 1074 | 1113 |
| Number of edges | 28,160 | 28,160 |
| Network diameter | 12 | 13 |
| Global transitivity | 0.6252799 | 0.5956005 |
| Edges similitude (by Jaccard index) | 68.39% | |
| Number of genes in largest connected component | 529 | 568 |
| Number of modules (Infomap partition) | 71 | 68 |
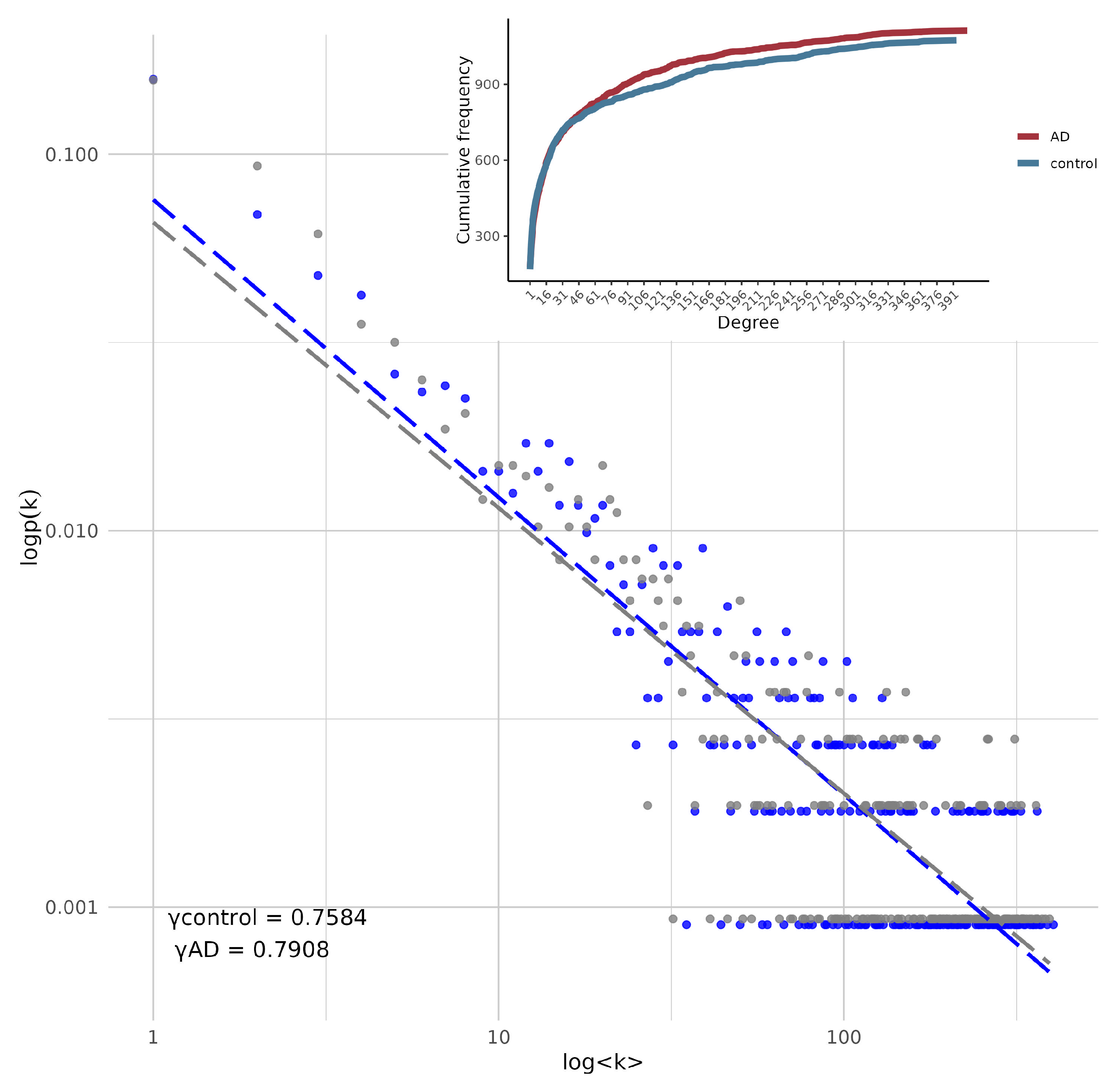
| Scaling exponent | 0.7584 | 0.7908 |
| Number of modules | 65 | 71 |
| Modularity (Q) | 0.2027528 | 0.2834913 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-González, A.P.; Anda-Jáuregui, G.d.; Hernández-Lemus, E. Differential Transcriptional Programs Reveal Modular Network Rearrangements Associated with Late-Onset Alzheimer’s Disease. Int. J. Mol. Sci. 2025, 26, 2361. https://doi.org/10.3390/ijms26052361
Pérez-González AP, Anda-Jáuregui Gd, Hernández-Lemus E. Differential Transcriptional Programs Reveal Modular Network Rearrangements Associated with Late-Onset Alzheimer’s Disease. International Journal of Molecular Sciences. 2025; 26(5):2361. https://doi.org/10.3390/ijms26052361
Chicago/Turabian StylePérez-González, Alejandra Paulina, Guillermo de Anda-Jáuregui, and Enrique Hernández-Lemus. 2025. "Differential Transcriptional Programs Reveal Modular Network Rearrangements Associated with Late-Onset Alzheimer’s Disease" International Journal of Molecular Sciences 26, no. 5: 2361. https://doi.org/10.3390/ijms26052361
APA StylePérez-González, A. P., Anda-Jáuregui, G. d., & Hernández-Lemus, E. (2025). Differential Transcriptional Programs Reveal Modular Network Rearrangements Associated with Late-Onset Alzheimer’s Disease. International Journal of Molecular Sciences, 26(5), 2361. https://doi.org/10.3390/ijms26052361







