Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi
Abstract
1. Introduction
2. Results
2.1. Mapping Reads
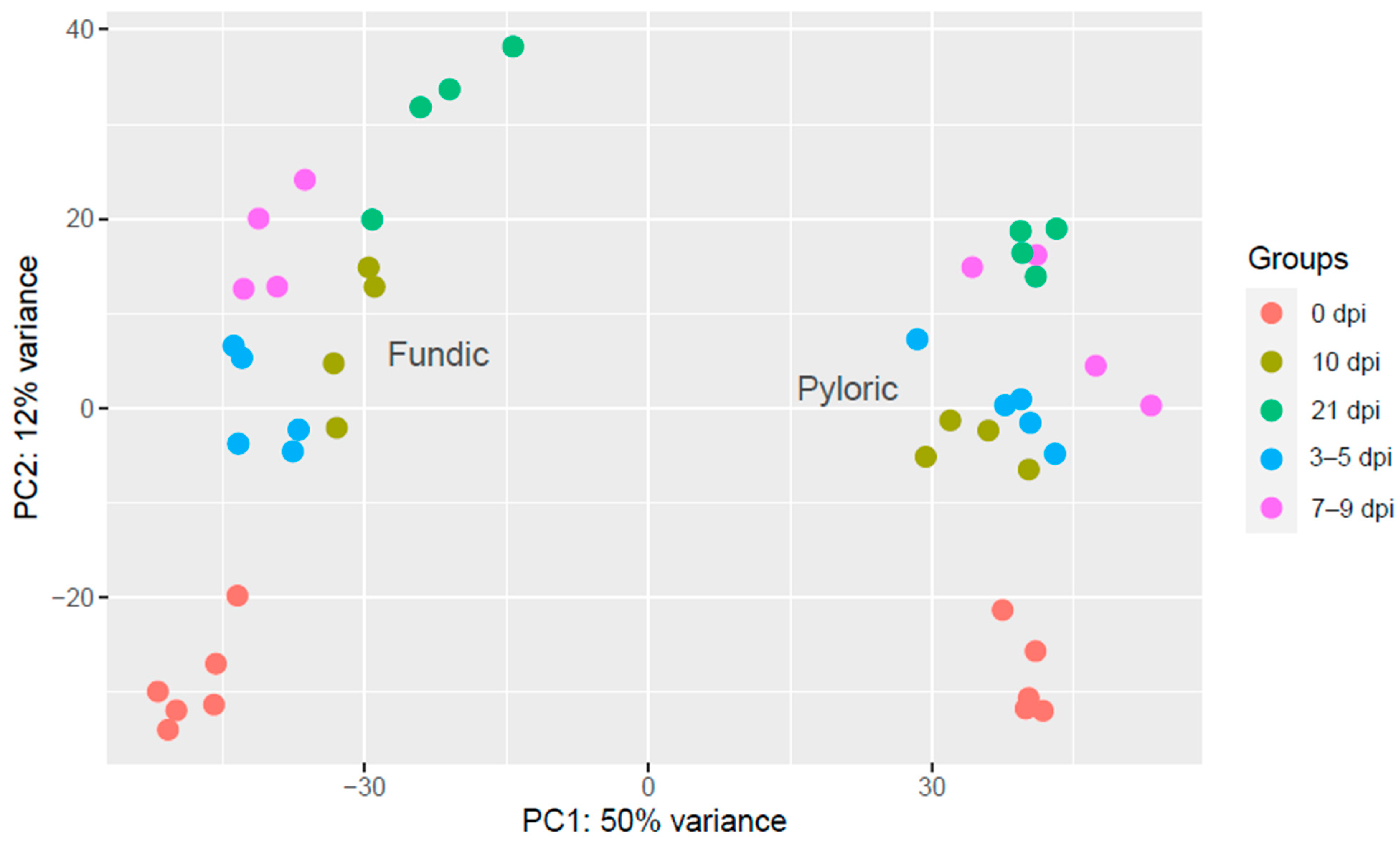
2.2. Quality Assessment of the Samples
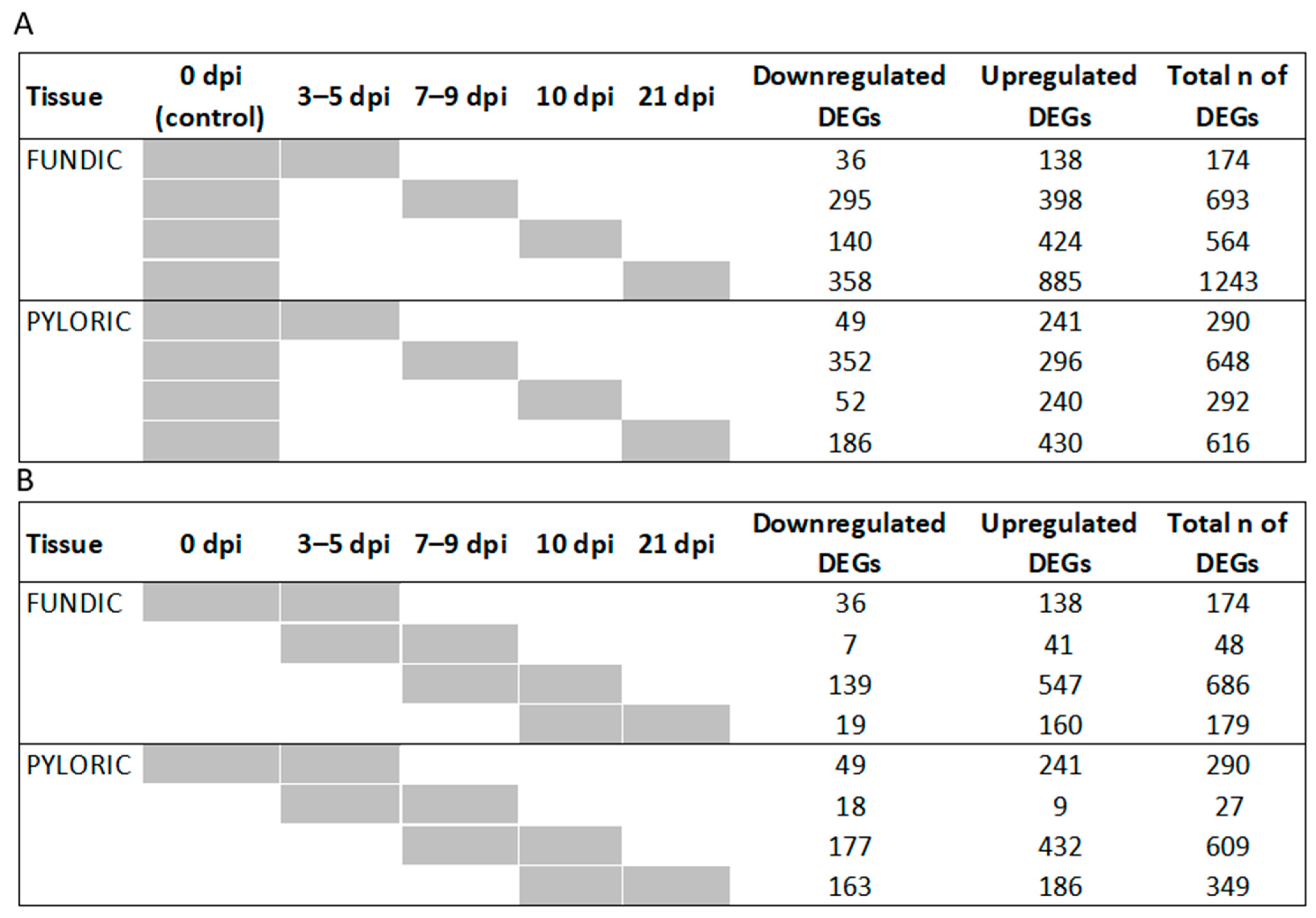
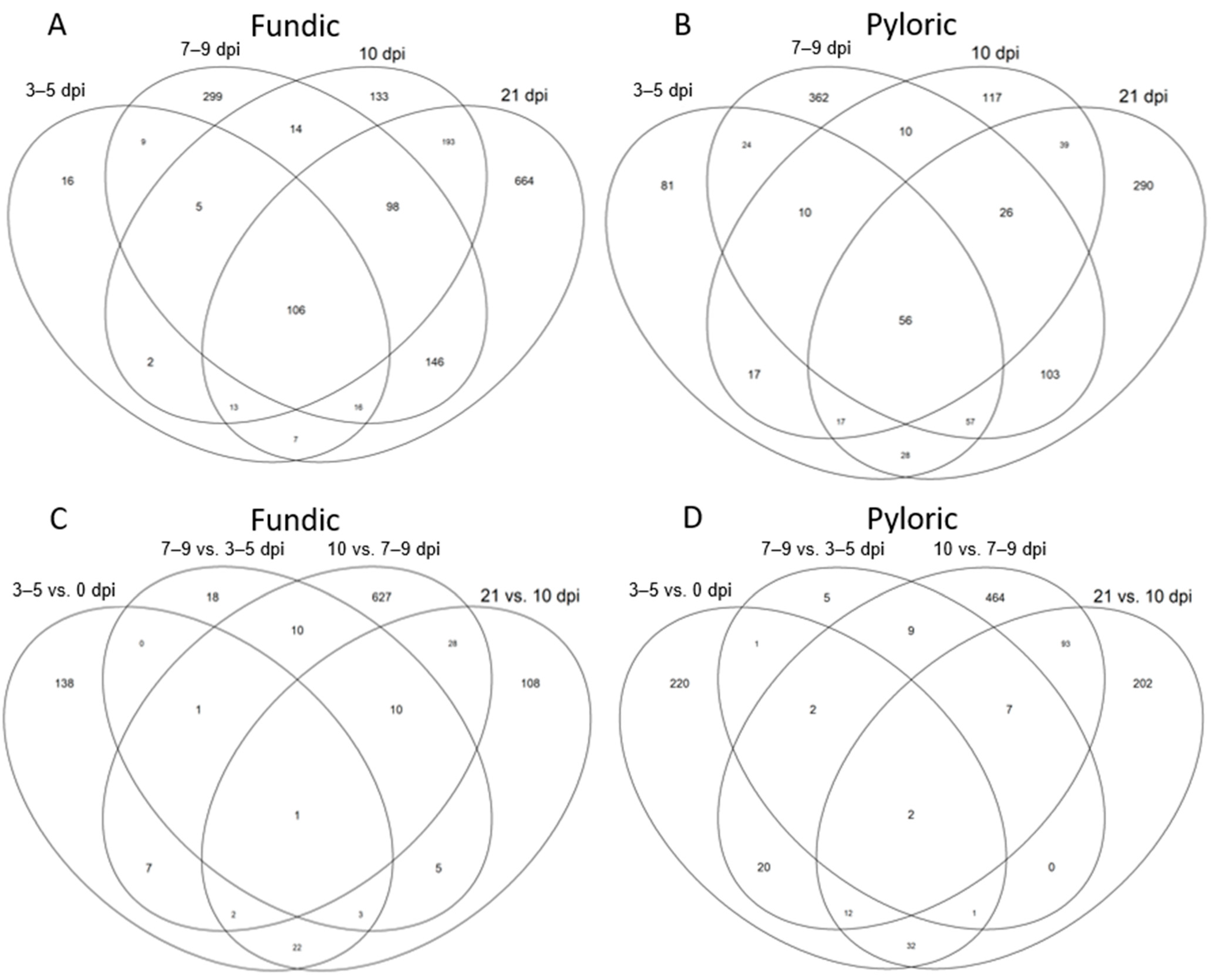
2.3. Differentially Expressed Genes (DEGs)
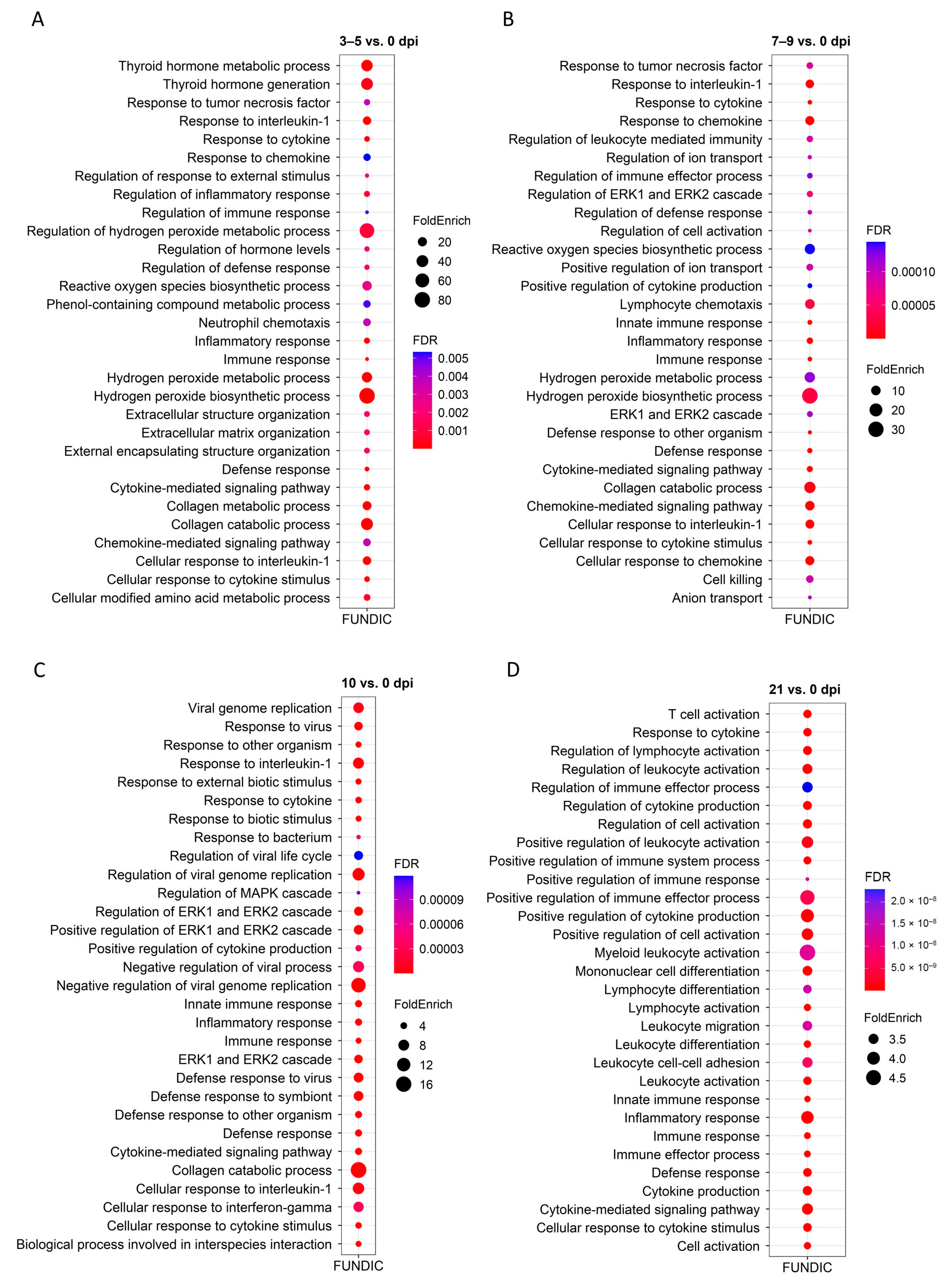
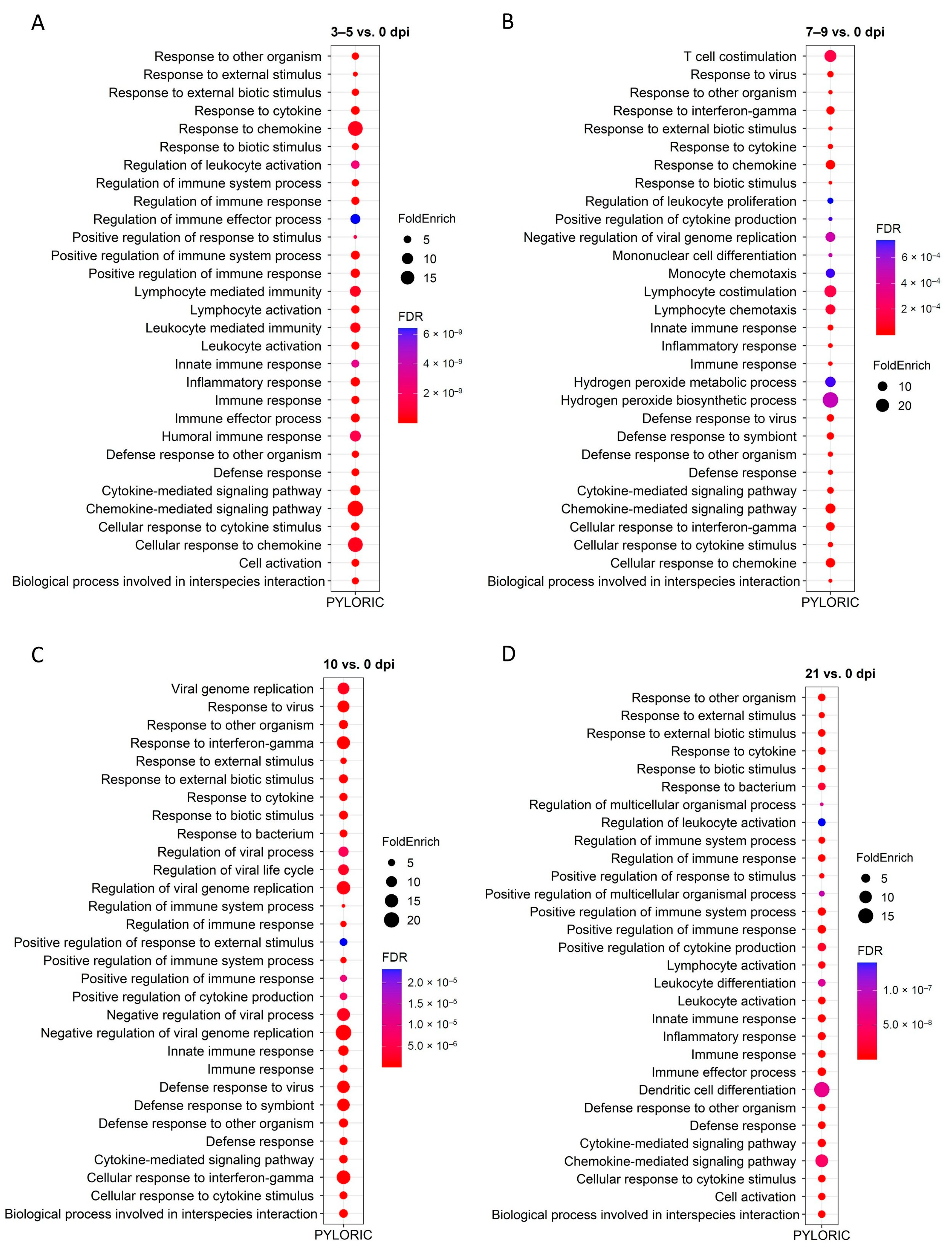
2.4. GO and KEGG Enrichment Analysis of DEGs
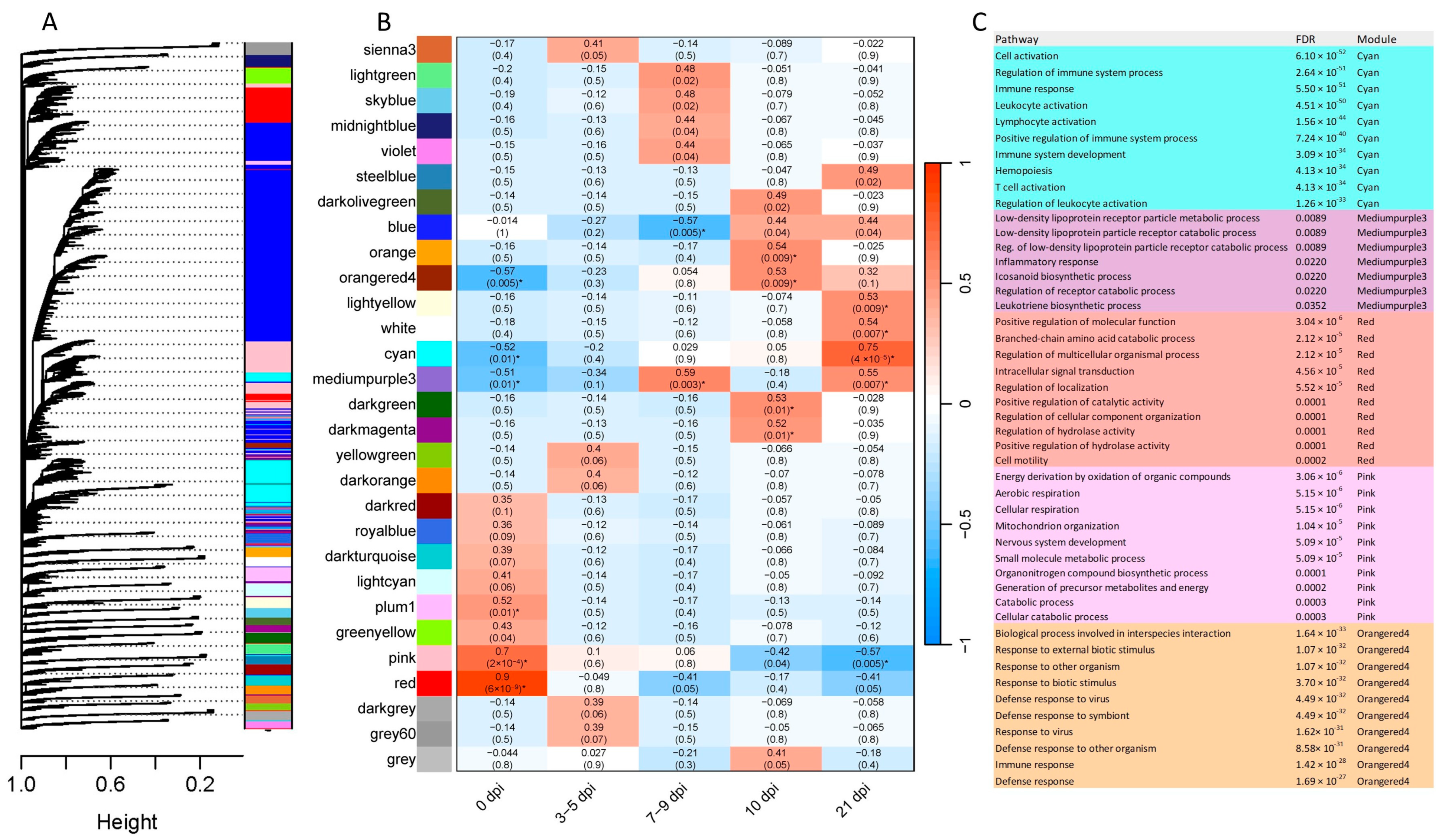
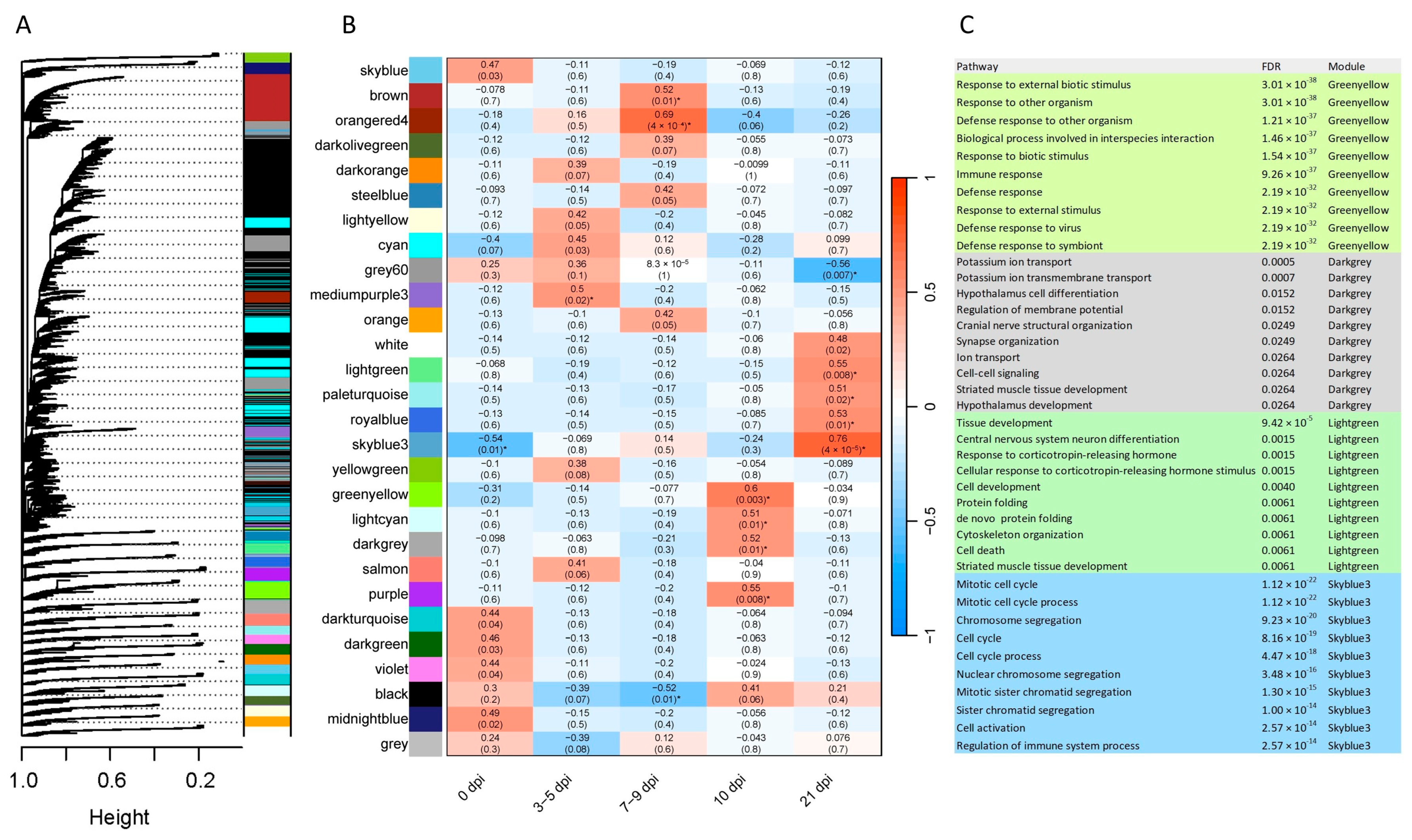
2.5. Gene Co-Expression
2.6. Identification of Networks and Canonical Pathways Using IPA
3. Discussion
3.1. Global Abomasal Responses to O. ostertagi Infection
3.2. Host Immune Response to Infection
3.3. Viral Infection Responses Induced by O. ostertagi
3.4. Co-Expression Analysis and Immune-Related Processes
4. Materials and Methods
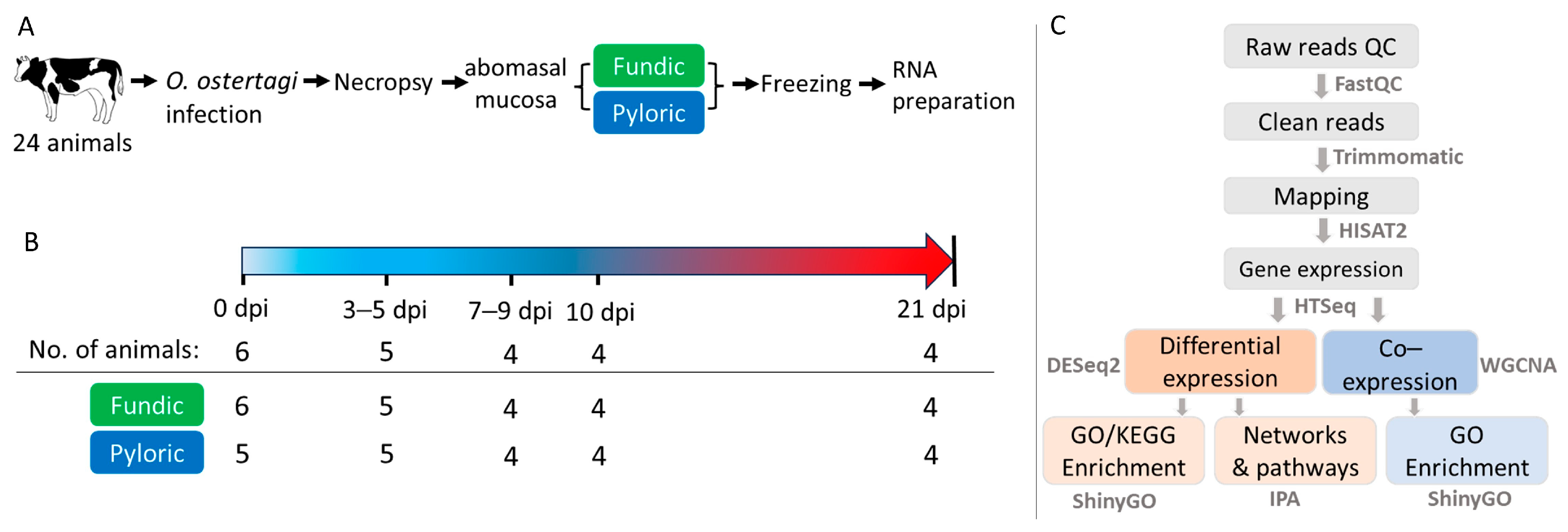
4.1. Experimental Animals
4.2. RNA Isolation, Library Construction, and Sequencing
4.3. Pre-Processing Sequence Data and Alignment
4.4. Gene Expression and Differential Expression Analysis
4.5. Co-Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BARC | Beltsville Agricultural Research Center |
| BP | biological processes |
| CCL or CXCL | C-C or C-X-C motif chemokine ligand |
| CCR or CXCR | C-C or C-X-C motif chemokine receptor |
| DEGs | differentially expressed genes |
| DPI | days post-infection |
| EPX | eosinophil peroxidase |
| ES | excretory/secretory |
| FUN | abomasal fundic mucosa |
| GI | gastrointestinal |
| GZMB | granzyme B |
| GO | gene ontology |
| IFNG | interferon-gamma |
| Ig | immunoglobulin |
| IL | interleukin |
| IPA | Ingenuity Pathway Analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| L3 | stage 3 larvae |
| MMPs | matrix metalloproteinases |
| PBMCs | peripheral blood monocytes |
| PCA | principal component analysis |
| PCR | polymerase chain reaction |
| PYL | abomasal pyloric mucosa |
| PRG3 | proteoglycan 3 |
| RNA-seq | RNA sequencing |
| SRA | Sequence Read Archive |
| TCE | T cell exhaustion |
| TCN1 | Transcobalamin I |
| Th | T helper |
| TOM | topological overlap matrix |
| WGCNA | weighted gene co-expression network analysis |
References
- Klesius, P.H. Immunity to Ostertagia ostertagi. Vet. Parasitol. 1988, 27, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.T. Pathophysiology of infection with Ostertagia ostertagi in cattle. Vet. Parasitol. 1993, 46, 143–158. [Google Scholar] [CrossRef]
- Mihi, B.; Van Meulder, F.; Rinaldi, M.; Van Coppernolle, S.; Chiers, K.; Van den Broeck, W.; Goddeeris, B.; Vercruysse, J.; Claerebout, E.; Geldhof, P. Analysis of cell hyperplasia and parietal cell dysfunction induced by Ostertagia ostertagi infection. Vet. Res. 2013, 44, 121. [Google Scholar] [CrossRef] [PubMed]
- Claerebout, E.; Vercauteren, I.; Geldhof, P.; Olbrechts, A.; Zarlenga, D.S.; Goddeeris, B.M.; Vercruysse, J. Cytokine responses in immunized and non-immunized calves after Ostertagia ostertagi infection. Parasite Immunol. 2005, 27, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Guzmán, M.A.; Cuenca-Verde, C.; Valdivia-Anda, G.; Cuéllar-Ordaz, J.A.; Alba-Hurtado, F. Differential immune response between fundic and pyloric abomasal regions upon experimental ovine infection with Haemonchus contortus. Vet. Parasitol. 2012, 185, 175–180. [Google Scholar] [CrossRef]
- Jacobs, K.; Charvat, R.; Arrizabalaga, G. Identification of Fis1 Interactors in Toxoplasma gondii Reveals a Novel Protein Required for Peripheral Distribution of the Mitochondrion. mBio 2020, 11, e02732-19. [Google Scholar] [CrossRef]
- Bakshi, M.; Tuo, W.; Aroian, R.V.; Zarlenga, D. Immune reactivity and host modulatory roles of two novel Haemonchus contortus cathepsin B-like proteases. Parasit. Vectors 2021, 14, 580. [Google Scholar] [CrossRef]
- Borji, H.; Haghparast, A.; Soleimani, N.; Azizzadeh, M.; Nazemshirazi, M.H. The effects of Ostertagia occidentalis somatic antigens on ovine TLR2 and TLR4 expression. Iran. J. Parasitol. 2015, 10, 498–504. [Google Scholar]
- Li, R.W.; Hou, Y.; Li, C.; Gasbarre, L.C. Localized complement activation in the development of protective immunity against Ostertagia ostertagi infections in cattle. Vet. Parasitol. 2010, 174, 247–256. [Google Scholar] [CrossRef]
- Li, R.W.; Sonstegard, T.S.; Van Tassell, C.P.; Gasbarre, L.C. Local inflammation as a possible mechanism of resistance to gastrointestinal nematodes in Angus heifers. Vet. Parasitol. 2007, 145, 100–107. [Google Scholar] [CrossRef]
- Mihi, B.; van Meulder, F.; Vancoppernolle, S.; Rinaldi, M.; Chiers, K.; van den Broeck, W.; Goddeeris, B.M.; Vercruysse, J.; Claerebout, E.; Geldhof, P. Analysis of the mucosal immune responses induced by single and trickle infections with the bovine abomasal nematode Ostertagia ostertagi. Parasite Immunol. 2014, 36, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Rinaldi, M.; Capuco, A.V. Characterization of the abomasal transcriptome for mechanisms of resistance to gastrointestinal nematodes in cattle. Vet. Res. 2011, 42, 114. [Google Scholar] [CrossRef]
- Van Meulder, F.; Ratman, D.; Van Coppernolle, S.; Borloo, J.; Li, R.W.; Chiers, K.; Van den Broeck, W.; De Bosscher, K.; Claerebout, E.; Geldhof, P. Analysis of the protective immune response following intramuscular vaccination of calves against the intestinal parasite Cooperia oncophora. Int. J. Parasitol. 2015, 45, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 2008, 9, 559. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Claerebout, E.; Vercruysse, J. The immune response and the evaluation of acquired immunity against gastrointestinal nematodes in cattle: A review. Parasitology 2000, 120, S25–S42. [Google Scholar] [CrossRef] [PubMed]
- Gasbarre, L.C.; Smith, L.L.; Hoberg, E.; Pilitt, P.A. Further characterization of a cattle nematode population with demonstrated resistance to current anthelmintics. Vet. Parasitol. 2009, 166, 275–280. [Google Scholar] [CrossRef]
- Sutherland, I.A.; Leathwick, D.M. Anthelmintic resistance in nematode parasites of cattle: A global issue? Trends Parasitol. 2011, 27, 176–181. [Google Scholar] [CrossRef]
- Tuo, W.; Li, L.; Lv, Y.; Carrillo, J.; Brown, D.; Davis, W.C.; Song, J.; Zarlenga, D.; Xiao, Z. Abomasal mucosal immune responses of cattle with limited or continuous exposure to pasture-borne gastrointestinal nematode parasite infection. Vet. Parasitol. 2016, 229, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Umair, S.; Savoian, M.S.; Simpson, H.V. Abomasal dysfunction and cellular and mucin changes during infection of sheep with larval or adult Teladorsagia circumcincta. PLoS ONE 2017, 12, e0186752. [Google Scholar] [CrossRef]
- Murugaiah, V.; Tsolaki, A.G.; Kishore, U. Collectins: Innate Immune Pattern Recognition Molecules. Adv. Exp. Med. Biol. 2020, 1204, 75–127. [Google Scholar]
- Holmskov, U.L. Collectins and collectin receptors in innate immunity. APMIS Suppl. 2000, 100, 1–59. [Google Scholar] [CrossRef] [PubMed]
- Nonnecke, E.B.; Castillo, P.A.; Johansson, M.E.V.; Hollox, E.J.; Shen, B.; Lönnerdal, B.; Bevins, C.L. Human intelectin-2 (ITLN2) is selectively expressed by secretory Paneth cells. FASEB J. 2022, 36, e22200. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Holm, D.; Moeller, V.; Vitved, L.; Bendixen, C.; Reid, K.B.; Skjoedt, K.; Holmskov, U. CL-46, a novel collectin highly expressed in bovine thymus and liver. J. Immunol. 2002, 169, 5726–5734. [Google Scholar] [CrossRef]
- Blanco Vázquez, C.; Balseiro, A.; Alonso-Hearn, M.; Juste, R.A.; Iglesias, N.; Canive, M.; Casais, R. Bovine Intelectin 2 Expression as a Biomarker of Paratuberculosis Disease Progression. Animals 2021, 11, 1370. [Google Scholar] [CrossRef]
- Pemberton, A.D.; Knight, P.A.; Wright, S.H.; Miller, H.R. Proteomic analysis of mouse jejunal epithelium and its response to infection with the intestinal nematode, Trichinella spiralis. Proteomics 2004, 4, 1101–1108. [Google Scholar] [CrossRef]
- Fingleton, B. Matrix metalloproteinases as regulators of inflammatory processes. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864 Pt A, 2036–2042. [Google Scholar] [CrossRef]
- Hong, S.Y.; Jiang, H.C.; Xu, W.C.; Zeng, H.S.; Wang, S.G.; Qin, B.L. Bioinformatics analysis reveals the potential role of matrix metalloproteinases in immunity and urolithiasis. Front. Immunol. 2023, 14, 1158379. [Google Scholar] [CrossRef] [PubMed]
- Salomão, R.; Assis, V.; de Sousa Neto, I.V.; Petriz, B.; Babault, N.; Durigan, J.L.Q.; de Cássia Marqueti, R. Involvement of Matrix Metalloproteinases in COVID-19: Molecular Targets, Mechanisms, and Insights for Therapeutic Interventions. Biology 2023, 12, 843. [Google Scholar] [CrossRef] [PubMed]
- Elkington, P.T.; O’Kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Bellac, C.L.; Dufour, A.; Krisinger, M.J.; Loonchanta, A.; Starr, A.E.; Auf dem Keller, U.; Lange, P.F.; Goebeler, V.; Kappelhoff, R.; Butler, G.S.; et al. Macrophage matrix metalloproteinase-12 dampens inflammation and neutrophil influx in arthritis. Cell Rep. 2014, 9, 618–632. [Google Scholar] [CrossRef]
- Amor, M.; Bianco, V.; Buerger, M.; Lechleitner, M.; Vujić, N.; Dobrijević, A.; Akhmetshina, A.; Pirchheim, A.; Schwarz, B.; Pessentheiner, A.R.; et al. Genetic deletion of MMP12 ameliorates cardiometabolic disease by improving insulin sensitivity, systemic inflammation, and atherosclerotic features in mice. Cardiovasc. Diabetol. 2023, 22, 327. [Google Scholar] [CrossRef]
- Collison, J. MMP12 makes the cut. Nat. Rev. Rheumatol. 2018, 14, 501. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.M.; Collins, M.H.; Kemme, K.A.; Sherrill, J.D.; Wen, T.; Rochman, M.; Stucke, E.M.; Amin, L.; Tai, H.; Putnam, P.E.; et al. Cadherin 26 is an alpha integrin-binding epithelial receptor regulated during allergic inflammation. Mucosal Immunol. 2017, 10, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Langhe, R.P.; Gudzenko, T.; Bachmann, M.; Becker, S.F.; Gonnermann, C.; Winter, C.; Abbruzzese, G.; Alfandari, D.; Kratzer, M.C.; Franz, C.M.; et al. Cadherin-11 localizes to focal adhesions and promotes cell-substrate adhesion. Nat. Commun. 2016, 7, 10909. [Google Scholar] [CrossRef]
- Derycke, L.D.; Bracke, M.E. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int. J. Dev. Biol. 2004, 48, 463–476. [Google Scholar] [CrossRef]
- Liu, C.L.; Shi, G.P. Calcium-activated chloride channel regulator 1 (CLCA1): More than a regulator of chloride transport and mucus production. World Allergy Organ. J. 2019, 12, 100077. [Google Scholar] [CrossRef]
- Jenvey, C.J.; Alenizi, D.; Almasi, F.; Cairns, C.; Holmes, A.; Sloan, S.; Stear, M.J. Bioinformatic analysis of eosinophil activity and its implications for model and target species. Parasitology 2020, 147, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Gurish, M.F.; Humbles, A.; Tao, H.; Finkelstein, S.; Boyce, J.A.; Gerard, C.; Friend, D.S.; Austen, K.F. CCR3 is required for tissue eosinophilia and larval cytotoxicity after infection with Trichinella spiralis. J. Immunol. 2002, 168, 5730–5736. [Google Scholar] [CrossRef]
- Immler, R.; Nussbaumer, K.; Doerner, A.; El Bounkari, O.; Huber, S.; Abisch, J.; Napoli, M.; Schmidt, S.; Margraf, A.; Pruenster, M.; et al. CCR3-dependent eosinophil recruitment is regulated by sialyltransferase ST3Gal-IV. Proc. Natl. Acad. Sci. USA 2024, 121, e2319057121. [Google Scholar] [CrossRef] [PubMed]
- Viola-Villegas, N.; Rabideau, A.E.; Bartholomä, M.; Zubieta, J.; Doyle, R.P. Targeting the cubilin receptor through the vitamin B(12) uptake pathway: Targeting the cubilin receptor through the vitamin B(12) uptake pathway: Cytotoxicity and mechanistic insight through fluorescent Re(I) delivery. J. Med. Chem. 2009, 52, 5253–5261. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.J.; Wang, Y.J.; Yue, M.; Zhao, L.M.; Guo, Y.D.; Liu, Y.P.; Yang, H.C.; Liu, F.; Zhang, X.; Zhi, L.H.; et al. High expression of TCN1 is a negative prognostic biomarker and can predict neoadjuvant chemosensitivity of colon cancer. Sci. Rep. 2020, 10, 11951. [Google Scholar] [CrossRef]
- Fox, M.T. Pathophysiology of infection with gastrointestinal nematodes in domestic ruminants: Recent developments. Vet. Parasitol. 1997, 72, 285–297; discussion 297–308. [Google Scholar] [CrossRef]
- Ray, U.; Pathoulas, C.L.; Thirusangu, P.; Purcell, J.W.; Kannan, N.; Shridhar, V. Exploiting LRRC15 as a Novel Therapeutic Target in Cancer. Cancer Res. 2022, 82, 1675–1681. [Google Scholar] [CrossRef]
- Loo, L.; Waller, M.A.; Moreno, C.L.; Cole, A.J.; Stella, A.O.; Pop, O.T.; Jochum, A.K.; Ali, O.H.; Denes, C.E.; Hamoudi, Z.; et al. Fibroblast-expressed LRRC15 is a receptor for SARS-CoV-2 spike and controls antiviral and antifibrotic transcriptional programs. PLoS Biol. 2023, 21, e3001967. [Google Scholar] [CrossRef]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- El-Naccache, D.W.; Haskó, G.; Gause, W.C. Early Events Triggering the Initiation of a Type 2 Immune Response. Trends Immunol. 2021, 42, 151–164. [Google Scholar] [CrossRef]
- Canals, A.; Zarlenga, D.S.; Almeria, S.; Gasbarre, L.C. Cytokine profile induced by a primary infection with Ostertagia ostertagi in cattle. Vet. Immunol. Immunopathol. 1997, 58, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Gasbarre, L.C.; Leighton, E.A.; Sonstegard, T. Role of the bovine immune system and genome in resistance to gastrointestinal nematodes. Vet. Parasitol. 2001, 98, 51–64. [Google Scholar] [CrossRef]
- Zaros, L.G.; Bricarello, P.A.; Amarante, A.F.; Rocha, R.A.; Kooyman, F.N.; De Vries, E.; Coutinho, L.L. Cytokine gene expression in response to Haemonchus placei infections in Nelore cattle. Vet. Parasitol. 2010, 171, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Herzig, C.T.; Baldwin, C.L. Genomic organization and classification of the bovine WC1 genes and expression by peripheral blood gamma delta T cells. BMC Genomics 2009, 10, 191. [Google Scholar] [CrossRef]
- Rogers, A.N.; Vanburen, D.G.; Hedblom, E.E.; Tilahun, M.E.; Telfer, J.C.; Baldwin, C.L. Gammadelta T cell function varies with the expressed WC1 coreceptor. J. Immunol. 2005, 174, 3386–3393. [Google Scholar] [CrossRef] [PubMed]
- Aubatin, A.; Sako, N.; Decrouy, X.; Donnadieu, E.; Molinier-Frenkel, V.; Castellano, F. IL4-induced gene 1 is secreted at the immune synapse and modulates TCR activation independently of its enzymatic activity. Eur. J. Immunol. 2018, 48, 106–119. [Google Scholar] [CrossRef]
- Tau, G.; Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef]
- Van Meulder, F.; Van Coppernolle, S.; Borloo, J.; Rinaldi, M.; Li, R.W.; Chiers, K.; Van den Broeck, W.; Vercruysse, J.; Claerebout, E.; Geldhof, P. Granule exocytosis of granulysin and granzyme B as a potential key mechanism in vaccine-induced immunity in cattle against the nematode Ostertagia ostertagi. Infect. Immun. 2013, 81, 1798–1809. [Google Scholar] [CrossRef]
- Varyani, F.; Löser, S.; Filbey, K.J.; Harcus, Y.; Drurey, C.; Poveda, M.C.; Rasid, O.; White, M.P.J.; Smyth, D.J.; Gerbe, F.; et al. The IL-25-dependent tuft cell circuit driven by intestinal helminths requires macrophage migration inhibitory factor (MIF). Mucosal Immunol. 2022, 15, 1243–1256. [Google Scholar] [CrossRef]
- Yi, J.S.; Cox, M.A.; Zajac, A.J. T-cell exhaustion: Characteristics, causes and conversion. Immunology 2010, 129, 474–481. [Google Scholar] [CrossRef]
- Jayaraman, P.; Jacques, M.K.; Zhu, C.; Steblenko, K.M.; Stowell, B.L.; Madi, A.; Anderson, A.C.; Kuchroo, V.K.; Behar, S.M. TIM3 Mediates T Cell Exhaustion during Mycobacterium tuberculosis Infection. PLoS Pathog. 2016, 12, e1005490. [Google Scholar] [CrossRef] [PubMed]
- Gasbarre, L.C. Ostertagia ostertagi: Changes in lymphoid populations in the local lymphoid tissues after primary or secondary infection. Vet. Parasitol. 1994, 55, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boschiero, C.; Beshah, E.; Bakshi, M.; Miramontes, E.; Hebert, D.; Thompson, P.C.; Li, C.-J.; Zhu, X.; Zarlenga, D.; Liu, G.E.; et al. Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi. Int. J. Mol. Sci. 2025, 26, 2264. https://doi.org/10.3390/ijms26052264
Boschiero C, Beshah E, Bakshi M, Miramontes E, Hebert D, Thompson PC, Li C-J, Zhu X, Zarlenga D, Liu GE, et al. Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi. International Journal of Molecular Sciences. 2025; 26(5):2264. https://doi.org/10.3390/ijms26052264
Chicago/Turabian StyleBoschiero, Clarissa, Ethiopia Beshah, Mariam Bakshi, Eliseo Miramontes, Deborah Hebert, Peter C. Thompson, Cong-Jun Li, Xiaoping Zhu, Dante Zarlenga, George E. Liu, and et al. 2025. "Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi" International Journal of Molecular Sciences 26, no. 5: 2264. https://doi.org/10.3390/ijms26052264
APA StyleBoschiero, C., Beshah, E., Bakshi, M., Miramontes, E., Hebert, D., Thompson, P. C., Li, C.-J., Zhu, X., Zarlenga, D., Liu, G. E., & Tuo, W. (2025). Transcriptional Profiling of Abomasal Mucosa from Young Calves Experimentally Infected with Ostertagia ostertagi. International Journal of Molecular Sciences, 26(5), 2264. https://doi.org/10.3390/ijms26052264





