The Impact of SNP Score on Low-Density Lipoprotein Cholesterol Concentration and Coronary Artery Disease
Abstract
1. Introduction
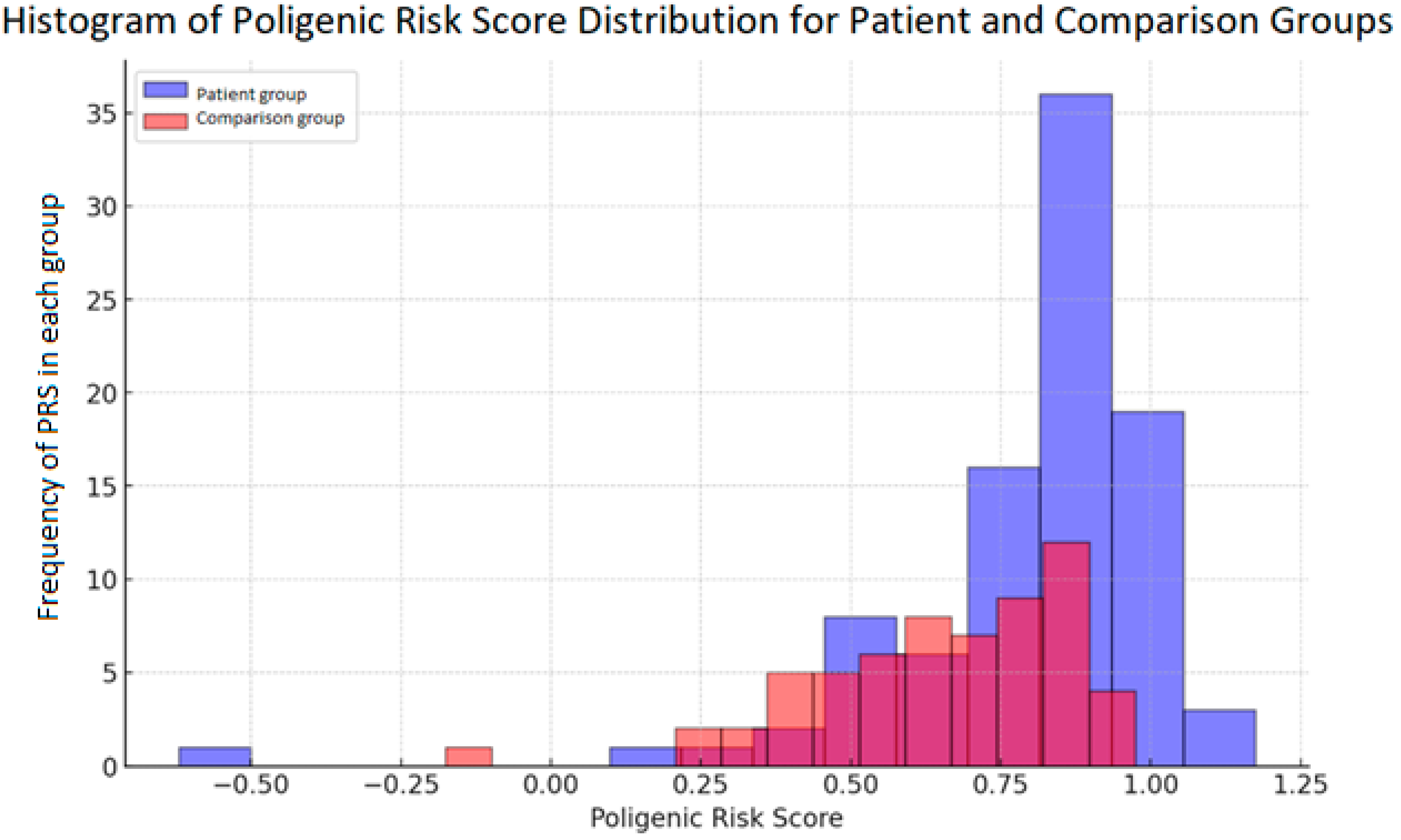
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient and Comparison Group Samples
4.2. Next Generation Sequencing
4.3. High-Resolution Melting Analysis
4.4. PRS Calculation
4.5. Sanger Sequencing
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mensah, G.A.; Roth, G.A.; Fuster, V. The Global Burden of Cardiovascular Diseases and Risk Factors: 2020 and Beyond. J. Am. Coll. Cardiol. 2019, 74, 2529–2532. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Hashim, M.J.; Mustafa, H.; Baniyas, M.Y.; Al Suwaidi, S.K.B.M.; AlKatheeri, R.; Alblooshi, F.M.K.; Almatrooshi, M.E.A.H.; Alzaabi, M.E.H.; Al Darmaki, R.S.; et al. Global Epidemiology of Ischemic Heart Disease: Results from the Global Burden of Disease Study. Cureus 2020, 12, e9349. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; Moorthy, M.V.; Cook, N.R.; Rifai, N.; Lee, I.M.; Buring, J.E. Inflammation, Cholesterol, Lipoprotein(a), and 30-Year Cardiovascular Outcomes in Women. N. Engl. J. Med. 2024, 391, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Linton, M.F.; Yancey, P.G.; Davies, S.S.; Jerome, W.G.; Linton, E.F.; Song, W.L.; Doran, A.C.; Vickers, K.C. The Role of Lipids and Lipoproteins in Atherosclerosis; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., Eds.; MDText.Com, Inc.: South Dartmouth, MA, USA, 2019. [Google Scholar] [PubMed]
- Nordestgaard, B.G.; Varbo, A. Triglycerides and Cardiovascular Disease. Lancet 2014, 384, 626–635. [Google Scholar] [CrossRef]
- Jensen, R.V.; Hjortbak, M.V.; Bøtker, H.E. Ischemic Heart Disease: An Update. Semin. Nucl. Med. 2020, 50, 195–207. [Google Scholar] [CrossRef]
- Heller, D.A.; de Faire, U.; Pedersen, N.L.; Dahlen, G.; McClearn, G.E. Genetic and Environmental Influences on Serum Lipid Levels in Twins. N. Engl. J. Med. 1993, 328, 1150–1156. [Google Scholar] [CrossRef]
- Warden, B.A.; Fazio, S.; Shapiro, M.D. Familial Hypercholesterolemia: Genes and Beyond; Feingold, K.R., Anawalt, B., Blackman, M.R., et al., Eds.; Endotext, MDText.Com, Inc.: South Dartmouth, MA, USA, 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK343488/ (accessed on 20 September 2024).
- Wu, Y.L.; Lin, Z.J.; Li, C.C.; Lin, X.; Shan, S.K.; Guo, B.; Zheng, M.H.; Li, F.; Yuan, L.Q.; Li, Z. Hong. Epigenetic Regulation in Metabolic Diseases: Mechanisms and Advances in Clinical Study. In Signal Transduction and Targeted Therapy; Springer Nature: London, UK, 2023. [Google Scholar] [CrossRef]
- Soufi, M.; Rust, S.; Walter, M.; Schaefer, J.R. A Combined LDL Receptor/LDL Receptor Adaptor Protein 1 Mutation as the Cause for Severe Familial Hypercholesterolemia. Gene 2013, 521, 200–203. [Google Scholar] [CrossRef]
- Bergeron, N.; Phan, B.A.P.; Ding, Y.; Fong, A.; Krauss, R.M. Proprotein Convertase Subtilisin/Kexin Type 9 Inhibition a New Therapeutic Mechanism for Reducing Cardiovascular Disease Risk. Circulation 2015, 132, 1648–1666. [Google Scholar] [CrossRef]
- Talmud, P.J.; Shah, S.; Whittall, R.; Futema, M.; Howard, P.; Cooper, J.A.; Harrison, S.C.; Li, K.; Drenos, F.; Karpe, F.; et al. Use of Low-Density Lipoprotein Cholesterol Gene Score to Distinguish Patients with Polygenic and Monogenic Familial Hypercholesterolaemia: A Case-Control Study. Lancet 2013, 381, 1293–1301. [Google Scholar] [CrossRef]
- Paththinige, C.S.; Sirisena, N.D.; Dissanayake, V.H.W. Genetic Determinants of Inherited Susceptibility to Hypercholesterolemia—A Comprehensive Literature Review. In Lipids in Health and Disease; BioMed Central Ltd.: London, UK, 2017. [Google Scholar] [CrossRef]
- Cupido, A.J.; Tromp, T.R.; Hovingh, G.K. The Clinical Applicability of Polygenic Risk Scores for LDL-Cholesterol: Considerations, Current Evidence and Future Perspectives. In Current Opinion in Lipidology; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2021; pp. 112–116. [Google Scholar] [CrossRef]
- Futema, M.; Bourbon, M.; Williams, M.; Humphries, S.E. Clinical Utility of the Polygenic LDL-C SNP Score in Familial Hypercholesterolemia. Atherosclerosis 2018, 277, 457–463. [Google Scholar] [CrossRef]
- Sharifi, M.; Futema, M.; Nair, D.; Humphries, S.E. Polygenic Hypercholesterolemia and Cardiovascular Disease Risk. In Current Cardiology Reports; Current Medicine Group LLC: Bethesda, MD, USA, 2019. [Google Scholar] [CrossRef]
- Futema, M.; Shah, S.; Cooper, J.A.; Li, K.; Whittall, R.A.; Sharifi, M.; Goldberg, O.; Drogari, E.; Mollaki, V.; Wiegman, A.; et al. Refinement of Variant Selection for the LDL Cholesterol Genetic Risk Score in the Diagnosis of the Polygenic Form of Clinical Familial Hypercholesterolemia and Replication in Samples from 6 Countries. Clin. Chem. 2015, 61, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Cardiero, G.; Ferrandino, M.; Calcaterra, I.L.; Iannuzzo, G.; Di Minno, M.N.D.; Buganza, R.; Guardamagna, O.; Auricchio, R.; Di Taranto, M.D.; Fortunato, G. Impact of 12-SNP and 6-SNP Polygenic Scores on Predisposition to High LDL-Cholesterol Levels in Patients with Familial Hypercholesterolemia. Genes 2024, 15, 462. [Google Scholar] [CrossRef]
- Xiang, R.; Kelemen, M.; Xu, Y.; Harris, L.W.; Parkinson, H.; Inouye, M.; Lambert, S.A. Recent Advances in Polygenic Scores: Translation, Equitability, Methods and FAIR Tools. Genome Med. 2024, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- NHLBI, NIH. Framingham Heart Study (FHS). Available online: https://www.nhlbi.nih.gov/science/framingham-heart-study-fhs (accessed on 31 October 2024).
- Ridker, P.M. How Common Is Residual Inflammatory Risk. In Circulation Research; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2017; pp. 617–619. [Google Scholar] [CrossRef]
- Brown, J.C.; Gerhardt, T.E.; Kwon, E.R.F. Risk Factors for Coronary Artery Disease. 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554410/ (accessed on 31 October 2024).
- Čereškevičius, D.; Zabiela, V.; Aldujeli, A.; Lesauskaitė, V.; Zubielienė, K.; Raškevičius, V.; Čiapienė, I.; Žaliaduonytė, D.; Giedraitienė, A.; Žvikas, V.; et al. Impact of CYP2C19 Gene Variants on Long-Term Treatment with Atorvastatin in Patients with Acute Coronary Syndromes. Int. J. Mol. Sci. 2024, 25, 5385. [Google Scholar] [CrossRef] [PubMed]
- Rott, D.; Klempfner, R.; Goldenberg, I.; Leibowitz, D. Cholesterol Levels Decrease soon after Acute Myocardial Infarction. Isr. Med. Assoc. J. 2015, 17, 370–373. [Google Scholar]
- Pitt, B.; Loscalzo, J.; Yčas, J.; Raichlen, J.S. Lipid Levels After Acute Coronary Syndromes. J. Am. Coll. Cardiol. 2008, 51, 1440–1445. [Google Scholar] [CrossRef]
- Kumar, N.; Kumar, S.; Kumar, A.; Shakoor, T.; Rizwan, A. Lipid Profile of Patients with Acute Myocardial Infarction (AMI). Cureus 2019, 11, e4265. [Google Scholar] [CrossRef]
- Kim, J.H.; Cha, J.J.; Lim, S.; An, J.; Kim, M.N.; Hong, S.J.; Joo, H.J.; Park, J.H.; Yu, C.W.; Lim, D.S.; et al. Target Low-Density Lipoprotein-Cholesterol and Secondary Prevention for Patients with Acute Myocardial Infarction: A Korean Nationwide Cohort Study. J. Clin. Med. 2022, 11, 2650. [Google Scholar] [CrossRef]
- Byrne, R.A.; Rossello, X.; Coughlan, J.J.; Barbato, E.; Berry, C.; Chieffo, A.; Claeys, M.J.; Dan, G.A.; Dweck, M.R.; Galbraith, M.; et al. 2023 ESC Guidelines for the management of acute coronary syndromes: Developed by the task force on the management of acute coronary syndromes of the European Society of Cardiology (ESC). Eur. Heart J. 2023, 44, 3720–3826. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Z.; Wu, T.; Liu, J.; Zhang, B.; Lai, W.; Tu, W.; Guo, Z.; Luo, T. SULT2B1b Inhibits Reverse Cholesterol Transport and Promotes Cholesterol Accumulation and Inflammation in Lymphocytes from AMI Patients with Low LDL-C Levels. Clin. Sci. 2020, 134, 273–287. [Google Scholar] [CrossRef]
- Sekimoto, T.; Koba, S.; Mori, H.; Sakai, R.; Arai, T.; Yokota, Y.; Sato, S.; Tanaka, H.; Masaki, R.; Oishi, Y.; et al. Small Dense Low-Density Lipoprotein Cholesterol: A Residual Risk for Rapid Progression of Non-Culprit Coronary Lesion in Patients with Acute Coronary Syndrome. J. Atheroscler. Thromb. 2021, 28, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Mora, S.; Caulfield, M.P.; Wohlgemuth, J.; Chen, Z.; Superko, H.R.; Rowland, C.M.; Glynn, R.J.; Ridker, P.M.; Krauss, R.M. Atherogenic Lipoprotein Subfractions Determined by Ion Mobility and First Cardiovascular Events after Random Allocation to High-Intensity Statin or Placebo: The Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin (JUPITER) Trial. Circulation 2015, 132, 2220–2229. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albers, J.J.; Slee, A.; Fleg, J.L.; O’Brien, K.D.; Marcovina, S.M. Relationship of Baseline HDL Subclasses, Small Dense LDL and LDL Triglyceride to Cardiovascular Events in the AIM-HIGH Clinical Trial. Atherosclerosis 2016, 251, 454–459. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Ma, X.; You, G.; Zhang, X.; Fu, Q. A Novel Multiplex HRM Assay to Detect Clopidogrel Resistance. Sci. Rep. 2017, 7, 16021. [Google Scholar] [CrossRef]
- Garritano, S.; Gemignani, F.; Voegele, C.; Nguyen-Dumont, T.; Le Calvez-Kelm, F.; De Silva, D.; Lesueur, F.; Landi, S.; Tavtigian, S.V. Determining the Effectiveness of High-Resolution Melting Analysis for SNP Genotyping and Mutation Scanning at the TP53 Locus. BMC Genet. 2009, 10, 5. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-Resolution DNA Melting Analysis for Simple and Efficient Molecular Diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
- Tan, J.; Che, Y.; Liu, Y.; Hu, J.; Wang, W.; Hu, L.; Zhou, Q.; Wang, H.; Li, J. CELSR2 deficiency suppresses lipid accumulation in hepatocyte by impairing the UPR and elevating ROS level. FASEB J. 2021, 35, e21908. [Google Scholar] [CrossRef]
- Noto, D.; Cefalù, A.B.; Martinelli, N.; Giammanco, A.; Spina, R.; Barbagallo, C.M.; Caruso, M.; Novo, S.; Sarullo, F.; Pernice, V.; et al. Rs629301 CELSR2 Polymorphism Confers a Ten-Year Equivalent Risk of Critical Stenosis Assessed by Coronary Angiography. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 1542–1547. [Google Scholar] [CrossRef]
- Hochner, H.; Allard, C.; Granot-Hershkovitz, E.; Chen, J.; Sitlani, C.M.; Sazdovska, S.; Lumley, T.; McKnight, B.; Rice, K.; Enquobahrie, D.A.; et al. Parent-of-Origin Effects of the APOB Gene on Adiposity in Young Adults. PLoS Genet. 2015, 11, e1005573. [Google Scholar] [CrossRef]
- Smalinskiene, A.; Petkeviciene, J.; Luksiene, D.; Jureniene, K.; Klumbiene, J.; Lesauskaite, V. Association Between APOE, SCARB1, PPARα Polymorphisms and Serum Lipids in a Population of Lithuanian Adults. Lipids Health Dis. 2013, 12, 120. [Google Scholar] [CrossRef]
- Fairoozy, R.H.; White, J.; Palmen, J.; Kalea, A.Z.; Humphries, S.E. Identification of the Functional Variant(s) That Explain the Low-Density Lipoprotein Receptor (LDLR) GWAS SNP Rs6511720 Association with Lower LDL-C and Risk of CHD. PLoS ONE 2016, 11, e0167676. [Google Scholar] [CrossRef]
- Feder, J.N.; Gnirke, A.; Thomas, W.; Tsuchihashi, Z.; Ruddy, D.A.; Basava, A.; Dormishian, F.; Domingo, R.; Ellis, M.C.; Fullan, A.; et al. A Novel MHC Class I-like Gene Is Mutated in Patients with Hereditary Haemochromatosis. Nat. Genet. 1996, 13, 399–408. [Google Scholar] [CrossRef]
- Demetz, E.; Tymoszuk, P.; Hilbe, R.; Volani, C.; Haschka, D.; Heim, C.; Auer, K.; Lener, D.; Zeiger, L.B.; Pfeifhofer-Obermair, C.; et al. The Haemochromatosis Gene Hfe and Kupffer Cells Control LDL Cholesterol Homeostasis and Impact on Atherosclerosis Development. Eur. Heart J. 2020, 41, 3949–3959B. [Google Scholar] [CrossRef]
- Chambers, J.C.; Zhang, W.; Sehmi, J.S.; Li, X.; Wass, M.N.; Van der Harst, P.; Holm, H.; Sanna, S.; Kavousi, M.; Baumeister, S.E.; et al. Genome-Wide Association Study Identifies Loci Influencing Concentrations of Liver Enzymes in Plasma. Nat. Genet. 2011, 43, 1131–1138. [Google Scholar] [CrossRef]
- Ference, B.A.; Braunwald, E.; Catapano, A.L. The LDL cumulative exposure hypothesis: Evidence and practical applications. Nat. Rev. Cardiol. 2024, 21, 701–716. [Google Scholar] [CrossRef]
- Schubert, J.; Lindahl, B.; Melhus, H.; Renlund, H.; Leosdottir, M.; Yari, A.; Ueda, P.; Jernberg, T.; Hagström, E. Elevated Low-Density Lipoprotein Cholesterol: An Inverse Marker of Morbidity and Mortality in Patients with Myocardial Infarction. J. Intern. Med. 2023, 294, 616–627. [Google Scholar] [CrossRef]
- Potter, J.M.; Hickman, P.E.; Cullen, L. Troponins in Myocardial Infarction and Injury. Aust. Prescr. 2022, 45, 53–57. [Google Scholar] [CrossRef]
- Peasey, A.; Bobak, M.; Kubinova, R.; Malyutina, S.; Pajak, A.; Tamosiunas, A.; Pikhart, H.; Nicholson, A.; Marmot, M. Determinants of Cardiovascular Disease and Other Non-Communicable Diseases in Central and Eastern Europe: Rationale and Design of the HAPIEE Study. BMC Public Health 2006, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. Available online: http://www.biomedcentral.com/1471-2105/13/134 (accessed on 8 December 2023). [CrossRef] [PubMed]
- Nassar, L.R.; Barber, G.P.; Benet-Pagès, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, B.T.; et al. The UCSC Genome Browser Database: 2023 Update. Nucleic Acids Res. 2023, 51, D1188–D1195. [Google Scholar] [CrossRef]
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A Suite for Analysis and Design of Nucleic Acid Oligomers. Nucleic Acids Res. 2008, 36, 163–169. [Google Scholar] [CrossRef]
- Dwight, Z.; Palais, R.; Wittwer, C.T. UMELT: Prediction of High-Resolution Melting Curves and Dynamic Melting Profiles of PCR Products in a Rich Web Application. Bioinformatics 2011, 27, 1019–1020. [Google Scholar] [CrossRef]
- Austin, M.A.; Hutter, C.M.; Zimmern, R.L.; Humphries, S.E. HUMAN GENOME EPIDEMIOLOGY (HuGE) REVIEWS Genetic Causes of Monogenic Heterozygous Familial Hypercholesterolemia: A HuGE Prevalence Review Downloaded From. Am. J. Epidemiol. 2004, 160, 407–420. [Google Scholar] [CrossRef]
- Stone, N.J.; Robinson, J.G.; Lichtenstein, A.H.; Bairey Merz, C.N.; Blum, C.B.; Eckel, R.H.; Goldberg, A.C.; Gordon, D.; Levy, D.; Lloyd-Jones, D.M.; et al. 2013 ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014, 129 (Suppl. S1), S1–S45, Erratum in Circulation 2015, 132, e396. https://doi.org/10.1161/01.cir.0000437738.63853.7a. [Google Scholar] [CrossRef]
- Benn, M.; Watts, G.F.; Tybjærg-Hansen, A.; Nordestgaard, B.G. Mutations Causative of Familial Hypercholesterolaemia: Screening of 98 098 Individuals from the Copenhagen General Population Study Estimated a Prevalence of 1 in 217. Eur. Heart J. 2016, 37, 1384–1394. [Google Scholar] [CrossRef]

| Variable | Patient Group n = 93 | Comparison Group n = 61 | p-Value Between Two Medians | ||
|---|---|---|---|---|---|
| Median | Min-Max | Median | Min-Max | ||
| Age in years | 68 | 42–96 | 71 | 45–72 | 0.259 |
| LDL-C concentration (mmol/L) at time of enrolment | 3.88 | 2.87–6.63 | 5.67 | 3.19–9.7 | <0.001 |
| N | % | N | % | χ2, p-value | |
| Sex Men Women | 48 45 | 51.6 48.4 | 34 27 | 55.7 44.3 | χ2 = 0.252, p = 0.625 |
| Smoking Current Past | 0 55 | 0 59.1 | 3 14 | 4.9 23 | p = 0.0114 # |
| Anterior STEMI | 38 | 40.9 | - | - | |
| Family history of ischemic heart disease | 39 | 41.9 | 0 | 0 | - |
| Hypertension | 48 | 51.6 | 45 | 75 | χ2 = 8.368, p = 0.004 |
| Diabetes mellitus | 15 | 16.1 | 0 | 0 | - |
| Renal insufficiency | 11 | 11.8 | - | - | - |
| Statins | 29 * | 31.2 * | 3 ** | 4.9 ** | p = 0.0001 # |
| dbSNP ID | HGVS | GnomAD Freq (%) | GnomAD EU FREQ (%) | Frequency (%) in the Patient Group (n = 93) | Frequency (%) in the Comparison Group (n = 61) |
|---|---|---|---|---|---|
| rs6511720 | NM_000527.5(LDLR): c.67 + 2015G > T | 11.38 | 11.49 | 16.13 | 36.07 |
| rs1800562 | NM_000410.4(HFE): c.845G > A (p.Cys282Tyr) | 5.698 | 7.102 | 4.301 | 4.92 |
| rs629301 | NM_001408.3(CELSR2): c.*1635G > T | 74.73 | 77.87 | 75.27 | 55.74 |
| rs11220462 | NM_006278.2(ST3GAL4): c.−61 + 17755G > A | 12.41 | 13.77 | 43.01 | 26.23 |
| rs1367117 | NM_000384.3(APOB): c.293C > T | 29.58 | 33.08 | 56.99 | 59.02 |
| rs7412 | NM_000041.2(APOE): c.526C > T | 7.42 | 7.83 | 15.05 | 19.67 |
| rs429358 | M_000041.4(APOE): c.388T > C | 14.85 | 15.06 | 22.58 | 16.39 |
| Name | Sequence | Amount Added (µM) | Reaction Mix |
|---|---|---|---|
| CELSR2_rs629301_F | TTGTACAGTTTGGTTGTTGCTG | 2.5 | 2 |
| CELSR2_rs629301_R | TACCACACAGAAGCGGACAG | 2.5 | 2 |
| APOB_rs1367117_F | CAGGGTTGAAGCCATACACC | 15 | 1 |
| APOB_rs1367117_R | TCTCAGGTTGAGCTGGAGGT | 15 | 1 |
| ST3GAL4_rs11220462_F | AGCGATGCTATCCGATGAAC | 7.5 | 2 |
| ST3GAL4_rs11220462_R | CAACTCCACACACCCAACAC | 7.5 | 2 |
| LDLR_rs6511720_F | TCACCAATCAACCTCTTCCTT | 16.25 | 1 |
| LDLR_rs6511720_R | GCCTTGCCTAAGACTTCCT | 16.25 | 1 |
| HFE_rs1800562_F | CTGGATAACCTTGGCTGTACC | 3.75 | 1 |
| HFE_rs1800562_R | GATCACAATGAGGGGCTGAT | 3.75 | 1 |
| dbSNP rs ID | Nearest Gene | Reference Allele | Risk Allele | Weight for Score Calculation |
|---|---|---|---|---|
| rs629301 | CELSR2 | G | T | 0.15 |
| rs1367117 | APOB | G | A | 0.1 |
| rs11220462 | ST3GAL4 | G | A | 0.05 |
| rs6511720 | LDLR | G | G | 0.18 |
| rs1800562 | HFE | G | G | 0.057 |
| APOE Haplotype | Weight for Score Calculation |
|---|---|
| ε2ε2 | −0.9 |
| ε2ε3 | −0.4 |
| ε2ε4 | −0.2 |
| ε3ε3 | 0 |
| ε3ε4 | 0.1 |
| ε4ε4 | 0.2 |
| Name | Sequence |
|---|---|
| LDLR_rs6511720_seq_F | TGCCACTCAGTTTTACAAAAGAA |
| LDLR_rs6511720_seq_R | TGGAGGAAAACATCAGGGGT |
| HFE_rs1800562_seq_F | CAATGGGGATGGGACCTAC |
| HFE_rs1800562_seq_R | CACCCCCTAACAAAGAGCAG |
| CELSR2_ rs629301_seq_F | TCTCCCCTCAGCAATTCCTG |
| CELSR2_ rs629301_seq_R | TACCACACAGAAGCGGACAG |
| ST3GAL4_rs11220462_seq_F | AGCGATGCTATCCGATGAAC |
| ST3GAL4_rs11220462_seq_R | CAGCTTCTCTACTTCCCAGCA |
| APOB_rs1367117_seq_F | TGACTTACCTGGACATGGCT |
| APOB_rs1367117_seq_R | CCTCAATGCTCTGCTACCCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čereškevičius, D.; Čiapienė, I.; Aldujeli, A.; Zabiela, V.; Lesauskaitė, V.; Zubielienė, K.; Raškevičius, V.; Žaliaduonytė, D.; Unikas, R.; Pranevičius, R.; et al. The Impact of SNP Score on Low-Density Lipoprotein Cholesterol Concentration and Coronary Artery Disease. Int. J. Mol. Sci. 2025, 26, 2337. https://doi.org/10.3390/ijms26052337
Čereškevičius D, Čiapienė I, Aldujeli A, Zabiela V, Lesauskaitė V, Zubielienė K, Raškevičius V, Žaliaduonytė D, Unikas R, Pranevičius R, et al. The Impact of SNP Score on Low-Density Lipoprotein Cholesterol Concentration and Coronary Artery Disease. International Journal of Molecular Sciences. 2025; 26(5):2337. https://doi.org/10.3390/ijms26052337
Chicago/Turabian StyleČereškevičius, Darius, Ieva Čiapienė, Ali Aldujeli, Vytautas Zabiela, Vaiva Lesauskaitė, Kristina Zubielienė, Vytautas Raškevičius, Diana Žaliaduonytė, Ramūnas Unikas, Robertas Pranevičius, and et al. 2025. "The Impact of SNP Score on Low-Density Lipoprotein Cholesterol Concentration and Coronary Artery Disease" International Journal of Molecular Sciences 26, no. 5: 2337. https://doi.org/10.3390/ijms26052337
APA StyleČereškevičius, D., Čiapienė, I., Aldujeli, A., Zabiela, V., Lesauskaitė, V., Zubielienė, K., Raškevičius, V., Žaliaduonytė, D., Unikas, R., Pranevičius, R., Simanauskas, I., Bakšytė, G., Tamošiūnas, A., Lukšienė, D., Šakalytė, G., & Tatarūnas, V. (2025). The Impact of SNP Score on Low-Density Lipoprotein Cholesterol Concentration and Coronary Artery Disease. International Journal of Molecular Sciences, 26(5), 2337. https://doi.org/10.3390/ijms26052337








