Nitroxidative Stress, Cell—Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain
Abstract
1. Introduction
2. Reactive Oxygen and Nitrogen Species
2.1. Reactive Oxygen Species: Chemistry and Sources
2.1.1. Singlet Oxygen
2.1.2. Superoxide Radical Anion
2.1.3. Hydrogen Peroxide
2.1.4. Hydroxyl Radical
2.2. Reactive Nitrogen Species: Shemistry and Sources
Nitric Oxide and Peroxynitrite
2.3. Endogenous Sources of Reactive Oxygen and Nitrogen Species (ROS/RNS)
2.3.1. Endoplasmic Reticulum
2.3.2. Mitochondria
2.3.3. Plasma Membrane
2.4. ROS/RNS-Mediated Cell Signaling
2.4.1. NRF2–ARE Pathway
2.4.2. NF-κB Signaling Pathway
2.4.3. MAPK/AP-1 Signaling Pathway
2.4.4. Phosphoinositide 3-Kinase (PI3K)/Akt Pathway
2.4.5. Calcium Signaling
2.4.6. Unfolded Protein Response (UPR) Pathway
3. Antioxidant Enzymes
3.1. Superoxide Dismutase
3.2. Catalase
3.3. Glutathione Peroxidase
3.4. Thioredoxin
4. Nitroxidative Stress in Neuronal Cells
5. MnPs: Mechanisms of Action and Potential Therapy for Neuropathic Pain
6. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP-1 | Activator Protein 1 |
| ATF6 | Activator Transcription Factor |
| BDNF | brain-derived neurotrophic factor |
| CAT | catalase |
| CHOP | C/EBP Homologous Protein |
| Cys | cysteine |
| EGF | epidermal growth factor |
| eNOS | Endothelial Nitric Oxide Synthase |
| ER | endoplasmic reticulum |
| ERK | Extracellular Signal-Regulated Kinase |
| ERO-1 | Endoplasmic Reticulum Oxidoreductin-1 |
| ETC | electron transfer chain |
| FAD | flavin adenine dinucleotide |
| GH | glutathione |
| GLUT1 | Glucose Transporter 1 |
| GPx | glutathione peroxidase |
| GR | glutathione reductase |
| GRP78 | Glucose-Regulated Protein 78 |
| GSH | reduced glutathione |
| GSSG | oxidized glutathione |
| H2O2 | hydrogen peroxide |
| HIF-1α | Hypoxia-Inducible Factor 1α |
| HO• | hydroxyl radical |
| HOCL | hypochlorous acid |
| HOO• | hydroperoxyl radical |
| IL | interleukin |
| iNOS | Inducible Nitric Oxide Synthase |
| IRE-1α | Inositol-Requiring Kinase 1α |
| IRI | ischemic reperfusion injury |
| JNK | c-Jun NH2 quinase terminal |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| LOOH | lipidic peroxides |
| M | Molar |
| MAPK | Mitogen-Activated Protein Kinase |
| MCU | Mitochondrial Calcium Uniporter |
| MDA | malondialdehyde |
| Met | methionine |
| MnP | manganese porphyrin |
| mtNOS | Mitochondrial Nitric Oxide Synthase |
| NAD | nicotinamide adenine dinucleotide |
| NCX | Na+/Ca2+ exchangers |
| NF-κB | Nuclear Factor kB |
| NGF | nerve growth Factor |
| nNOS | Neuronal Nitric Oxide Synthase |
| NO | nitric oxide |
| NOX | NADPH oxidase |
| NRF2 | Nuclear factor (erythroid-derived 2)-like 2 |
| O2•− | superoxide radical anion |
| ONOO− | peroxynitrite |
| PDI | Protein Disulfide Isomerase |
| PERK | Protein Kinase (PKR)-Like Endoplasmic Reticulum Kinase |
| PG | prostaglandin |
| PI3K | phosphatidylinositol 3 kinase |
| PIP | phosphatidylinositol phosphate |
| PIP2 | phosphatidylinositol biphosphate |
| PIP3 | phosphatidylinositol triphosphate |
| PMCA | the plasma membrane calcium ATPase |
| PPP | pentose phosphate pathway |
| RNS | reactive nitrogen species |
| ROO• | peroxyl radical |
| ROS | reactive oxygen species |
| RS• | thiyl radical |
| SERCA | Endoplasmic Reticulum Calcium ATPase |
| SOD | superoxide dismutase |
| Tyr | tyrosine |
| VGF | vascular growth factor |
| XO | xanthine oxidase |
References
- Burma, N.E.; Leduc-Pessah, H.; Fan, C.Y.; Trang, T. Animal Models of Chronic Pain: Advances and Challenges for Clinical Translation. J. Neurosci. Res. 2017, 95, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, D.S.; McGee, S.J. Pain as a Global Public Health Priority. BMC Public Health 2011, 11, 770. [Google Scholar] [CrossRef]
- Racine, M. Chronic Pain and Suicide Risk: A Comprehensive Review. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 87, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Finnerup, N.B.; Attal, N.; Haroutounian, S.; McNicol, E.; Baron, R.; Dworkin, R.H.; Gilron, I.; Haanpää, M.; Hansson, P.; Jensen, T.S.; et al. Pharmacotherapy for Neuropathic Pain in Adults: A Systematic Review and Meta-Analysis. Lancet Neurol. 2015, 14, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Teixeira-Santos, L.; Albino-Teixeira, A.; Pinho, D. Neuroinflammation, Oxidative Stress and Their Interplay in Neuropathic Pain: Focus on Specialized pro-Resolving Mediators and NADPH Oxidase Inhibitors as Potential Therapeutic Strategies. Pharmacol. Res. 2020, 162, 105280. [Google Scholar] [CrossRef]
- Mobasheri, A.; Simon, F.; Carrasco, C.; Nazirŏglu, M.; Nazirŏglu, N.; Rodríguez, A.B.; Pariente, J.A. Neuropathic Pain: Delving into the Oxidative Origin and the Possible Implication of Transient Receptor Potential Channels. Front. Physiol. 2018, 9, 95. [Google Scholar] [CrossRef]
- IASP. IASP Terminology. Available online: https://www.iasp-pain.org/terminology?navItemNumber=576#Pain (accessed on 10 April 2021).
- Colloca, L.; Ludman, T.; Bouhassira, D.; Baron, R.; Dickenson, A.H.; Yarnitsky, D.; Freeman, R.; Truini, A.; Attal, N.; Finnerup, N.B.; et al. Neuropathic Pain. Nat. Rev. Dis. Primers 2017, 3, 17002. [Google Scholar] [CrossRef]
- Ji, R.R.; Nackley, A.; Huh, Y.; Terrando, N.; Maixner, W. Neuroinflammation and Central Sensitization in Chronic and Widespread Pain. Anesthesiology 2018, 129, 343–366. [Google Scholar] [CrossRef]
- Singh, P.; Bansal, S.; Kuhad, A.; Kumar, A.; Chopra, K. Naringenin Ameliorates Diabetic Neuropathic Pain by Modulation of Oxidative-Nitrosative Stress, Cytokines and MMP-9 Levels. Food Funct. 2020, 11, 4548–4560. [Google Scholar] [CrossRef]
- Zamani, K.; Fakhri, S.; Kiani, A.; Abbaszadeh, F.; Farzaei, M.H. Rutin Engages Opioid/Benzodiazepine Receptors towards Anti-Neuropathic Potential in a Rat Model of Chronic Constriction Injury: Relevance to Its Antioxidant and Anti-Inflammatory Effects. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2025, 1–15. [Google Scholar] [CrossRef]
- Hashemi, B.; Fakhri, S.; Kiani, A.; Abbaszadeh, F.; Miraghaee, S.; Mohammadi, M.; Echeverría, J. Anti-Neuropathic Effects of Astaxanthin in a Rat Model of Chronic Constriction Injury: Passing through Opioid/Benzodiazepine Receptors and Relevance to Its Antioxidant and Anti-Inflammatory Effects. Front. Pharmacol. 2024, 15, 1467788. [Google Scholar] [CrossRef] [PubMed]
- Ilari, S.; Giancotti, L.A.; Lauro, F.; Gliozzi, M.; Malafoglia, V.; Palma, E.; Tafani, M.; Russo, M.A.; Tomino, C.; Fini, M.; et al. Natural Antioxidant Control of Neuropathic Pain-Exploring the Role of Mitochondrial SIRT3 Pathway. Antioxidants 2020, 9, 1103. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Venditti, P. Evolution of the Knowledge of Free Radicals and Other Oxidants. Oxid. Med. Cell. Longev. 2020, 2020, 9829176. [Google Scholar] [CrossRef] [PubMed]
- Teleanu, D.M.; Niculescu, A.G.; Lungu, I.I.; Radu, C.I.; Vladâcenco, O.; Roza, E.; Costăchescu, B.; Grumezescu, A.M.; Teleanu, R.I. An Overview of Oxidative Stress, Neuroinflammation, and Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 5938. [Google Scholar] [CrossRef]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free Radicals, Antioxidants and Functional Foods: Impact on Human Health. Pharmacogn. Rev. 2010, 4, 118. [Google Scholar] [CrossRef]
- Kim, D.; You, B.; Jo, E.K.; Han, S.K.; Simon, M.I.; Lee, S.J. NADPH Oxidase 2-Derived Reactive Oxygen Species in Spinal Cord Microglia Contribute to Peripheral Nerve Injury-Induced Neuropathic Pain. Proc. Natl. Acad. Sci. USA 2010, 107, 14851–14856. [Google Scholar] [CrossRef]
- Osmanlıoğlu, H.Ö.; Nazıroğlu, M. Resveratrol Modulates Diabetes-Induced Neuropathic Pain, Apoptosis, and Oxidative Neurotoxicity in Mice Through TRPV4 Channel Inhibition. Mol. Neurobiol. 2025, 61, 7269–7286. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive Oxygen Species (ROS) as Pleiotropic Physiological Signalling Agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Massaad, C.A.; Klann, E. Reactive Oxygen Species in the Regulation of Synaptic Plasticity and Memory. Antioxid. Redox Signal. 2011, 14, 2013. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS Signalling: Adaptive Redox Switches through Oxidative/Nitrosative Protein Modifications. Free Radic. Res. 2018, 52, 507–543. [Google Scholar] [CrossRef]
- Trevisan, G.; Benemei, S.; Materazzi, S.; De Logu, F.; De Siena, G.; Fusi, C.; Fortes Rossato, M.; Coppi, E.; Marone, I.M.; Ferreira, J.; et al. TRPA1 Mediates Trigeminal Neuropathic Pain in Mice Downstream of Monocytes/Macrophages and Oxidative Stress. Brain 2016, 139, 1361–1377. [Google Scholar] [CrossRef] [PubMed]
- Batinic-Haberle, I.; Spasojevic, I. Complex Chemistry and Biology of Redox-Active Compounds, Commonly Known as SOD Mimics, Affect Their Therapeutic Effects. Antioxid. Redox Signal. 2014, 20, 2323–2325. [Google Scholar] [CrossRef] [PubMed]
- Carballal, S.; Valez, V.; Alvarez-Paggi, D.; Tovmasyan, A.; Batinic-Haberle, I.; Ferrer-Sueta, G.; Murgida, D.H.; Radi, R. Manganese Porphyrin Redox State in Endothelial Cells: Resonance Raman Studies and Implications for Antioxidant Protection towards Peroxynitrite. Free Radic. Biol. Med. 2018, 126, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Ali, D.K.; Oriowo, M.; Tovmasyan, A.; Batinic-Haberle, I.; Benov, L. Late Administration of Mn Porphyrin-Based SOD Mimic Enhances Diabetic Complications. Redox Biol. 2013, 1, 457–466. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Spasojevic, I.; Tse, H.M.; Tovmasyan, A.; Rajic, Z.; Clair, D.K.S.; Vujaskovic, Z.; Dewhirst, M.W.; Piganelli, J.D. Design of Mn Porphyrins for Treating Oxidative Stress Injuries and Their Redox-Based Regulation of Cellular Transcriptional Activities. In Amino Acids; Springer: Vienna, Austria, 2012; Volume 42, pp. 95–113. [Google Scholar] [CrossRef]
- Ratcliffe, P.; Koivunen, P.; Myllyharju, J.; Ragoussis, J.; Bovée, J.V.; Batinic-Haberle, I.; Vinatier, C.; Trichet, V.; Robriquet, F.; Oliver, L.; et al. Hypoxia Dovepress Update on Hypoxia-Inducible Factors and Hydroxylases in Oxygen Regulatory Pathways: From Physiology to Therapeutics. Hypoxia 2017, 5, 11–20. [Google Scholar] [CrossRef]
- Rausaria, S.; Ghaffari, M.M.E.; Kamadulski, A.; Rodgers, K.; Bryant, L.; Chen, Z.; Doyle, T.; Shaw, M.J.; Salvemini, D.; Neumann, W.L. Retooling Manganese(III) Porphyrin-Based Peroxynitrite Decomposition Catalysts for Selectivity and Oral Activity: A Potential New Strategy for Treating Chronic Pain. J. Med. Chem. 2011, 54, 8658–8669. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Murphy, M.P. How Mitochondria Produce Reactive Oxygen Species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Miller, D.M.; Buettner, G.R.; Aust, S.D. Transition Metals as Catalysts of “Autoxidation” Reactions. Free Radic. Biol. Med. 1990, 8, 95–108. [Google Scholar] [CrossRef]
- Zheng, M.; Storz, G. Redox Sensing by Prokaryotic Transcription Factors. Biochem. Pharmacol. 2000, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Morris, H.; Cronin, M. Metals, Toxicity and Oxidative Stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef] [PubMed]
- Park, E.S.; Gao, X.; Chung, J.M.; Chung, K. Levels of Mitochondrial Reactive Oxygen Species Increase in Rat Neuropathic Spinal Dorsal Horn Neurons. Neurosci. Lett. 2006, 391, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Salvemini, D.; Little, J.W.; Doyle, T.; Neumann, W.L. Roles of Reactive Oxygen and Nitrogen Species in Pain. Free Radic. Biol. Med. 2011, 51, 951–966. [Google Scholar] [CrossRef]
- Grace, P.M.; Gaudet, A.D.; Staikopoulos, V.; Maier, S.F.; Hutchinson, M.R.; Salvemini, D.; Watkins, L.R. Nitroxidative Signaling Mechanisms in Pathological Pain. Trends Neurosci. 2016, 39, 862–879. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Dröge, W. Free Radicals in the Physiological Control of Cell Function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Klotz, L.-O.; Briviba, K.; Sies, H. Signaling by Singlet Oxygen in Biological Systems. In Antioxidant and Redox Regulation of Genes; Academic Press: Cambridge, MA, USA, 2000; pp. 3–20. [Google Scholar] [CrossRef]
- Al-Shehri, S.S. Reactive Oxygen and Nitrogen Species and Innate Immune Response. Biochimie 2021, 181, 52–64. [Google Scholar] [CrossRef]
- Turrens, J.F. Mitochondrial Formation of Reactive Oxygen Species. J. Physiol. 2003, 552, 335. [Google Scholar] [CrossRef]
- Kovacic, P.; Pozos, R.S.; Somanathan, R.; Shangari, N.; O’Brien, P.J. Mechanism of Mitochondrial Uncouplers, Inhibitors, and Toxins: Focus on Electron Transfer, Free Radicals, and Structure-Activity Relationships. Curr. Med. Chem. 2005, 12, 2601–2623. [Google Scholar] [CrossRef]
- Valko, M.; Izakovic, M.; Mazur, M.; Rhodes, C.J.; Telser, J. Role of Oxygen Radicals in DNA Damage and Cancer Incidence. Mol. Cell. Biochem. 2004, 266, 37–56. [Google Scholar] [CrossRef]
- De Grey, A.D.N.J. HO2•: The Forgotten Radical. DNA Cell Biol. 2004, 21, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Raptova, R.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Reactive Oxygen Species, Toxicity, Oxidative Stress, and Antioxidants: Chronic Diseases and Aging. Arch. Toxicol. 2023, 97, 2499–2574. [Google Scholar] [CrossRef]
- Siniscalco, D.; Fuccio, C.; Giordano, C.; Ferraraccio, F.; Palazzo, E.; Luongo, L.; Rossi, F.; Roth, K.A.; Maione, S.; de Novellis, V. Role of Reactive Oxygen Species and Spinal Cord Apoptotic Genes in the Development of Neuropathic Pain. Pharmacol. Res. 2007, 55, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.S.; Lee, I.; Chung, K.; Chung, J.M. Oxidative Stress in the Spinal Cord Is an Important Contributor in Capsaicin-Induced Mechanical Secondary Hyperalgesia in Mice. Pain 2008, 138, 514. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, H.K.; Kim, J.H.; Chung, K.; Chung, J.M. The Role of Reactive Oxygen Species in Capsaicin-Induced Mechanical Hyperalgesia and in the Activities of Dorsal Horn Neurons. Pain 2007, 133, 9. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Park, S.K.; Zhou, J.L.; Taglialatela, G.; Chung, K.; Coggeshall, R.E.; Chung, J.M. Reactive Oxygen Species (ROS) Play an Important Role in a Rat Model of Neuropathic Pain. Pain 2004, 111, 116–124. [Google Scholar] [CrossRef]
- Yowtak, J.; Lee, K.Y.; Kim, H.Y.; Wang, J.; Kim, H.K.; Chung, K.; Chung, J.M. Reactive Oxygen Species Contribute to Neuropathic Pain by Reducing Spinal GABA Release. Pain 2011, 152, 844–852. [Google Scholar] [CrossRef]
- Gwak, Y.S.; Hassler, S.E.; Hulsebosch, C.E. Reactive Oxygen Species Contribute to Neuropathic Pain and Locomotor Dysfunction via Activation of CamKII in Remote Segments Following Spinal Cord Contusion Injury in Rats. Pain 2013, 154, 1699–1708. [Google Scholar] [CrossRef]
- Kallenborn-Gerhardt, W.; Schröder, K.; Schmidtko, A. NADPH Oxidases in Pain Processing. Antioxidants 2022, 11, 1162. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Juan, C.A.; Plou, F.J.; Pérez-Lebeña, E. Chemistry of Hydrogen Peroxide Formation and Elimination in Mammalian Cells, and Its Role in Various Pathologies. Stresses 2022, 2, 256–274. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen Peroxide—Production, Fate and Role in Redox Signaling of Tumor Cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen Peroxide as a Central Redox Signaling Molecule in Physiological Oxidative Stress: Oxidative Eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, P.A.; Chaney, M.O. Mechanism of GAPDH Redox Signaling by H2O2 Activation of a Two-Cysteine Switch. Int. J. Mol. Sci. 2022, 23, 4604. [Google Scholar] [CrossRef]
- Day, A.M.; Brown, J.D.; Taylor, S.R.; Rand, J.D.; Morgan, B.A.; Veal, E.A. Inactivation of a Peroxiredoxin by Hydrogen Peroxide Is Critical for Thioredoxin-Mediated Repair of Oxidized Proteins and Cell Survival. Mol. Cell 2012, 45, 398–408. [Google Scholar] [CrossRef]
- Sözbir, E.; Nazıroğlu, M. Diabetes Enhances Oxidative Stress-Induced TRPM2 Channel Activity and Its Control by N-Acetylcysteine in Rat Dorsal Root Ganglion and Brain. Metab. Brain Dis. 2016, 31, 385–393. [Google Scholar] [CrossRef]
- Viggiano, A.; Monda, M.; Viggiano, A.; Viggiano, D.; Viggiano, E.; Chiefari, M.; Aurilio, C.; De Luca, B. Trigeminal Pain Transmission Requires Reactive Oxygen Species Production. Brain Res. 2005, 1050, 72–78. [Google Scholar] [CrossRef]
- Lorenz, J.E.; Kallenborn-Gerhardt, W.; Lu, R.; Syhr, K.M.J.; Eaton, P.; Geisslinger, G.; Schmidtko, A. Oxidant-Induced Activation of CGMP-Dependent Protein Kinase Iα Mediates Neuropathic Pain After Peripheral Nerve Injury. Antioxid. Redox Signal. 2014, 21, 1504–1515. [Google Scholar] [CrossRef]
- Chen, Z.; Muscoli, C.; Doyle, T.; Bryant, L.; Cuzzocrea, S.; Mollace, V.; Mastroianni, R.; Masini, E.; Salvemini, D. NMDA-Receptor Activation and Nitroxidative Regulation of the Glutamatergic Pathway during Nociceptive Processing. Pain 2010, 149, 100–106. [Google Scholar] [CrossRef]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Halliwell, B.; Clement, M.V.; Ramalingam, J.; Long, L.H. Hydrogen Peroxide. Ubiquitous in Cell Culture and in Vivo? IUBMB Life 2000, 50, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Kadiiska, M.B.; Mason, R.P. In Vivo Copper-Mediated Free Radical Production: An ESR Spin-Trapping Study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2002, 58, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Zastawny, T.H.; Altman, S.A.; Randers-Eichhorn, L.; Madurawe, R.; Lumpkin, J.A.; Dizdaroglu, M.; Rao, G. DNA Base Modifications and Membrane Damage in Cultured Mammalian Cells Treated with Iron Ions. Free Radic. Biol. Med. 1995, 18, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Kroth, A.; do Carmo Quevedo Santos, M.; da Silva, T.C.B.; Silveira, E.M.S.; Trapp, M.; Bezzerra, R.M.N.; Simabuco, F.; Niero, R.; Partata, W.A. Aqueous Extract from Luehea divaricata Mart. Leaves Reduces Nociception in Rats with Neuropathic Pain. J. Ethnopharmacol. 2020, 256, 112761. [Google Scholar] [CrossRef] [PubMed]
- Bittar, A.; Jun, J.; La, J.H.; Wang, J.; Leem, J.W.; Chung, J.M. Reactive Oxygen Species Affect Spinal Cell Type-Specific Synaptic Plasticity in a Model of Neuropathic Pain. Pain 2017, 158, 2137. [Google Scholar] [CrossRef]
- Ghafourifar, P.; Cadenas, E. Mitochondrial Nitric Oxide Synthase. Trends Pharmacol. Sci. 2005, 26, 190–195. [Google Scholar] [CrossRef]
- Bergendi, L.; Beneš, L.; Ďuracková, Z.; Ferenčik, M. Chemistry, Physiology and Pathology of Free Radicals. Life Sci. 1999, 65, 1865–1874. [Google Scholar] [CrossRef]
- Ignarro, L.J. Biosynthesis and Metabolism of Endothelium-Derived Nitric Oxide. Annu. Rev. Pharmacol. Toxicol. 1990, 30, 535–560. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef]
- Nathan, C.; Xie, Q.W. Nitric Oxide Synthases: Roles, Tolls, and Controls. Cell 1994, 78, 915–918. [Google Scholar] [CrossRef]
- Cury, Y.; Picolo, G.; Gutierrez, V.P.; Ferreira, S.H. Pain and Analgesia: The Dual Effect of Nitric Oxide in the Nociceptive System. Nitric Oxide 2011, 25, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Nitric Oxide, Oxidants, and Protein Tyrosine Nitration. Proc. Natl. Acad. Sci. USA 2004, 101, 4003–4008. [Google Scholar] [CrossRef]
- Bartesaghi, S.; Radi, R. Fundamentals on the Biochemistry of Peroxynitrite and Protein Tyrosine Nitration. Redox Biol. 2018, 14, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.C.; McCall, M.R.; Frei, B. Oxidation of LDL by Myeloperoxidase and Reactive Nitrogen Species: Reaction Pathways and Antioxidant Protection. Arter. Thromb. Vasc. Biol. 2000, 20, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Sueta, G.; Radi, R. Chemical Biology of Peroxynitrite: Kinetics, Diffusion, and Radicals. ACS Chem. Biol. 2009, 4, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Radi, R.; Peluffo, G.; Alvarez, M.N.; Naviliat, M.; Cayota, A. Unraveling Peroxynitrite Formation in Biological Systems. Free Radic. Biol. Med. 2001, 30, 463–488. [Google Scholar] [CrossRef]
- Szabó, C.; Ischiropoulos, H.; Radi, R. Peroxynitrite: Biochemistry, Pathophysiology and Development of Therapeutics. Nat. Rev. Drug Discov. 2007, 6, 662–680. [Google Scholar] [CrossRef]
- Muscoli, C.; Cuzzocrea, S.; Riley, D.P.; Zweier, J.L.; Thiemermann, C.; Wang, Z.Q.; Salvemini, D. On the Selectivity of Superoxide Dismutase Mimetics and Its Importance in Pharmacological Studies. Br. J. Pharmacol. 2003, 140, 445–460. [Google Scholar] [CrossRef]
- Julius, D. TRP Channels and Pain. Annu. Rev. Cell Dev. Biol. 2013, 29, 355–384. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. The Endoplasmic Reticulum. In Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Helenius, A.; Marquardt, T.; Braakman, I. The Endoplasmic Reticulum as a Protein-Folding Compartment. Trends Cell Biol. 1992, 2, 227–231. [Google Scholar] [CrossRef]
- De Sousa, M.; Oliveira, S.J.; Pinto, J.P. ER Stress and Iron Homeostasis: A New Frontier for the UPR. Biochem. Res. Int. 2011, 2011, 896474. [Google Scholar] [CrossRef]
- Napier, J.A.; Michaelson, L.V.; Sayanova, O. The Role of Cytochrome b 5 Fusion Desaturases in the Synthesis of Polyunsaturated Fatty Acids. Prostaglandins Leukot. Essent. Fat. Acids 2003, 68, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wiercinska, P.; Lou, Y.; Squires, E.J. The Roles of Different Porcine Cytochrome P450 Enzymes and Cytochrome B5A in Skatole Metabolism. Animal 2012, 6, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ. Res. 2018, 122, 877. [Google Scholar] [CrossRef]
- Zangar, R.C.; Davydov, D.R.; Verma, S. Mechanisms That Regulate Production of Reactive Oxygen Species by Cytochrome P450. Toxicol. Appl. Pharmacol. 2004, 199, 316–331. [Google Scholar] [CrossRef]
- Veith, A.; Moorthy, B. Role of Cytochrome P450s in the Generation and Metabolism of Reactive Oxygen Species. Curr. Opin. Toxicol. 2018, 7, 44–51. [Google Scholar] [CrossRef]
- Bhandary, B.; Marahatta, A.; Kim, H.R.; Chae, H.J. An Involvement of Oxidative Stress in Endoplasmic Reticulum Stress and Its Associated Diseases. Int. J. Mol. Sci. 2012, 14, 434–456. [Google Scholar] [CrossRef]
- Santos, C.X.C.; Tanaka, L.Y.; Wosniak, J.; Laurindo, F.R.M. Mechanisms and Implications of Reactive Oxygen Species Generation during the Unfolded Protein Response: Roles of Endoplasmic Reticulum Oxidoreductases, Mitochondrial Electron Transport, and NADPH Oxidase. Antioxid. Redox Signal. 2009, 11, 2409–2427. [Google Scholar] [CrossRef]
- Higa, A.; Chevet, E. Redox Signaling Loops in the Unfolded Protein Response. Cell Signal. 2012, 24, 1548–1555. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, D.; Shen, S. Endoplasmic Reticular Stress as an Emerging Therapeutic Target for Chronic Pain: A Narrative Review. Br. J. Anaesth. 2024, 132, 707–724. [Google Scholar] [CrossRef]
- Lei, J.; Paul, J.; Wang, Y.; Gupta, M.; Vang, D.; Thompson, S.; Jha, R.; Nguyen, J.; Valverde, Y.; Lamarre, Y.; et al. Heme Causes Pain in Sickle Mice via Toll-Like Receptor 4-Mediated Reactive Oxygen Species- and Endoplasmic Reticulum Stress-Induced Glial Activation. Antiox. Redox Signal. 2021, 34, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Kawanaka, R.; Jin, H.; Aoe, T. Unraveling the Connection: Pain and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2024, 25, 4995. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. Mitochondrial Generation of Superoxide and Hydrogen Peroxide as the Source of Mitochondrial Redox Signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Boveris, A. The Mitochondrial Energy Transduction System and the Aging Process. Am. J. Physiol. Cell Physiol. 2007, 292, 670–686. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free Radicals and Antioxidants in Normal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Brand, M.D.; Nicholls, D.G. Assessing Mitochondrial Dysfunction in Cells. Biochem. J. 2011, 435, 297. [Google Scholar] [CrossRef]
- Li, B.; Yu, K.; Zhou, X.; Sun, J.; Qi, L.; Li, W.; Yang, T.; Li, W.; Wang, N.; Gu, X.; et al. Increased TSPO Alleviates Neuropathic Pain by Preventing Pyroptosis via the AMPK-PGC-1α Pathway. J. Headache Pain 2025, 26, 16. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, X.C. Effects of Mitochondrial Dysfunction via AMPK/PGC-1 α Signal Pathway on Pathogenic Mechanism of Diabetic Peripheral Neuropathy and the Protective Effects of Chinese Medicine. Chin. J. Integr. Med. 2019, 25, 386–394. [Google Scholar] [CrossRef]
- Kim, H.Y.; Chung, J.M.; Chung, K. Increased Production of Mitochondrial Superoxide in the Spinal Cord Induces Pain Behaviors in Mice: The Effect of Mitochondrial Electron Transport Complex Inhibitors. Neurosci. Lett. 2008, 447, 87–91. [Google Scholar] [CrossRef]
- Silva Santos Ribeiro, P.; Willemen, H.L.D.M.; Eijkelkamp, N. Mitochondria and Sensory Processing in Inflammatory and Neuropathic Pain. Front. Pain Res. 2022, 3, 1013577. [Google Scholar] [CrossRef]
- Skulachev, V.P. Why Are Mitochondria Involved in Apoptosis? Permeability Transition Pores and Apoptosis as Selective Mechanisms to Eliminate Superoxide-Producing Mitochondria and Cell. FEBS Lett. 1996, 397, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Vignais, P.V. The Superoxide-Generating NADPH Oxidase: Structural Aspects and Activation Mechanism. Cell. Mol. Life Sci. 2002, 59, 1428–1459. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.D. NOX Enzymes and the Biology of Reactive Oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiao, B.; Yu, S.; Zhang, K.; Sun, J.; Liu, B.; Zhang, X. Spinal AT1R Contributes to Neuroinflammation and Neuropathic Pain via NOX2-Dependent Redox Signaling in Microglia. Free Radic. Biol. Med. 2025, 227, 143–156. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, S.; Jiao, B.; Zhang, C.; Zhang, K.; Liu, B.; Zhang, X. Vitamin D3 Attenuates Neuropathic Pain via Suppression of Mitochondria-Associated Ferroptosis by Inhibiting PKCα/NOX4 Signaling Pathway. CNS Neurosci. Ther. 2024, 30. [Google Scholar] [CrossRef]
- Sorce, S.; Krause, K.H. NOX Enzymes in the Central Nervous System: From Signaling to Disease. Antioxid. Redox Signal. 2009, 11, 2481–2504. [Google Scholar] [CrossRef]
- De Logu, F.; Nassini, R.; Materazzi, S.; Carvalho Gonçalves, M.; Nosi, D.; Rossi Degl’Innocenti, D.; Marone, I.M.; Ferreira, J.; Li Puma, S.; Benemei, S.; et al. Schwann Cell TRPA1 Mediates Neuroinflammation That Sustains Macrophage-Dependent Neuropathic Pain in Mice. Nat. Commun. 2017, 8, 1887. [Google Scholar] [CrossRef]
- Kallenborn-Gerhardt, W.; Hohmann, S.W.; Syhr, K.M.J.; Schröder, K.; Sisignano, M.; Weigert, A.; Lorenz, J.E.; Lu, R.; Brüne, B.; Brandes, R.P.; et al. Nox2-Dependent Signaling between Macrophages and Sensory Neurons Contributes to Neuropathic Pain Hypersensitivity. Pain 2014, 155, 2161–2170. [Google Scholar] [CrossRef]
- Berger, J.V.; Deumens, R.; Goursaud, S.; Schäfer, S.; Lavand’homme, P.; Joosten, E.A.; Hermans, E. Enhanced Neuroinflammation and Pain Hypersensitivity after Peripheral Nerve Injury in Rats Expressing Mutated Superoxide Dismutase 1. J. Neuroinflamm. 2011, 8, 33. [Google Scholar] [CrossRef]
- Geis, C.; Geuss, E.; Sommer, C.; Schmidt, H.H.H.W.; Kleinschnitz, C. NOX4 Is an Early Initiator of Neuropathic Pain. Exp. Neurol. 2017, 288, 94–103. [Google Scholar] [CrossRef]
- Fu, C.N.; Wei, H.; Gao, W.S.; Song, S.S.; Yue, S.W.; Qu, Y.J. Obesity Increases Neuropathic Pain via the AMPK-ERK-NOX4 Pathway in Rats. Aging 2021, 13, 18606–18619. [Google Scholar] [CrossRef] [PubMed]
- Kallenborn-Gerhardt, W.; Schröder, K.; del Turco, D.; Lu, R.; Kynast, K.; Kosowski, J.; Niederberger, E.; Shah, A.M.; Brandes, R.P.; Geisslinger, G.; et al. NADPH Oxidase-4 Maintains Neuropathic Pain after Peripheral Nerve Injury. J. Neurosci. 2012, 32, 10136–10145. [Google Scholar] [CrossRef] [PubMed]
- Rius-Pérez, S.; Pérez, S.; Toledano, M.B.; Sastre, J. Mitochondrial Reactive Oxygen Species and Lytic Programmed Cell Death in Acute Inflammation. Antioxid. Redox Signal. 2023, 39, 708–727. [Google Scholar] [CrossRef] [PubMed]
- Tousoulis, D.; Kampoli, A.-M.; Tentolouris Nikolaos Papageorgiou, C.; Stefanadis, C. The Role of Nitric Oxide on Endothelial Function. Curr. Vasc. Pharmacol. 2012, 10, 4–18. [Google Scholar] [CrossRef]
- Kasai, S.; Shimizu, S.; Tatara, Y.; Mimura, J.; Itoh, K. Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology. Biomolecules 2020, 10, 320. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen Peroxide Sensing, Signaling and Regulation of Transcription Factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Shi, T.; Dansen, T.B. Reactive Oxygen Species Induced P53 Activation: DNA Damage, Redox Signaling, or Both? Antioxid. Redox Signal. 2020, 33, 839–859. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Vikash, V.; Ye, Q.; Wu, D.; Liu, Y.; Dong, W. ROS and ROS-Mediated Cellular Signaling. Oxid. Med. Cell. Longev. 2016, 2016, 4350965. [Google Scholar] [CrossRef]
- Bak, D.W.; Bechtel, T.J.; Falco, J.A.; Weerapana, E. Cysteine Reactivity Across the Sub-Cellular Universe. Curr. Opin. Chem. Biol. 2019, 48, 96. [Google Scholar] [CrossRef]
- Behring, J.B.; van der Post, S.; Mooradian, A.D.; Egan, M.J.; Zimmerman, M.I.; Clements, J.L.; Bowman, G.R.; Held, J.M. Spatial and Temporal Alterations in Protein Structure by EGF Regulate Cryptic Cysteine Oxidation. Sci. Signal. 2020, 13, eaay7315. [Google Scholar] [CrossRef]
- Roos, G.; Messens, J. Protein Sulfenic Acid Formation: From Cellular Damage to Redox Regulation. Free Radic. Biol. Med. 2011, 51, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K. Cellular Thiols and Redox-Regulated Signal Transduction. Curr. Top. Cell Regul. 2001, 36, 1–30. [Google Scholar] [CrossRef]
- Chen, H.; Xie, K.; Chen, Y.; Wang, Y.; Wang, Y.; Lian, N.; Zhang, K.; Yu, Y. Nrf2/HO-1 Signaling Pathway Participated in the Protection of Hydrogen Sulfide on Neuropathic Pain in Rats. Int. Immunopharmacol. 2019, 75, 105746. [Google Scholar] [CrossRef] [PubMed]
- Yardim, A.; Kandemir, F.M.; Ozdemir, S.; Kucukler, S.; Comakli, S.; Gur, C.; Celik, H. Quercetin Provides Protection against the Peripheral Nerve Damage Caused by Vincristine in Rats by Suppressing Caspase 3, NF-ΚB, ATF-6 Pathways and Activating Nrf2, Akt Pathways. Neurotoxicology 2020, 81, 137–146. [Google Scholar] [CrossRef]
- Guo, X.; Kang, J.; Wang, Z.; Wang, Y.; Liu, M.; Zhu, D.; Yang, F.; Kang, X. Nrf2 Signaling in the Oxidative Stress Response After Spinal Cord Injury. Neuroscience 2022, 498, 311–324. [Google Scholar] [CrossRef]
- Zhou, Y.Q.; Mei, W.; Tian, X.B.; Tian, Y.K.; Liu, D.Q.; Ye, D.W. The Therapeutic Potential of Nrf2 Inducers in Chronic Pain: Evidence from Preclinical Studies. Pharmacol. Ther. 2021, 225, 107846. [Google Scholar] [CrossRef]
- Parvez, S.; Long, M.J.C.; Poganik, J.R.; Aye, Y. Redox Signaling by Reactive Electrophiles and Oxidants. Chem. Rev. 2018, 118, 8798–8888. [Google Scholar] [CrossRef]
- Singh, S.; Vrishni, S.; Singh, B.K.; Rahman, I.; Kakkar, P. Nrf2-ARE Stress Response Mechanism: A Control Point in Oxidative Stress-Mediated Dysfunctions and Chronic Inflammatory Diseases. Free Radic. Res. 2010, 44, 1267–1288. [Google Scholar] [CrossRef]
- Fourquet, S.; Guerois, R.; Biard, D.; Toledano, M.B. Activation of NRF2 by Nitrosative Agents and H2O2 Involves KEAP1 Disulfide Formation. J. Biol. Chem. 2010, 285, 8463. [Google Scholar] [CrossRef]
- Lee, J.H.; Khor, T.O.; Shu, L.; Su, Z.Y.; Fuentes, F.; Kong, A.N.T. Dietary Phytochemicals and Cancer Prevention: Nrf2 Signaling, Epigenetics, and Cell Death Mechanisms in Blocking Cancer Initiation and Progression. Pharmacol. Ther. 2013, 137, 153–171. [Google Scholar] [CrossRef]
- Vasconcelos, A.R.; Dos Santos, N.B.; Scavone, C.; Munhoz, C.D. Nrf2/ARE Pathway Modulation by Dietary Energy Regulation in Neurological Disorders. Front. Pharmacol. 2019, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.-I.; Watai, Y.; Tong, K.I.; Shibata, T.; Uchida, K.; Yamamoto, M. Oxidative and Electrophilic Stresses Activate Nrf2 through Inhibition of Ubiquitination Activity of Keap1. Mol. Cell Biol. 2006, 26, 221–229. [Google Scholar] [CrossRef]
- Motohashi, H.; Yamamoto, M. Nrf2-Keap1 Defines a Physiologically Important Stress Response Mechanism. Trends Mol. Med. 2004, 10, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169. [Google Scholar] [CrossRef] [PubMed]
- Kaidery, N.A.; Banerjee, R.; Yang, L.; Smirnova, N.A.; Hushpulian, D.M.; Liby, K.T.; Williams, C.R.; Yamamoto, M.; Kensler, T.W.; Ratan, R.R.; et al. Targeting Nrf2-Mediated Gene Transcription by Extremely Potent Synthetic Triterpenoids Attenuate Dopaminergic Neurotoxicity in the MPTP Mouse Model of Parkinson’s Disease. Antioxid. Redox Signal. 2012, 18, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Vollmer, M.K.; Kelly, M.G.; Fernandez, V.M.; Fernandez, T.G.; Kim, H.; Doré, S. Reactive Gliosis Contributes to Nrf2-Dependent Neuroprotection by Pretreatment with Dimethyl Fumarate or Korean Red Ginseng against Hypoxic-Ischemia: Focus on Hippocampal Injury. Mol. Neurobiol. 2020, 57, 105. [Google Scholar] [CrossRef]
- Itoh, K.; Mochizuki, M.; Ishii, Y.; Ishii, T.; Shibata, T.; Kawamoto, Y.; Kelly, V.; Sekizawa, K.; Uchida, K.; Yamamoto, M. Transcription Factor Nrf2 Regulates Inflammation by Mediating the Effect of 15-Deoxy-Δ12,14-Prostaglandin J2. Mol. Cell Biol. 2004, 24, 36. [Google Scholar] [CrossRef]
- Li, S.; Yang, C.; Fang, X.; Zhan, G.; Huang, N.; Gao, J.; Xu, H.; Hashimoto, K.; Luo, A. Role of Keap1-Nrf2 Signaling in Anhedonia Symptoms in a Rat Model of Chronic Neuropathic Pain: Improvement with Sulforaphane. Front. Pharmacol. 2018, 9, 887. [Google Scholar] [CrossRef]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-Kappa B and Rel Proteins: Evolutionarily Conserved Mediators of Immune Responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef]
- Popiolek-Barczyk, K.; Mika, J. Targeting the Microglial Signaling Pathways: New Insights in the Modulation of Neuropathic Pain. Curr. Med. Chem. 2016, 23, 2908. [Google Scholar] [CrossRef]
- Bakalkin, G.Y.; Yakovleva, T.; Terenius, L. NF-Kappa B-like Factors in the Murine Brain. Developmentally-Regulated and Tissue-Specific Expression. Brain Res. Mol. Brain Res. 1993, 20, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Massa, P.T.; Aleyasin, H.; Park, D.S.; Mao, X.; Barger, S.W. NFκB in Neurons? The Uncertainty Principle in Neurobiology. J. Neurochem. 2006, 97, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Shih, R.H.; Wang, C.Y.; Yang, C.M. NF-KappaB Signaling Pathways in Neurological Inflammation: A Mini Review. Front. Mol. Neurosci. 2015, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Gloire, G.; Legrand-Poels, S.; Piette, J. NF-ΚB Activation by Reactive Oxygen Species: Fifteen Years Later. Biochem. Pharmacol. 2006, 72, 1493–1505. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Basic Principles and Emerging Concepts in the Redox Control of Transcription Factors. Antioxid. Redox Signal. 2011, 15, 2335–2381. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira-Marques, V.; Cyrne, L.; Marinho, H.S.; Antunes, F. A Quantitative Study of NF-KappaB Activation by H2O2: Relevance in Inflammation and Synergy with TNF-Alpha. J. Immunol. 2007, 178, 3893–3902. [Google Scholar] [CrossRef]
- Basak, S.; Hoffmann, A. Crosstalk via the NF-ΚB Signaling System. Cytokine Growth Factor Rev. 2008, 19, 187–197. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L. GDF11 Mitigates Neuropathic Pain via Regulation of Microglial Polarization and Neuroinflammation through TGF-ΒR1/SMAD2/NF-ΚB Pathway in Male Mice. J. Neuroimmune Pharmacol. 2025, 20, 20. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Y.Q.; Qadri, Y.J.; Serhan, C.N.; Ji, R.R. Microglia in Pain: Detrimental and Protective Roles in Pathogenesis and Resolution of Pain. Neuron 2018, 100, 1292–1311. [Google Scholar] [CrossRef]
- Wang, L.; Yin, C.; Liu, T.; Abdul, M.; Zhou, Y.; Cao, J.L.; Lu, C. Pellino1 Regulates Neuropathic Pain as Well as Microglial Activation through the Regulation of MAPK/NF-ΚB Signaling in the Spinal Cord. J. Neuroinflamm. 2020, 17, 83. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Veron, A.S.; Weiner, J.; Robinson-Rechavi, M.; Bornberg-Bauer, E.; Oliver, S.G.; Robertson, D.L. One Billion Years of BZIP Transcription Factor Evolution: Conservation and Change in Dimerization and DNA-Binding Site Specificity. Mol. Biol. Evol. 2007, 24, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Boiti, A.; Vallone, D.; Foulkes, N.S. Reactive Oxygen Species Signaling and Oxidative Stress: Transcriptional Regulation and Evolution. Antioxidants 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Liu, Z.G.; Zandi, E. AP-1 Function and Regulation. Curr. Opin. Cell Biol. 1997, 9, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, P.B.; Kim, H.H.; Abusaliya, A.; Vetrivel, P.; Ha, S.E.; Park, M.Y.; Lee, H.J.; Kim, G.S. Structural and Functional Properties of Activator Protein-1 in Cancer and Inflammation. Evid. Based Complement. Altern. Med. 2022, 2022, 9797929. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 as a Regulator of Cell Life and Death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef]
- Wisdom, R. AP-1: One Switch for Many Signals. Exp. Cell Res. 1999, 253, 180–185. [Google Scholar] [CrossRef]
- Gius, D.; Botero, A.; Shah, S.; Curry, H.A. Intracellular Oxidation/Reduction Status in the Regulation of Transcription Factors NF-KappaB and AP-1. Toxicol. Lett. 1999, 106, 93–106. [Google Scholar] [CrossRef]
- Xanthoudakis’, S.; Miao, G.; Wang, F.; Pan, Y.-C.E.; Curran, T. Redox Activation of Fos-Jun DNA Binding Activity Is Mediated by a DNA Repair Enzyme. EMBO J. 1992, 11, 3323. [Google Scholar] [CrossRef]
- Tanos, T.; Marinissen, M.J.; Leskow, F.C.; Hochbaum, D.; Martinetto, H.; Gutkind, J.S.; Coso, O.A. Phosphorylation of C-Fos by Members of the P38 MAPK Family. Role in the AP-1 Response to UV Light. J. Biol. Chem. 2005, 280, 18842–18852. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 in Cell Proliferation and Survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Zhuang, Z.Y.; Gerner, P.; Woolf, C.J.; Ji, R.R. ERK Is Sequentially Activated in Neurons, Microglia, and Astrocytes by Spinal Nerve Ligation and Contributes to Mechanical Allodynia in This Neuropathic Pain Model. Pain 2005, 114, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.X.; Zhuang, Z.Y.; Woolf, C.J.; Ji, R.R. P38 Mitogen-Activated Protein Kinase Is Activated after a Spinal Nerve Ligation in Spinal Cord Microglia and Dorsal Root Ganglion Neurons and Contributes to the Generation of Neuropathic Pain. J. Neurosci. 2003, 23, 4017–4022. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.J.; Li, Y.L.; Zhu, Z.L.; Jia, D.M.; Fan, M.L.; Li, T.; Wang, X.J.; Li, Z.G.; Ma, H.S. Inhibition of Activator Protein 1 Attenuates Neuroinflammation and Brain Injury after Experimental Intracerebral Hemorrhage. CNS Neurosci. Ther. 2019, 25, 1182. [Google Scholar] [CrossRef] [PubMed]
- Chinenov, Y.; Kerppola, T.K. Close Encounters of Many Kinds: Fos-Jun Interactions That Mediate Transcription Regulatory Specificity. Oncogene 2001, 20, 2438–2452. [Google Scholar] [CrossRef]
- Satoh, T.; Nakatsuka, D.; Watanabe, Y.; Nagata, I.; Kikuchi, H.; Namura, S. Neuroprotection by MAPK/ERK Kinase Inhibition with U0126 against Oxidative Stress in a Mouse Neuronal Cell Line and Rat Primary Cultured Cortical Neurons. Neurosci. Lett. 2000, 288, 163–166. [Google Scholar] [CrossRef]
- Samanta, S.; Perkinton, M.S.; Morgan, M.; Williams, R.J. Hydrogen Peroxide Enhances Signal-Responsive Arachidonic Acid Release from Neurons: Role of Mitogen-Activated Protein Kinase. J. Neurochem. 1998, 70, 2082–2090. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Compromised MAPK Signaling in Human Diseases: An Update. Arch. Toxicol. 2015, 89, 867–882. [Google Scholar] [CrossRef]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK Cascades: Signaling Components, Nuclear Roles and Mechanisms of Nuclear Translocation. Biochim. Biophys. Acta 2011, 1813, 1619–1633. [Google Scholar] [CrossRef]
- Whitmarsh, A.J. Regulation of Gene Transcription by Mitogen-Activated Protein Kinase Signaling Pathways. Biochim. Biophys. Acta 2007, 1773, 1285–1298. [Google Scholar] [CrossRef]
- Niault, T.S.; Baccarini, M. Targets of Raf in Tumorigenesis. Carcinogenesis 2010, 31, 1165–1174. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Pathological Roles of MAPK Signaling Pathways in Human Diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Seger, R. The Extracellular Signal-Regulated Kinase: Multiple Substrates Regulate Diverse Cellular Functions. Growth Factors 2006, 24, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.P.; Wang, B.R.; Liu, H.L.; You, S.W.; Huang, W.J.; Jiao, X.Y.; Ju, G. Phosphorylation of Extracellular Signal-Regulated Kinases 1/2 Is Predominantly Enhanced in the Microglia of the Rat Spinal Cord Following Dorsal Root Transection. Neuroscience 2003, 119, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Befort, K.; Brenner, G.J.; Woolf, C.J. ERK MAP Kinase Activation in Superficial Spinal Cord Neurons Induces Prodynorphin and NK-1 Upregulation and Contributes to Persistent Inflammatory Pain Hypersensitivity. J. Neurosci. 2002, 22, 478–485. [Google Scholar] [CrossRef]
- Wang, H.; Dai, Y.; Fukuoka, T.; Yamanaka, H.; Obata, K.; Tokunaga, A.; Noguchi, K. Enhancement of Stimulation-Induced ERK Activation in the Spinal Dorsal Horn and Gracile Nucleus Neurons in Rats with Peripheral Nerve Injury. Eur. J. Neurosci. 2004, 19, 884–890. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, S.; Kang, X.; Chen, S.; Zhang, L.; Tian, Z.K.; Liang, X.; Meng, C. Isoliquiritigenin Alleviates Neuropathic Pain by Reducing Microglia Inflammation through Inhibition of the ERK Signaling Pathway and Decreasing CEBPB Transcription Expression. Int. Immunopharmacol. 2024, 143, 113536. [Google Scholar] [CrossRef]
- Weston, C.R.; Davis, R.J. The JNK Signal Transduction Pathway. Curr. Opin. Cell Biol. 2007, 19, 142–149. [Google Scholar] [CrossRef]
- Johnson, G.L.; Nakamura, K. The C-Jun Kinase/Stress-Activated Pathway: Regulation, Function and Role in Human Disease. Biochim. Biophys. Acta 2007, 1773, 1341–1348. [Google Scholar] [CrossRef]
- Haeusgen, W.; Boehm, R.; Zhao, Y.; Herdegen, T.; Waetzig, V. Specific Activities of Individual C-Jun N-Terminal Kinases in the Brain. Neuroscience 2009, 161, 951–959. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, G.; He, J.; Xu, Y. The Protective Effects of Orexin B in Neuropathic Pain by Suppressing Inflammatory Response. Neuropeptides 2024, 108, 102458. [Google Scholar] [CrossRef]
- Zarubin, T.; Han, J. Activation and Signaling of the P38 MAP Kinase Pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Whitmarsh, A.J.; Davis, R.J. Role of Mitogen-Activated Protein Kinase Kinase 4 in Cancer. Oncogene 2007, 26, 3172–3184. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Suter, M.R. P38 MAPK, Microglial Signaling, and Neuropathic Pain. Mol. Pain 2007, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T.; Yu, L.E.; Wang, J.Y. Nitroxide Antioxidant as a Potential Strategy to Attenuate the Oxidative/Nitrosative Stress Induced by Hydrogen Peroxide plus Nitric Oxide in Cultured Neurons. Nitric Oxide 2016, 54, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Whitehead, M.A.; Piñeiro, R. Molecules in Medicine Mini-Review: Isoforms of PI3K in Biology and Disease. J. Mol. Med. 2016, 94, 5–11. [Google Scholar] [CrossRef]
- Hawkins, P.T.; Stephens, L.R. Emerging Evidence of Signalling Roles for PI(3,4)P2 in Class I and II PI3K-Regulated Pathways. Biochem. Soc. Trans. 2016, 44, 307–314. [Google Scholar] [CrossRef]
- Deane, J.A.; Fruman, D.A. Phosphoinositide 3-Kinase: Diverse Roles in Immune Cell Activation. Annu. Rev. Immunol. 2004, 22, 563–598. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Chen, S.-P.; Zhou, Y.-Q.; Liu, D.-Q.; Zhang, W.; Manyande, A.; Guan, X.-H.; Tian, Y.; Ye, D.-W.; Omar, D.M. PI3K/Akt Pathway: A Potential Therapeutic Target for Chronic Pain. Curr. Pharm. Des. 2017, 23, 1860–1868. [Google Scholar] [CrossRef]
- Cianciulli, A.; Porro, C.; Calvello, R.; Trotta, T.; Lofrumento, D.D.; Panaro, M.A. Microglia Mediated Neuroinflammation: Focus on PI3K Modulation. Biomolecules 2020, 10, 137. [Google Scholar] [CrossRef]
- Saponaro, C.; Cianciulli, A.; Calvello, R.; Dragone, T.; Iacobazzi, F.; Panaro, M.A. The PI3K/Akt Pathway Is Required for LPS Activation of Microglial Cells. Immunopharmacol. Immunotoxicol. 2012, 34, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.B.; Choi, J.S.; Kang, B.; Herr, S.; Pereira, C.; Moraes, C.T.; Al-Ali, H.; Lee, J.K. PI3K Signaling Promotes Formation of Lipid-Laden Foamy Macrophages at the Spinal Cord Injury Site. Neurobiol. Dis. 2024, 190, 106370. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, M.; Cao, B.; Hou, T.; Mao, X. Targeting the Phosphatidylinositol 3-Kinase/AKT Pathway for the Treatment of Multiple Myeloma. Curr. Med. Chem. 2014, 21, 3173–3187. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, M.A.; Bombardieri, M.; Pitzalis, C.; Vanhaesebroeck, B. Isoform-Selective Induction of Human P110δ PI3K Expression by TNFα: Identification of a New and Inducible PIK3CD Promoter. Biochem. J. 2012, 443, 857. [Google Scholar] [CrossRef]
- Amzel, L.M.; Huang, C.H.; Mandelker, D.; Lengauer, C.; Gabelli, S.B.; Vogelstein, B. Structural Comparisons of Class I Phosphoinositide 3-Kinases. Nat. Rev. Cancer 2008, 8, 665–669. [Google Scholar] [CrossRef]
- Katso, R.; Okkenhaug, K.; Ahmadi, K.; White, S.; Timms, J.; Waterfield, M.D. Cellular Function of Phosphoinositide 3-Kinases: Implications for Development, Homeostasis, and Cancer. Annu. Rev. Cell Dev. Biol. 2001, 17, 615–675. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The Emerging Mechanisms of Isoform-Specific PI3K Signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef]
- Hawkins, P.T.; Stephens, L.R. PI3K Signalling in Inflammation. Biochim. Biophys. Acta 2015, 1851, 882–897. [Google Scholar] [CrossRef]
- Fritsch, R.; Downward, J. SnapShot: Class I PI3K Isoform Signaling. Cell 2013, 154, 940.e1. [Google Scholar] [CrossRef]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef]
- Rauf, A.; Khalil, A.A.; Awadallah, S.; Khan, S.A.; Abu-Izneid, T.; Kamran, M.; Hemeg, H.A.; Mubarak, M.S.; Khalid, A.; Wilairatana, P. Reactive Oxygen Species in Biological Systems: Pathways, Associated Diseases, and Potential Inhibitors—A Review. Food Sci. Nutr. 2023, 12, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Kma, L.; Baruah, T.J. The Interplay of ROS and the PI3K/Akt Pathway in Autophagy Regulation. Biotechnol. Appl. Biochem. 2022, 69, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Troutman, T.D.; Bazan, J.F.; Pasare, C. Toll-like Receptors, Signaling Adapters and Regulation of the pro-Inflammatory Response by PI3K. Cell Cycle 2012, 11, 3559. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Wada, Y.; Kitagishi, Y.; Matsuda, S. Link between PI3K/AKT/PTEN Pathway and NOX Proteinin Diseases. Aging Dis. 2014, 5, 203. [Google Scholar] [CrossRef]
- Leslie, N.R.; Downes, C.P. PTEN: The down Side of PI 3-Kinase Signalling. Cell Signal. 2002, 14, 285–295. [Google Scholar] [CrossRef]
- Innocenti, M.; Frittoli, E.; Ponzanelli, I.; Falck, J.R.; Brachmann, S.M.; Di Fiore, P.P.; Scita, G. Phosphoinositide 3-Kinase Activates Rac by Entering in a Complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 2003, 160, 17. [Google Scholar] [CrossRef]
- Lee, S.R.; Yang, K.S.; Kwon, J.; Lee, C.; Jeong, W.; Rhee, S.G. Reversible Inactivation of the Tumor Suppressor PTEN by H2O2. J. Biol. Chem. 2002, 277, 20336–20342. [Google Scholar] [CrossRef]
- Murata, H.; Ihara, Y.; Nakamura, H.; Yodoi, J.; Sumikawa, K.; Kondo, T. Glutaredoxin Exerts an Antiapoptotic Effect by Regulating the Redox State of Akt. J. Biol. Chem. 2003, 278, 50226–50233. [Google Scholar] [CrossRef]
- Arcaro, A.; Khanzada, U.K.; Vanhaesebroeck, B.; Tetley, T.D.; Waterfield, M.D.; Seckl, M.J. Two Distinct Phosphoinositide 3-Kinases Mediate Polypeptide Growth Factor-Stimulated PKB Activation. EMBO J. 2002, 21, 5097. [Google Scholar] [CrossRef]
- Seitz, C.; Hugle, M.; Cristofanon, S.; Tchoghandjian, A.; Fulda, S. The Dual PI3K/MTOR Inhibitor NVP-BEZ235 and Chloroquine Synergize to Trigger Apoptosis via Mitochondrial-Lysosomal Cross-Talk. Int. J. Cancer 2013, 132, 2682–2693. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Y.; Sternberg, P.; Cai, J. Essential Roles of the PI3 Kinase/Akt Pathway in Regulating Nrf2-Dependent Antioxidant Functions in the RPE. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Guan, X.H.; Yu, J.X.; Lv, J.; Zhang, H.X.; Fu, Q.C.; Xiang, H.B.; Bu, H.L.; Shi, D.; Shu, B.; et al. Activation of Spinal Phosphatidylinositol 3-Kinase/Protein Kinase B Mediates Pain Behavior Induced by Plantar Incision in Mice. Exp. Neurol. 2014, 255, 71–82. [Google Scholar] [CrossRef]
- Xu, J.T.; Tu, H.Y.; Xin, W.J.; Liu, X.G.; Zhang, G.H.; Zhai, C.H. Activation of Phosphatidylinositol 3-Kinase and Protein Kinase B/Akt in Dorsal Root Ganglia and Spinal Cord Contributes to the Neuropathic Pain Induced by Spinal Nerve Ligation in Rats. Exp. Neurol. 2007, 206, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, T.T.; Flauzino, T.; Otaguiri, E.S.; Batistela, A.P.; Zarpelon, A.C.; Cunha, T.M.; Ferreira, S.H.; Cunha, F.Q.; Verri, W.A. Granulocyte-Colony Stimulating Factor (G-CSF) Induces Mechanical Hyperalgesia via Spinal Activation of MAP Kinases and PI3K in Mice. Pharmacol. Biochem. Behav. 2011, 98, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.H.; Lu, X.F.; Zhang, H.X.; Wu, J.R.; Yuan, Y.; Bao, Q.; Ling, D.Y.; Cao, J.L. Phosphatidylinositol 3-Kinase Mediates Pain Behaviors Induced by Activation of Peripheral EphrinBs/EphBs Signaling in Mice. Pharmacol. Biochem. Behav. 2010, 95, 315–324. [Google Scholar] [CrossRef]
- Song, X.J.; Zheng, J.H.; Cao, J.L.; Liu, W.T.; Song, X.S.; Huang, Z.J. EphrinB-EphB Receptor Signaling Contributes to Neuropathic Pain by Regulating Neural Excitability and Spinal Synaptic Plasticity in Rats. Pain 2008, 139, 168–180. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, P.; Yang, Y.; Cai, M.-T.; Gan, L.; Qu, K.; Cheng, Y.-Y.; Dong, M. PI3K-Mediated Kif1a DNA Methylation Contributes to Neuropathic Pain: An in Vivo Study. Pain 2025, 1–15. [Google Scholar] [CrossRef]
- Gleichmann, M.; Mattson, M.P. Neuronal Calcium Homeostasis and Dysregulation. Antioxid. Redox Signal. 2011, 14, 1261. [Google Scholar] [CrossRef]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The Versatility and Universality of Calcium Signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef]
- Brini, M.; Calì, T.; Ottolini, D.; Carafoli, E. Neuronal Calcium Signaling: Function and Dysfunction. Cell. Mol. Life Sci. 2014, 71, 2787. [Google Scholar] [CrossRef]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative Genomics Identifies MCU as an Essential Component of the Mitochondrial Calcium Uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria, Oxidative Stress and Cell Death. Apoptosis 2007, 12, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Angelova, P.R. Sources and Triggers of Oxidative Damage in Neurodegeneration. Free Radic. Biol. Med. 2021, 173, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Zu, W.; Rawe, V.Y.; Pelto-Huikko, M.; Flickinger, C.J.; Sutovsky, P.; Gustafsson, J.Å.; Oko, R.; Miranda-Vizuete, A. Spermatocyte/Spermatid-Specific Thioredoxin-3, a Novel Golgi Apparatus-Associated Thioredoxin, Is a Specific Marker of Aberrant Spermatogenesis. J. Biol. Chem. 2004, 279, 34971–34982. [Google Scholar] [CrossRef]
- Cosentino-Gomes, D.; Rocco-Machado, N.; Meyer-Fernandes, J.R. Cell Signaling through Protein Kinase C Oxidation and Activation. Int. J. Mol. Sci. 2012, 13, 10697. [Google Scholar] [CrossRef]
- Angelova, P.R.; Abramov, A.Y. Functional Role of Mitochondrial Reactive Oxygen Species in Physiology. Free Radic. Biol. Med. 2016, 100, 81–85. [Google Scholar] [CrossRef]
- Kania, E.; Roest, G.; Vervliet, T.; Parys, J.B.; Bultynck, G. IP3 Receptor-Mediated Calcium Signaling and Its Role in Autophagy in Cancer. Front. Oncol. 2017, 7, 140. [Google Scholar] [CrossRef]
- Novikova, I.N.; Manole, A.; Zherebtsov, E.A.; Stavtsev, D.D.; Vukolova, M.N.; Dunaev, A.V.; Angelova, P.R.; Abramov, A.Y. Adrenaline Induces Calcium Signal in Astrocytes and Vasoconstriction via Activation of Monoamine Oxidase. Free Radic. Biol. Med. 2020, 159, 15–22. [Google Scholar] [CrossRef]
- Görlach, A.; Bertram, K.; Hudecova, S.; Krizanova, O. Calcium and ROS: A Mutual Interplay. Redox Biol. 2015, 6, 260. [Google Scholar] [CrossRef]
- Todorovic, S.M.; Jevtovic-Todorovic, V. Redox Regulation of Neuronal Voltage-Gated Calcium Channels. Antioxid. Redox Signal. 2014, 21, 880. [Google Scholar] [CrossRef]
- Bogeski, I.; Kummerow, C.; Al-Ansary, D.; Schwarz, E.C.; Koehler, R.; Kozai, D.; Takahashi, N.; Peinelt, C.; Griesemer, D.; Bozem, M.; et al. Differential Redox Regulation of ORAI Ion Channels: A Mechanism to Tune Cellular Calcium Signaling. Sci. Signal. 2010, 3, 2000672. [Google Scholar] [CrossRef] [PubMed]
- Perraud, A.L.; Takanishi, C.L.; Shen, B.; Kang, S.; Smith, M.K.; Schmitz, C.; Knowles, H.M.; Ferraris, D.; Li, W.; Zhang, J.; et al. Accumulation of Free ADP-Ribose from Mitochondria Mediates Oxidative Stress-Induced Gating of TRPM2 Cation Channels. J. Biol. Chem. 2005, 280, 6138–6148. [Google Scholar] [CrossRef] [PubMed]
- Susankova, K.; Tousova, K.; Vyklicky, L.; Teisinger, J.; Vlachova, V. Reducing and Oxidizing Agents Sensitize Heat-Activated Vanilloid Receptor (TRPV1) Current. Mol. Pharmacol. 2006, 70, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Bogeski, I.; Kilch, T.; Niemeyer, B.A. ROS and SOCE: Recent Advances and Controversies in the Regulation of STIM and Orai. J. Physiol. 2012, 590, 4193–4200. [Google Scholar] [CrossRef]
- Li, W.W.; Zhao, Y.; Liu, H.C.; Liu, J.; Chan, S.O.; Zhong, Y.F.; Zhang, T.Y.; Liu, Y.; Zhang, W.; Xia, Y.Q.; et al. Roles of Thermosensitive Transient Receptor Channels TRPV1 and TRPM8 in Paclitaxel-Induced Peripheral Neuropathic Pain. Int. J. Mol. Sci. 2024, 25, 5813. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Y. Transient Receptor Potential Channels: Multiple Modulators of Peripheral Neuropathic Pain in Several Rodent Models. Neurochem. Res. 2024, 49, 872–886. [Google Scholar] [CrossRef]
- Chu, W.G.; Wang, F.D.; Sun, Z.C.; Ma, S.B.; Wang, X.; Han, W.J.; Wang, F.; Bai, Z.T.; Wu, S.X.; Freichel, M.; et al. TRPC1/4/5 Channels Contribute to Morphine-Induced Analgesic Tolerance and Hyperalgesia by Enhancing Spinal Synaptic Potentiation and Structural Plasticity. FASEB J. 2020, 34, 8526–8543. [Google Scholar] [CrossRef]
- Alessandri-Haber, N.; Dina, O.A.; Chen, X.; Levine, J.D. TRPC1 and TRPC6 Channels Cooperate with TRPV4 to Mediate Mechanical Hyperalgesia and Nociceptor Sensitization. J. Neurosci. 2009, 29, 6217. [Google Scholar] [CrossRef]
- Godoy, J.A.; Rios, J.A.; Picón-Pagès, P.; Herrera-Fernández, V.; Swaby, B.; Crepin, G.; Vicente, R.; Fernández-Fernández, J.M.; Muñoz, F.J. Mitostasis, Calcium and Free Radicals in Health, Aging and Neurodegeneration. Biomolecules 2021, 11, 1012. [Google Scholar] [CrossRef]
- Gong, J.; Wang, X.Z.; Wang, T.; Chen, J.J.; Xie, X.Y.; Hu, H.; Yu, F.; Liu, H.L.; Jiang, X.Y.; Fan, H.D. Molecular Signal Networks and Regulating Mechanisms of the Unfolded Protein Response. J. Zhejiang Univ. Sci. B 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Malhotra, J.D.; Kaufman, R.J. Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Cycle or a Double-Edged Sword? Antioxid. Redox Signal. 2007, 9, 2277–2293. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; d’Hellencourt, C.L.; Ravanan, P. A Molecular Web: Endoplasmic Reticulum Stress, Inflammation, and Oxidative Stress. Front. Cell. Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Sokka, A.L.; Putkonen, N.; Mudo, G.; Pryazhnikov, E.; Reijonen, S.; Khiroug, L.; Belluardo, N.; Lindholm, D.; Korhonen, L. Endoplasmic Reticulum Stress Inhibition Protects against Excitotoxic Neuronal Injury in the Rat Brain. J. Neurosci. 2007, 27, 901–908. [Google Scholar] [CrossRef]
- Niella, R.V.; Corrêa, J.M.X.; dos Santos, J.F.R.; Lima, L.F.; da Costa Marques, C.S.; Santos, L.C.; Santana, L.R.; Silva, Á.J.C.; Farias, K.S.; Pirovani, C.P.; et al. Post-Treatment with Maropitant Reduces Oxidative Stress, Endoplasmic Reticulum Stress and Neuroinflammation on Peripheral Nerve Injury in Rats. PLoS ONE 2024, 19, e287390. [Google Scholar] [CrossRef]
- Zhou, F.; Zhang, W.; Zhou, J.; Li, M.; Zhong, F.; Zhang, Y.; Liu, Y.; Wang, Y. Involvement of Endoplasmic Reticulum Stress in Formalin-Induced Pain Is Attenuated by 4-Phenylbutyric Acid. J. Pain Res. 2017, 10, 653–662. [Google Scholar] [CrossRef]
- Patel, S.; Pangarkar, A.; Mahajan, S.; Majumdar, A. Therapeutic Potential of Endoplasmic Reticulum Stress Inhibitors in the Treatment of Diabetic Peripheral Neuropathy. Metab. Brain Dis. 2023, 38, 1841–1856. [Google Scholar] [CrossRef]
- Read, A.; Schröder, M. The Unfolded Protein Response: An Overview. Biology 2021, 10, 384. [Google Scholar] [CrossRef]
- Borges, C.R.; Lake, D.F. Oxidative Protein Folding: Nature’s Knotty Challenge. Antioxid. Redox Signal. 2014, 21, 392–395. [Google Scholar] [CrossRef]
- Kozlov, G.; Määttänen, P.; Thomas, D.Y.; Gehring, K. A Structural Overview of the PDI Family of Proteins. FEBS J. 2010, 277, 3924–3936. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Zhou, L.; Lei, Y.; Zhang, Y.; Huang, C. Redox Signaling and Unfolded Protein Response Coordinate Cell Fate Decisions under ER Stress. Redox Biol. 2019, 25, 101047. [Google Scholar] [CrossRef]
- Klappa, P.; Ruddock, L.W.; Darby, N.J.; Freedman, R.B. The b’ Domain Provides the Principal Peptide-Binding Site of Protein Disulfide Isomerase but All Domains Contribute to Binding of Misfolded Proteins. EMBO J. 1998, 17, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Iemura, S.I.; Kamiya, Y.; Ron, D.; Kato, K.; Natsume, T.; Nagata, K. Ero1-α and PDIs Constitute a Hierarchical Electron Transfer Network of Endoplasmic Reticulum Oxidoreductases. J. Cell Biol. 2013, 202, 861. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.R.; Riaz, T.A.; Kim, H.R.; Chae, H.J. The Aftermath of the Interplay between the Endoplasmic Reticulum Stress Response and Redox Signaling. Exp. Mol. Med. 2021, 53, 151. [Google Scholar] [CrossRef]
- Sevier, C.S. Erv2 and Quiescin Sulfhydryl Oxidases: Erv-Domain Enzymes Associated with the Secretory Pathway. Antioxid. Redox Signal. 2012, 16, 800–808. [Google Scholar] [CrossRef]
- Zeeshan, H.M.A.; Lee, G.H.; Kim, H.R.; Chae, H.J. Endoplasmic Reticulum Stress and Associated ROS. Int. J. Mol. Sci. 2016, 17, 327. [Google Scholar] [CrossRef]
- Eletto, D.; Chevet, E.; Argon, Y.; Appenzeller-Herzog, C. Redox Controls UPR to Control Redox. J. Cell Sci. 2014, 127, 3649–3658. [Google Scholar] [CrossRef]
- Uehara, T.; Nakamura, T.; Yao, D.; Shi, Z.Q.; Gu, Z.; Ma, Y.; Masliah, E.; Nomura, Y.; Lipton, S.A. S-Nitrosylated Protein-Disulphide Isomerase Links Protein Misfolding to Neurodegeneration. Nature 2006, 441, 513–517. [Google Scholar] [CrossRef]
- Vinokurov, A.Y.; Stelmashuk, O.A.; Ukolova, P.A.; Zherebtsov, E.A.; Abramov, A.Y. Brain Region Specificity in Reactive Oxygen Species Production and Maintenance of Redox Balance. Free Radic. Biol. Med. 2021, 174, 195–201. [Google Scholar] [CrossRef]
- Lee, K.H.; Cha, M.; Lee, B.H. Neuroprotective Effect of Antioxidants in the Brain. Int. J. Mol. Sci. 2020, 21, 7152. [Google Scholar] [CrossRef]
- Niedzielska, E.; Smaga, I.; Gawlik, M.; Moniczewski, A.; Stankowicz, P.; Pera, J.; Filip, M. Oxidative Stress in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 4094. [Google Scholar] [CrossRef]
- Halliwell, B. How to Characterize a Biological Antioxidant. Free Radic. Res. Commun. 1990, 9, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A. The Role of Antioxidants in the Chemistry of Oxidative Stress: A Review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.; Di Mascio, P.; Wagner, J.R. Formation and Repair of Oxidatively Generated Damage in Cellular DNA. Free Radic. Biol. Med. 2017, 107, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Several Lines of Antioxidant Defense against Oxidative Stress: Antioxidant Enzymes, Nanomaterials with Multiple Enzyme-Mimicking Activities, and Low-Molecular-Weight Antioxidants. Arch. Toxicol. 2024, 98, 1323–1367. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First Line Defence Antioxidants-Superoxide Dismutase (SOD), Catalase (CAT) and Glutathione Peroxidase (GPX): Their Fundamental Role in the Entire Antioxidant Defence Grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide Dismutases: Dual Roles in Controlling ROS Damage and Regulating ROS Signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Davis, S.M.; Pennypacker, K.R. Targeting Antioxidant Enzyme Expression as a Therapeutic Strategy for Ischemic Stroke. Neurochem. Int. 2017, 107, 23. [Google Scholar] [CrossRef]
- Adachi, T.; Wang, X.L. Association of Extracellular-Superoxide Dismutase Phenotype with the Endothelial Constitutive Nitric Oxide Synthase Polymorphism. FEBS Lett. 1998, 433, 166–168. [Google Scholar] [CrossRef]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide Dismutase Multigene Family: A Comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) Gene Structures, Evolution, and Expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Agrahari, G.; Sah, S.K.; Bang, C.H.; Kim, Y.H.; Kim, T.Y. Superoxide Dismutase 3 Controls the Activation and Differentiation of CD4+T Cells. Front. Immunol. 2021, 12, 628117. [Google Scholar] [CrossRef]
- Mccords, J.M.; Fridovich, I. Superoxide Dismutase: An Enzymic Function for Erythrocuprein (Hemocuprein). J. Biol. Chem. 1969, 244, 6049–6065. [Google Scholar] [CrossRef]
- Quist, D.A.; Diaz, D.E.; Liu, J.J.; Karlin, K.D. Activation of Dioxygen by Copper Metalloproteins and Insights from Model Complexes. J. Biol. Inorg. Chem. 2017, 22, 253. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Bai, Q.; Yang, Y.; Shi, X.; Du, G.; Yan, J.; Shi, J.; Wang, D. The Key Roles of Reactive Oxygen Species in Microglial Inflammatory Activation: Regulation by Endogenous Antioxidant System and Exogenous Sulfur-Containing Compounds. Eur. J. Pharmacol. 2023, 956, 175966. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.D.; Wang, J.A.; Miller, D.M.; Cebak, J.E.; Hill, R.L. Newer Pharmacological Approaches for Antioxidant Neuroprotection in Traumatic Brain Injury. Neuropharmacology 2019, 145, 247–258. [Google Scholar] [CrossRef]
- Murakami, K.; Murata, N.; Noda, Y.; Tahara, S.; Kaneko, T.; Kinoshita, N.; Hatsuta, H.; Murayama, S.; Barnham, K.J.; Irie, K.; et al. SOD1 (Copper/Zinc Superoxide Dismutase) Deficiency Drives Amyloid β Protein Oligomerization and Memory Loss in Mouse Model of Alzheimer Disease. J. Biol. Chem. 2011, 286, 44557. [Google Scholar] [CrossRef]
- Kirkman, H.N.; Gaetani, G.F. Catalase: A Tetrameric Enzyme with Four Tightly Bound Molecules of NADPH. Proc. Natl. Acad. Sci. USA 1984, 81, 4343–4347. [Google Scholar] [CrossRef]
- Amir Aslani, B.; Ghobadi, S. Studies on Oxidants and Antioxidants with a Brief Glance at Their Relevance to the Immune System. Life Sci. 2016, 146, 163–173. [Google Scholar] [CrossRef]
- Usui, S.; Komeima, K.; Lee, S.Y.; Jo, Y.J.; Ueno, S.; Rogers, B.S.; Wu, Z.; Shen, J.; Lu, L.; Oveson, B.C.; et al. Increased Expression of Catalase and Superoxide Dismutase 2 Reduces Cone Cell Death in Retinitis Pigmentosa. Mol. Ther. 2009, 17, 778–786. [Google Scholar] [CrossRef]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative Stress and Neurodegenerative Diseases: A Review of Upstream and Downstream Antioxidant Therapeutic Options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef]
- Baker, A.; Lin, C.C.; Lett, C.; Karpinska, B.; Wright, M.H.; Foyer, C.H. Catalase: A Critical Node in the Regulation of Cell Fate. Free Radic. Biol. Med. 2023, 199, 56–66. [Google Scholar] [CrossRef]
- Nandi, A.; Yan, L.J.; Jana, C.K.; Das, N. Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases. Oxid. Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef]
- Dutta, R.K.; Lee, J.N.; Maharjan, Y.; Park, C.; Choe, S.K.; Ho, Y.S.; Park, R. Catalase Deficiency Facilitates the Shuttling of Free Fatty Acid to Brown Adipose Tissue through Lipolysis Mediated by ROS during Sustained Fasting. Cell Biosci. 2021, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Kumar Kalvala, A.; Bagde, A.; Arthur, P.; Kumar Surapaneni, S.; Ramesh, N.; Nathani, A.; Singh, M. Role of Cannabidiol and Tetrahydrocannabivarin on Paclitaxel-Induced Neuropathic Pain in Rodents. Int. Immunopharmacol. 2022, 107, 108693. [Google Scholar] [CrossRef] [PubMed]
- Luangwattananun, P.; Yainoy, S.; Eiamphungporn, W.; Songtawee, N.; Bülow, L.; Ayudhya, C.I.N.; Prachayasittikul, V. Engineering of a Novel Tri-Functional Enzyme with MnSOD, Catalase and Cell-Permeable Activities. Int. J. Biol. Macromol. 2016, 85, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research Progress of Glutathione Peroxidase Family (GPX) in Redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Loscalzo, J. The Role of Glutathione Peroxidase-1 in Health and Disease. Free Radic. Biol. Med. 2022, 188, 146–161. [Google Scholar] [CrossRef]
- Weaver, K.; Skouta, R. The Selenoprotein Glutathione Peroxidase 4: From Molecular Mechanisms to Novel Therapeutic Opportunities. Biomedicines 2022, 10, 891. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Regulatory Phenomena in the Glutathione Peroxidase Superfamily. Antioxid. Redox Signal. 2020, 33, 498–516. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, Present and Future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Hazzaa, S.M.; Abdou, A.G.; Ibraheim, E.O.; Salem, E.A.; Hassan, M.H.A.; Abdel-Razek, H.A.D. Effect of L-Carnitine and Atorvastatin on a Rat Model of Ischemia-Reperfusion Injury of Spinal Cord. J. Immunoass. Immunochem. 2021, 42, 596–619. [Google Scholar] [CrossRef]
- Drummond, I.S.A.; de Oliveira, J.N.S.; Niella, R.V.; Silva, Á.J.C.; de Oliveira, I.S.; de Souza, S.S.; da Costa Marques, C.S.; Corrêa, J.M.X.; Silva, J.F.; de Lavor, M.S.L. Evaluation of the Therapeutic Potential of Amantadine in a Vincristine-Induced Peripheral Neuropathy Model in Rats. Animals 2024, 14, 1941. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, R.; Qi, W.; Jia, L.; Ma, K.; Si, J.; Yin, J.; Zhao, Y.; Dai, Z.; Yin, J. Methyl Ferulic Acid Alleviates Neuropathic Pain by Inhibiting Nox4-Induced Ferroptosis in Dorsal Root Ganglia Neurons in Rats. Mol. Neurobiol. 2023, 60, 3175–3189. [Google Scholar] [CrossRef]
- Zhang, X.; Song, T.; Zhao, M.; Tao, X.; Zhang, B.; Sun, C.; Wang, P.; Wang, K.; Zhao, L. Sirtuin 2 Alleviates Chronic Neuropathic Pain by Suppressing Ferroptosis in Rats. Front. Pharmacol. 2022, 13, 827016. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huo, X.; Han, C.; Ning, J.; Chen, H.; Li, B.; Liu, J.; Ma, W.; Li, Q.; Yu, Y.; et al. Ferroptosis Is Involved in the Development of Neuropathic Pain and Allodynia. Mol. Cell. Biochem. 2021, 476, 3149–3161. [Google Scholar] [CrossRef]
- Guo, Y.; Du, J.; Xiao, C.; Xiang, P.; Deng, Y.; Hei, Z.; Li, X. Inhibition of Ferroptosis-like Cell Death Attenuates Neuropathic Pain Reactions Induced by Peripheral Nerve Injury in Rats. Eur. J. Pain 2021, 25, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, K.; Zhang, T.; Deng, D.; Shen, J.; Zhao, B.; Fu, D.; Chen, X. Mechanisms and Therapeutic Potential of GPX4 in Pain Modulation. Pain. Ther. 2025, 14, 21–45. [Google Scholar] [CrossRef]
- Bjørklund, G.; Zou, L.; Peana, M.; Chasapis, C.T.; Hangan, T.; Lu, J.; Maes, M. The Role of the Thioredoxin System in Brain Diseases. Antioxidants 2022, 11, 2161. [Google Scholar] [CrossRef]
- Burke-Gaffney, A.; Callister, M.E.J.; Nakamura, H. Thioredoxin: Friend or Foe in Human Disease? Trends Pharmacol. Sci. 2005, 26, 398–404. [Google Scholar] [CrossRef]
- Arnér, E.S.J.; Holmgren, A. Physiological Functions of Thioredoxin and Thioredoxin Reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Hasan, A.A.; Kalinina, E.; Tatarskiy, V.; Shtil, A. The Thioredoxin System of Mammalian Cells and Its Modulators. Biomedicines 2022, 10, 1757. [Google Scholar] [CrossRef]
- Monteiro, H.P.; Ogata, F.T.; Stern, A. Thioredoxin Promotes Survival Signaling Events under Nitrosative/Oxidative Stress Associated with Cancer Development. Biomed. J. 2017, 40, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Silva-Adaya, D.; Gonsebatt, M.E.; Guevara, J. Thioredoxin System Regulation in the Central Nervous System: Experimental Models and Clinical Evidence. Oxid. Med. Cell. Longev. 2014, 2014, 590808. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zou, L.; Zhang, X.; Branco, V.; Wang, J.; Carvalho, C.; Holmgren, A.; Lu, J. Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System. Antioxid. Redox Signal. 2017, 27, 989–1010. [Google Scholar] [CrossRef] [PubMed]
- Vilhardt, F.; Haslund-Vinding, J.; Jaquet, V.; McBean, G. Microglia Antioxidant Systems and Redox Signalling. Br. J. Pharmacol. 2017, 174, 1719–1732. [Google Scholar] [CrossRef] [PubMed]
- Valek, L.; Kanngießer, M.; Häussler, A.; Agarwal, N.; Lillig, C.H.; Tegeder, I. Redoxins in Peripheral Neurons after Sciatic Nerve Injury. Free Radic. Biol. Med. 2015, 89, 581–592. [Google Scholar] [CrossRef]
- Ji, R.R.; Chamessian, A.; Zhang, Y.Q. Pain Regulation by Non-Neuronal Cells and Inflammation. Science 1979 2016, 354, 572–577. [Google Scholar] [CrossRef]
- Fernandes, V.; Sharma, D.; Vaidya, S.; Shantanu, P.A.; Guan, Y.; Kalia, K.; Tiwari, V. Cellular and Molecular Mechanisms Driving Neuropathic Pain: Recent Advancements and Challenges. Expert. Opin. Ther. Targets 2018, 22, 131–142. [Google Scholar] [CrossRef]
- Jensen, T.S.; Baron, R.; Haanpää, M.; Kalso, E.; Loeser, J.D.; Rice, A.S.C.; Treede, R.D. A New Definition of Neuropathic Pain. Pain 2011, 152, 2204–2205. [Google Scholar] [CrossRef]
- Bouhassira, D. Neuropathic Pain: Definition, Assessment and Epidemiology. Rev. Neurol. 2019, 175, 16–25. [Google Scholar] [CrossRef]
- Bouhassira, D.; Lantéri-Minet, M.; Attal, N.; Laurent, B.; Touboul, C. Prevalence of Chronic Pain with Neuropathic Characteristics in the General Population. Pain 2008, 136, 380–387. [Google Scholar] [CrossRef]
- Ji, R.R.; Berta, T.; Nedergaard, M. Glia and Pain: Is Chronic Pain a Gliopathy? Pain 2013, 154, S10–S28. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Xu, Z.Z.; Gao, Y.J. Emerging Targets in Neuroinflammation-Driven Chronic Pain. Nat. Rev. Drug Discov. 2014, 13, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Q.; Liu, Z.; Liu, Z.H.; Chen, S.P.; Li, M.; Shahveranov, A.; Ye, D.W.; Tian, Y.K. Interleukin-6: An Emerging Regulator of Pathological Pain. J. Neuroinflamm. 2016, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Gwak, Y.S.; Kang, J.; Unabia, G.C.; Hulsebosch, C.E. Spatial and Temporal Activation of Spinal Glial Cells: Role of Gliopathy in Central Neuropathic Pain Following Spinal Cord Injury in Rats. Exp. Neurol. 2012, 234, 362–372. [Google Scholar] [CrossRef]
- Raghavendra, V.; Tanga, F.; Deleo, J.A. Inhibition of Microglial Activation Attenuates the Development but Not Existing Hypersensitivity in a Rat Model of Neuropathy. J. Pharmacol. Exp. Ther. 2003, 306, 624–630. [Google Scholar] [CrossRef]
- Peng, J.; Gu, N.; Zhou, L.; Eyo, U.B.; Murugan, M.; Gan, W.B.; Wu, L.J. Microglia and Monocytes Synergistically Promote the Transition from Acute to Chronic Pain after Nerve Injury. Nat. Commun. 2016, 7, 12029. [Google Scholar] [CrossRef]
- Ellis, A.; Bennett, D.L.H. Neuroinflammation and the Generation of Neuropathic Pain. Br. J. Anaesth. 2013, 111, 26–37. [Google Scholar] [CrossRef]
- Town, T.; Nikolic, V.; Tan, J. The Microglial “Activation” Continuum: From Innate to Adaptive Responses. J. Neuroinflamm. 2005, 2, 24. [Google Scholar] [CrossRef]
- Hu, X.; Leak, R.K.; Shi, Y.; Suenaga, J.; Gao, Y.; Zheng, P.; Chen, J. Microglial and Macrophage Polarization—New Prospects for Brain Repair. Nat. Rev. Neurol. 2015, 11, 56–64. [Google Scholar] [CrossRef]
- Schreiner, B.; Romanelli, E.; Liberski, P.; Ingold-Heppner, B.; Sobottka-Brillout, B.; Hartwig, T.; Chandrasekar, V.; Johannssen, H.; Zeilhofer, H.U.; Aguzzi, A.; et al. Astrocyte Depletion Impairs Redox Homeostasis and Triggers Neuronal Loss in the Adult CNS. Cell Rep. 2015, 12, 1377–1384. [Google Scholar] [CrossRef]
- Gan, L.; Vargas, M.R.; Johnson, D.A.; Johnson, J.A. Astrocyte-Specific Overexpression of Nrf2 Delays Motor Pathology and Synuclein Aggregation throughout the CNS in the Alpha-Synuclein Mutant (A53T) Mouse Model. J. Neurosci. 2012, 32, 17775. [Google Scholar] [CrossRef]
- Wang, D.; Couture, R.; Hong, Y. Activated Microglia in the Spinal Cord Underlies Diabetic Neuropathic Pain. Eur. J. Pharmacol. 2014, 728, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Gao, H.M.; Wang, J.Y.; Jeohn, G.H.; Cooper, C.L.; Hong, J.S. Role of Nitric Oxide in Inflammation-Mediated Neurodegeneration. Ann. N. Y. Acad. Sci. 2002, 962, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Cha, M.; Lee, B.H. Crosstalk between Neuron and Glial Cells in Oxidative Injury and Neuroprotection. Int. J. Mol. Sci. 2021, 22, 13315. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Aid, S.; Kim, H.W.; Jackson, S.H.; Bosetti, F. Inhibition of NADPH Oxidase Promotes Alternative and Anti-Inflammatory Microglial Activation during Neuroinflammation. J. Neurochem. 2012, 120, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Lijia, Z.; Zhao, S.; Wang, X.; Wu, C.; Yang, J. A Self-Propelling Cycle Mediated by Reactive Oxide Species and Nitric Oxide Exists in LPS-Activated Microglia. Neurochem. Int. 2012, 61, 1220–1230. [Google Scholar] [CrossRef]
- Abramov, A.Y.; Canevari, L.; Duchen, M.R. Beta-Amyloid Peptides Induce Mitochondrial Dysfunction and Oxidative Stress in Astrocytes and Death of Neurons through Activation of NADPH Oxidase. J. Neurosci. 2004, 24, 565–575. [Google Scholar] [CrossRef]
- Teismann, P.; Schulz, J.B. Cellular Pathology of Parkinson’s Disease: Astrocytes, Microglia and Inflammation. Cell Tissue Res. 2004, 318, 149–161. [Google Scholar] [CrossRef]
- Markiewicz, I.; Lukomska, B. The Role of Astrocytes in the Physiology and Pathology of the Central Nervous System. Acta Neurobiol. Exp. 2006, 66, 343–358. [Google Scholar] [CrossRef]
- Wang, A.; Xu, C. The Role of Connexin43 in Neuropathic Pain Induced by Spinal Cord Injury. Acta Biochim. Biophys. Sin. 2019, 51, 555–561. [Google Scholar] [CrossRef]
- Shih, E.K.; Robinson, M.B. Role of Astrocytic Mitochondria in Limiting Ischemic Brain Injury? Physiology 2018, 33, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guan, T.; Li, C.; Shang, H.; Cui, L.; Li, X.M.; Kong, J. SOD1 Aggregation in Astrocytes Following Ischemia/Reperfusion Injury: A Role of NO-Mediated S-Nitrosylation of Protein Disulfide Isomerase (PDI). J. Neuroinflamm. 2012, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, F.; Vivacqua, G.; Lazzeri, G.; Ferese, R.; Iannacone, S.; Onori, P.; Morini, S.; D’Este, L.; Fornai, F. Chronic MPTP in Mice Damage-Specific Neuronal Phenotypes within Dorsal Laminae of the Spinal Cord. Neurotoxic. Res. 2021, 39, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Liu, Y.; Wang, T.; Wei, S.J.; Block, M.L.; Wilson, B.; Liu, B.; Hong, J.S. NADPH Oxidase Mediates Lipopolysaccharide-Induced Neurotoxicity and Proinflammatory Gene Expression in Activated Microglia. J. Biol. Chem. 2004, 279, 1415–1421. [Google Scholar] [CrossRef]
- Andersson, D.A.; Filipovic, M.R.; Gentry, C.; Eberhardt, M.; Vastani, N.; Leffler, A.; Reeh, P.; Bevan, S. Streptozotocin Stimulates the Ion Channel TRPA1 Directly: Involvement of Peroxynitrite. J. Biol. Chem. 2015, 290, 15185–15196. [Google Scholar] [CrossRef]
- Chen, S.J.; Zhang, W.; Tong, Q.; Conrad, K.; Hirschler-Laszkiewicz, I.; Bayerl, M.; Kim, J.K.; Cheung, J.Y.; Miller, B.A. Role of TRPM2 in Cell Proliferation and Susceptibility to Oxidative Stress. Am. J. Physiol. Cell Physiol. 2013, 304, 548–560. [Google Scholar] [CrossRef]
- Özdemir, Ü.S.; Nazıroğlu, M.; Şenol, N.; Ghazizadeh, V. Hypericum Perforatum Attenuates Spinal Cord Injury-Induced Oxidative Stress and Apoptosis in the Dorsal Root Ganglion of Rats: Involvement of TRPM2 and TRPV1 Channels. Mol. Neurobiol. 2016, 53, 3540–3551. [Google Scholar] [CrossRef]
- Haraguchi, K.; Kawamoto, A.; Isami, K.; Maeda, S.; Kusano, A.; Asakura, K.; Shirakawa, H.; Mori, Y.; Nakagawa, T.; Kaneko, S. TRPM2 Contributes to Inflammatory and Neuropathic Pain through the Aggravation of Pronociceptive Inflammatory Responses in Mice. J. Neurosci. 2012, 32, 3931–3941. [Google Scholar] [CrossRef]
- Dangi, A.; Sharma, S.S. Pharmacological Agents Targeting Transient Receptor Potential (TRP) Channels in Neuropathic Pain: Preclinical and Clinical Status. Eur. J. Pharmacol. 2024, 980, 176845. [Google Scholar] [CrossRef]
- Rodrigues, P.; Ruviaro, N.A.; Trevisan, G. TRPV4 Role in Neuropathic Pain Mechanisms in Rodents. Antioxidants 2022, 12, 24. [Google Scholar] [CrossRef]
- Antoniazzi, C.T.D.D.; Nassini, R.; Rigo, F.K.; Milioli, A.M.; Bellinaso, F.; Camponogara, C.; Silva, C.R.; de Almeida, A.S.; Rossato, M.F.; De Logu, F.; et al. Transient Receptor Potential Ankyrin 1 (TRPA1) Plays a Critical Role in a Mouse Model of Cancer Pain. Int. J. Cancer 2019, 144, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Mobasheri, A.; Simon, F.; Bello, A.; Soriano, S.; Nazıro, M.; Braidy, N. Thermo-Sensitive TRP Channels: Novel Targets for Treating Chemotherapy-Induced Peripheral Pain. Front. Physiol. 2017, 8, 1040. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Petropoulos, I.; Grimm, S.; Georgila, K.; Catalgol, B.; Friguet, B.; Grune, T.; Gonos, E.S. Protein Damage, Repair and Proteolysis. Mol. Asp. Med. 2014, 35, 1–71. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Hampton, M.B. Thiol Chemistry and Specificity in Redox Signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef]
- Go, Y.M.; Chandler, J.D.; Jones, D.P. The Cysteine Proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef]
- Chandran, S.; Binninger, D. Role of Oxidative Stress, Methionine Oxidation and Methionine Sulfoxide Reductases (MSR) in Alzheimer’s Disease. Antioxidants 2023, 13, 21. [Google Scholar] [CrossRef]
- Rayner, B.S.; Love, D.T.; Hawkins, C.L. Comparative Reactivity of Myeloperoxidase-Derived Oxidants with Mammalian Cells. Free Radic. Biol. Med. 2014, 71, 240–255. [Google Scholar] [CrossRef]
- Fujiwara, N.; Nakano, M.; Kato, S.; Yoshihara, D.; Ookawara, T.; Eguchi, H.; Taniguchi, N.; Suzuki, K. Oxidative Modification to Cysteine Sulfonic Acid of Cys111 in Human Copper-Zinc Superoxide Dismutase. J. Biol. Chem. 2007, 282, 35933–35944. [Google Scholar] [CrossRef]
- Rubio, M.A.; Herrando-Grabulosa, M.; Vilches, J.J.; Navarro, X. Involvement of Sensory Innervation in the Skin of SOD1G93A ALS Mice. J. Peripher. Nerv. Syst. 2016, 21, 88–95. [Google Scholar] [CrossRef]
- Sharp, P.S.; Dick, J.R.T.; Greensmith, L. The Effect of Peripheral Nerve Injury on Disease Progression in the SOD1 (G93A) Mouse Model of Amyotrophic Lateral Sclerosis. Neuroscience 2005, 130, 897–910. [Google Scholar] [CrossRef]
- Bombaci, A.; Lupica, A.; Pozzi, F.E.; Remoli, G.; Manera, U.; Di Stefano, V. Sensory Neuropathy in Amyotrophic Lateral Sclerosis: A Systematic Review. J. Neurol. 2023, 270, 5677–5691. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Jozsa, F.; Al-Chalabi, A. Nonmotor Symptoms in Amyotrophic Lateral Sclerosis: A Systematic Review. Int. Rev. Neurobiol. 2017, 134, 1409–1441. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Uys, J.D.; Tew, K.D.; Townsend, D.M. S-Glutathionylation: From Molecular Mechanisms to Health Outcomes. Antioxid. Redox Signal. 2011, 15, 233. [Google Scholar] [CrossRef]
- Tu, C.; Wang, S.-C.; Dai, M.-X.; Lai, S.-Q.; Huang, Z.-W.; Yu, Y.-P.; Chen, Y.-B.; Zeng, J.-H.; Wang, L.; Zhong, Z.-M. Accumulation of Advanced Oxidative Protein Products Exacerbate Satellite Glial Cells Activation and Neuropathic Pain. Mol. Med. 2025, 31, 25. [Google Scholar] [CrossRef] [PubMed]
- Nuriel, T.; Hansler, A.; Gross, S.S. Protein Nitrotryptophan: Formation, Significance and Identification. J. Proteom. 2011, 74, 2300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salvemini, D.; Doyle, T.; Chen, Z.; Muscoli, C.; Bryant, L.; Esposito, E.; Cuzzocrea, S.; Dagostino, C.; Ryerse, J.; Rausaria, S.; et al. Targeting the Overproduction of Peroxynitrite for the Prevention and Reversal of Paclitaxel-Induced Neuropathic Pain. J. Neurosci. 2012, 32, 6149–6160. [Google Scholar] [CrossRef]
- Salvemini, D.; Neumann, W. Targeting Peroxynitrite Driven Nitroxidative Stress with Synzymes: A Novel Therapeutic Approach in Chronic Pain Management. Life Sci. 2010, 86, 604–614. [Google Scholar] [CrossRef]
- Salvemini, D.; Kim, S.F.; Mollace, V. Reciprocal Regulation of the Nitric Oxide and Cyclooxygenase Pathway in Pathophysiology: Relevance and Clinical Implications. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, 473–487. [Google Scholar] [CrossRef]
- Ilari, S.; Giancotti, L.A.; Lauro, F.; Dagostino, C.; Gliozzi, M.; Malafoglia, V.; Sansone, L.; Palma, E.; Tafani, M.; Russo, M.A.; et al. Antioxidant Modulation of Sirtuin 3 during Acute Inflammatory Pain: The ROS Control. Pharmacol. Res. 2020, 157, 104851. [Google Scholar] [CrossRef]
- Liao, M.F.; Lu, K.T.; Hsu, J.L.; Lee, C.H.; Cheng, M.Y.; Ro, L.S. The Role of Autophagy and Apoptosis in Neuropathic Pain Formation. Int. J. Mol. Sci. 2022, 23, 2685. [Google Scholar] [CrossRef]
- Valluru, L.; Diao, Y.; Hachmeister, J.E.; Liu, D. Mn (III) Tetrakis (4-Benzoic Acid) Porphyrin Protects against Neuronal and Glial Oxidative Stress and Death after Spinal Cord Injury. CNS Neurol. Disord. Drug Targets 2012, 11, 774–790. [Google Scholar] [CrossRef] [PubMed]
- Ntalouka, F.; Tsirivakou, A. Luteolin: A Promising Natural Agent in Management of Pain in Chronic Conditions. Front. Pain Res. 2023, 4, 1114428. [Google Scholar] [CrossRef] [PubMed]
- Frediani, J.K.; Lal, A.A.; Kim, E.; Leslie, S.L.; Boorman, D.W.; Singh, V. The Role of Diet and Non-Pharmacologic Supplements in the Treatment of Chronic Neuropathic Pain: A Systematic Review. Pain Pract. 2024, 24, 186–210. [Google Scholar] [CrossRef] [PubMed]
- Batinic-Haberle, I.; Tovmasyan, A.; Roberts, E.R.H.; Vujaskovic, Z.; Leong, K.W.; Spasojevic, I. SOD Therapeutics: Latest Insights into Their Structure-Activity Relationships and Impact on the Cellular Redox-Based Signaling Pathways. Antioxid. Redox Signal. 2014, 20, 2372–2415. [Google Scholar] [CrossRef]
- Bonetta, R. Potential Therapeutic Applications of MnSODs and SOD-Mimetics. Chem.—A Eur. J. 2018, 24, 5032–5041. [Google Scholar] [CrossRef]
- Buettner, G.R.; Wagner, B.A.; Rodgers, V.G.J. Quantitative Redox Biology: An Approach to Understand the Role of Reactive Species in Defining the Cellular Redox Environment. Cell Biochem. Biophys. 2013, 67, 477–483. [Google Scholar] [CrossRef]
- Policar, C.; Bouvet, J.; Bertrand, H.C.; Delsuc, N. SOD Mimics: From the Tool Box of the Chemists to Cellular Studies. Curr. Opin. Chem. Biol. 2022, 67, 102109. [Google Scholar] [CrossRef]
- Eckshtain, M.; Zilbermann, I.; Mahammed, A.; Saltsman, I.; Okun, Z.; Maimon, E.; Cohen, H.; Meyerstein, D.; Gross, Z. Superoxide Dismutase Activity of Corrole Metal Complexes. Dalton Trans. 2009, 38, 7879–7882. [Google Scholar] [CrossRef]
- Franke, A.; Scheitler, A.; Kenkel, I.; Lippert, R.; Zahl, A.; Balbinot, D.; Jux, N.; Ivanović-Burmazović, I. Positive Charge on Porphyrin Ligand and Nature of Metal Center Define Basic Physicochemical Properties of Cationic Manganese and Iron Porphyrins in Aqueous Solution. Inorg. Chem. 2019, 58, 9618–9630. [Google Scholar] [CrossRef]
- Batinić-Haberle, I.; Rebouças, J.S.; Spasojević, I. Superoxide Dismutase Mimics: Chemistry, Pharmacology, and Therapeutic Potential. Antioxid. Redox Signal. 2010, 13, 877–918. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Tome, M.E. Thiol Regulation by Mn Porphyrins, Commonly Known as SOD Mimics. Redox Biol. 2019, 25, 101139. [Google Scholar] [CrossRef] [PubMed]
- Day, B.J. Catalase and Glutathione Peroxidase Mimics. Biochem. Pharmacol. 2009, 77, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Rebouças, J.S.; Spasojević, I.; Batinić-Haberle, I. Quality of Potent Mn Porphyrin-Based SOD Mimics and Peroxynitrite Scavengers for Pre-Clinical Mechanistic/Therapeutic Purposes. J. Pharm. Biomed. Anal. 2008, 48, 1046. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.M.; Reboucas, J.S.; Durazo, A.; Kos, I.; Fike, F.; Panni, M.; Gralla, E.B.; Valentine, J.S.; Batinic-Haberle, I.; Gatti, R.A. Radio-Protective Effects of Manganese-Containing Superoxide Dismutase Mimics on Ataxia Telangiectasia Cells. Free Radic. Biol. Med. 2009, 47, 250. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Hannibal, L.; Batinic-Haberle, I.; Radi, R. Reduction of Manganese Porphyrins by Flavoenzymes and Submitochondrial Particles: A Catalytic Cycle for the Reduction of Peroxynitrite. Free Radic. Biol. Med. 2006, 41, 503–512. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Shang, D.J.; Wang, X.L.; You, Z.L.; Li, H.B. Antihyperglycemic and Neuroprotective Effects of One Novel Cu-Zn SOD Mimetic. Bioorg Med. Chem. Lett. 2011, 21, 4320–4324. [Google Scholar] [CrossRef]
- Celic, T.; Španjol, J.; Bobinac, M.; Tovmasyan, A.; Vukelic, I.; Reboucas, J.S.; Batinic-Haberle, I.; Bobinac, D. Mn Porphyrin-Based SOD Mimic, MnTnHex-2-PyP(5+), and Non-SOD Mimic, MnTBAP(3−), Suppressed Rat Spinal Cord Ischemia/Reperfusion Injury via NF-ΚB Pathways. Free Radic. Res. 2014, 48, 1426–1442. [Google Scholar] [CrossRef]
- Tse, H.M.; Milton, M.J.; Piganelli, J.D. Mechanistic Analysis of the Immunomodulatory Effects of a Catalytic Antioxidant on Antigen-Presenting Cells: Implication for Their Use in Targeting Oxidation–Reduction Reactions in Innate Immunity. Free Radic. Biol. Med. 2004, 36, 233–247. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Vitturi, D.; Batinić-Haberle, I.; Fridovich, I.; Goldstein, S.; Czapski, G.; Radi, R. Reactions of Manganese Porphyrins with Peroxynitrite and Carbonate Radical Anion. J. Biol. Chem. 2003, 278, 27432–27438. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Batinić-Haberle, I.; Spasojević, I.; Fridovich, I.; Radi, R. Catalytic Scavenging of Peroxynitrite by Isomeric Mn(III) N-Methylpyridylporphyrins in the Presence of Reductants. Chem. Res. Toxicol. 1999, 12, 442–449. [Google Scholar] [CrossRef]
- Campo, G.M.; Avenoso, A.; D’Ascola, A.; Scuruchi, M.; Nastasi, G.; Micali, A.; Puzzolo, D.; Pisani, A.; Calatroni, A.; Campo, S. The SOD Mimic MnTM-2-PyP(5+) Reduces Hyaluronan Degradation-Induced Inflammation in Mouse Articular Chondrocytes Stimulated with Fe (II) plus Ascorbate. Int. J. Biochem. Cell Biol. 2013, 45, 1610–1619. [Google Scholar] [CrossRef] [PubMed]
- Janes, K.; Doyle, T.; Bryant, L.; Esposito, E.; Cuzzocrea, S.; Ryerse, J.; Bennett, G.J.; Salvemini, D. Bioenergetic Deficits in Peripheral Nerve Sensory Axons during Chemotherapy-Induced Neuropathic Pain Resulting from Peroxynitrite-Mediated Post-Translational Nitration of Mitochondrial Superoxide Dismutase. Pain 2013, 154, 2432–2440. [Google Scholar] [CrossRef] [PubMed]
- Negi, G.; Kumar, A.; Sharma, S.S. Concurrent Targeting of Nitrosative Stress-PARP Pathway Corrects Functional, Behavioral and Biochemical Deficits in Experimental Diabetic Neuropathy. Biochem. Biophys. Res. Commun. 2010, 391, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Kumar, A.; Kaundal, R.K.; Sharma, S.S. Amelioration of Neurological and Biochemical Deficits by Peroxynitrite Decomposition Catalysts in Experimental Diabetic Neuropathy. Eur. J. Pharmacol. 2008, 596, 77–83. [Google Scholar] [CrossRef]
- Drel, V.R.; Pacher, P.; Vareniuk, I.; Pavlov, I.; Ilnytska, O.; Lyzogubov, V.V.; Tibrewala, J.; Groves, J.T.; Obrosova, I.G. A Peroxynitrite Decomposition Catalyst Counteracts Sensory Neuropathy in Streptozotocin-Diabetic Mice. Eur. J. Pharmacol. 2007, 569, 48–58. [Google Scholar] [CrossRef][Green Version]
- Kwak, K.H.; Jung, H.; Park, J.M.; Yeo, J.S.; Kim, H.; Lee, H.C.; Byun, S.H.; Kim, J.C.; Park, S.S.; Lim, D.G. A Peroxynitrite Decomposition Catalyst Prevents Mechanical Allodynia and NMDA Receptor Activation in the Hind-Paw Ischemia Reperfusion Injury Rats. Exp. Ther. Med. 2014, 7, 508–512. [Google Scholar] [CrossRef][Green Version]
- Komirishetty, P.; Areti, A.; Arruri, V.K.; Sistla, R.; Gogoi, R.; Kumar, A. FeTMPyP a Peroxynitrite Decomposition Catalyst Ameliorated Functional and Behavioral Deficits in Chronic Constriction Injury Induced Neuropathic Pain in Rats. Free Radic. Res. 2021, 55, 1005–1017. [Google Scholar] [CrossRef]
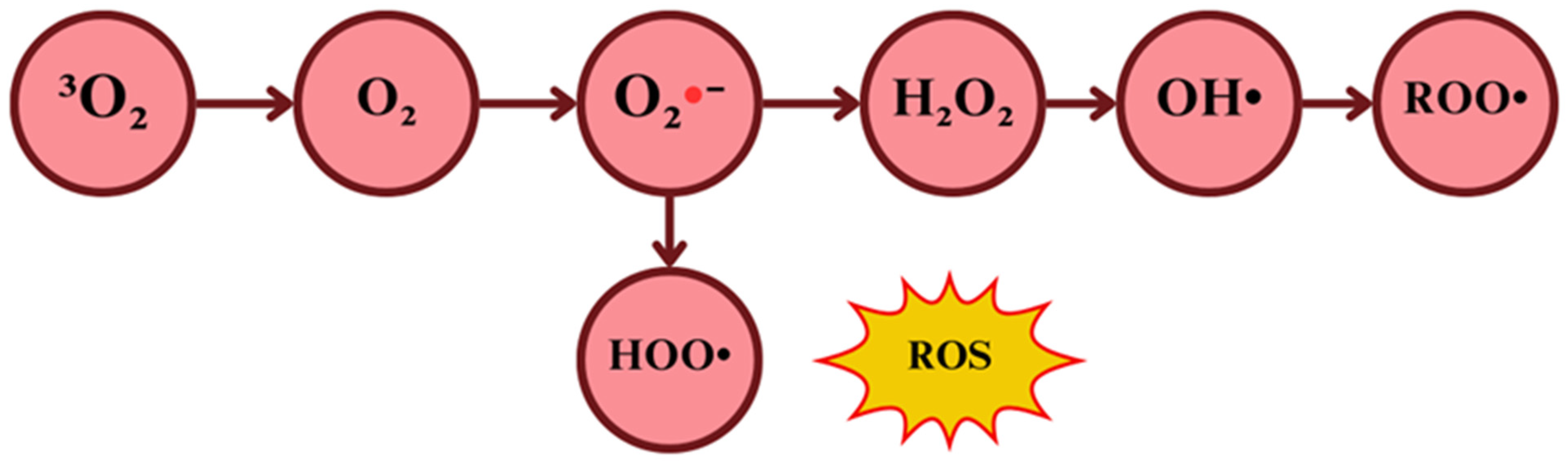
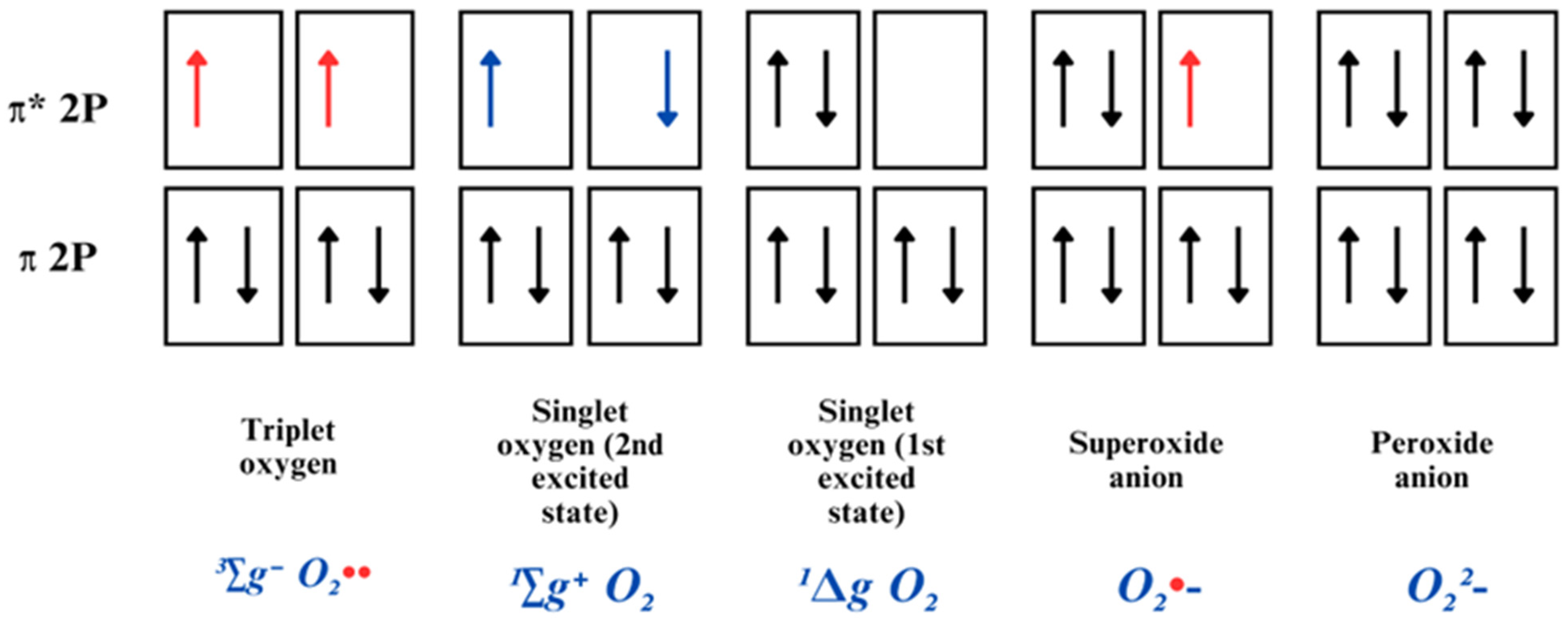
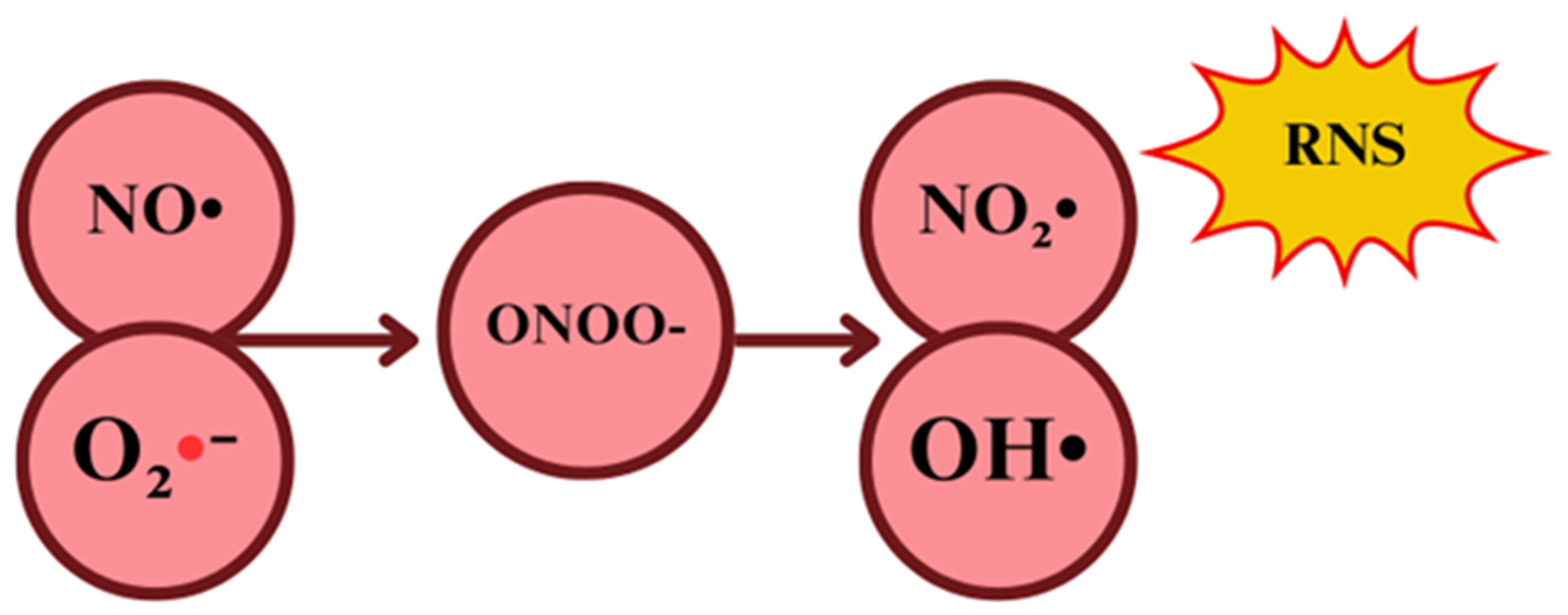
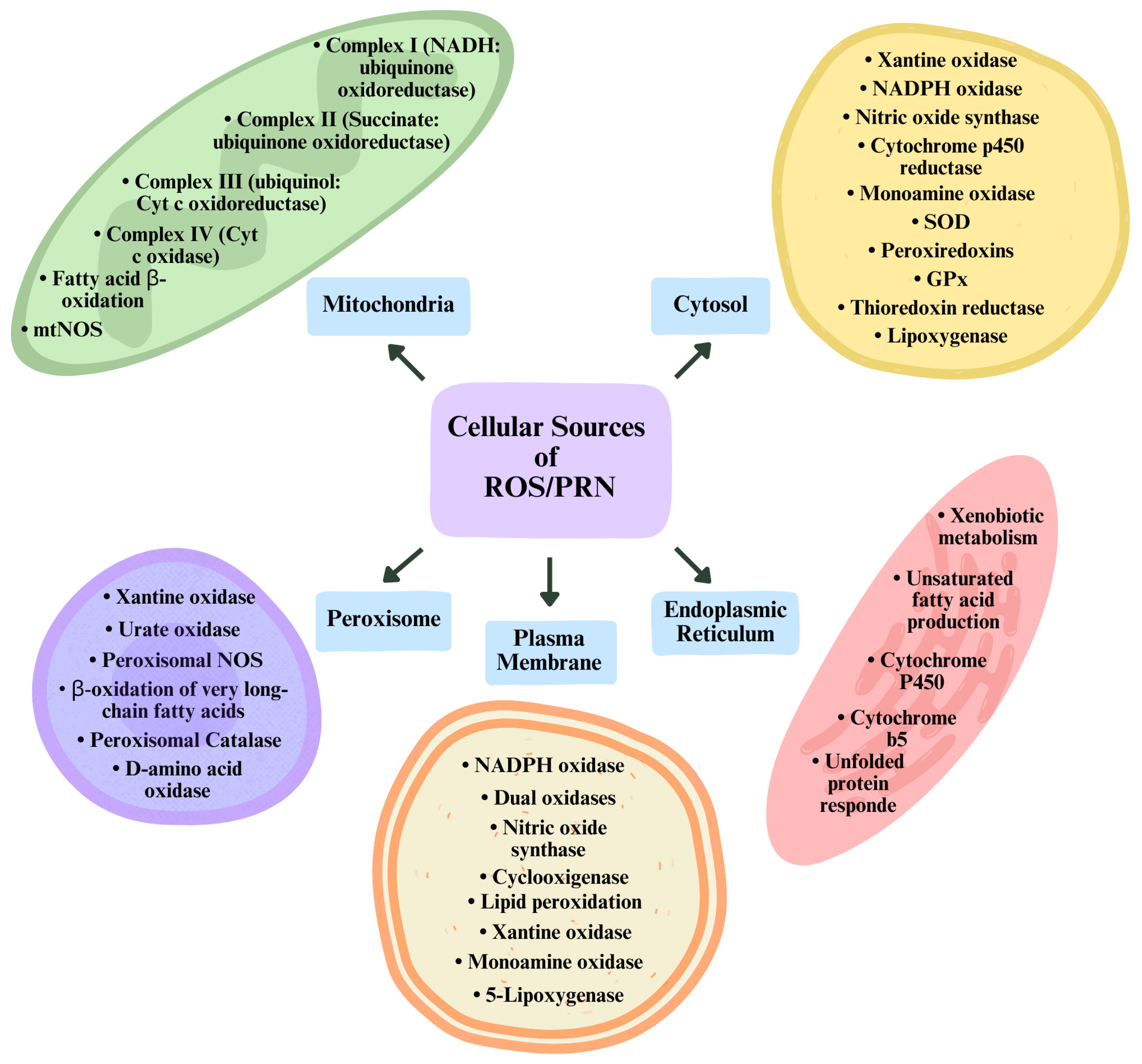
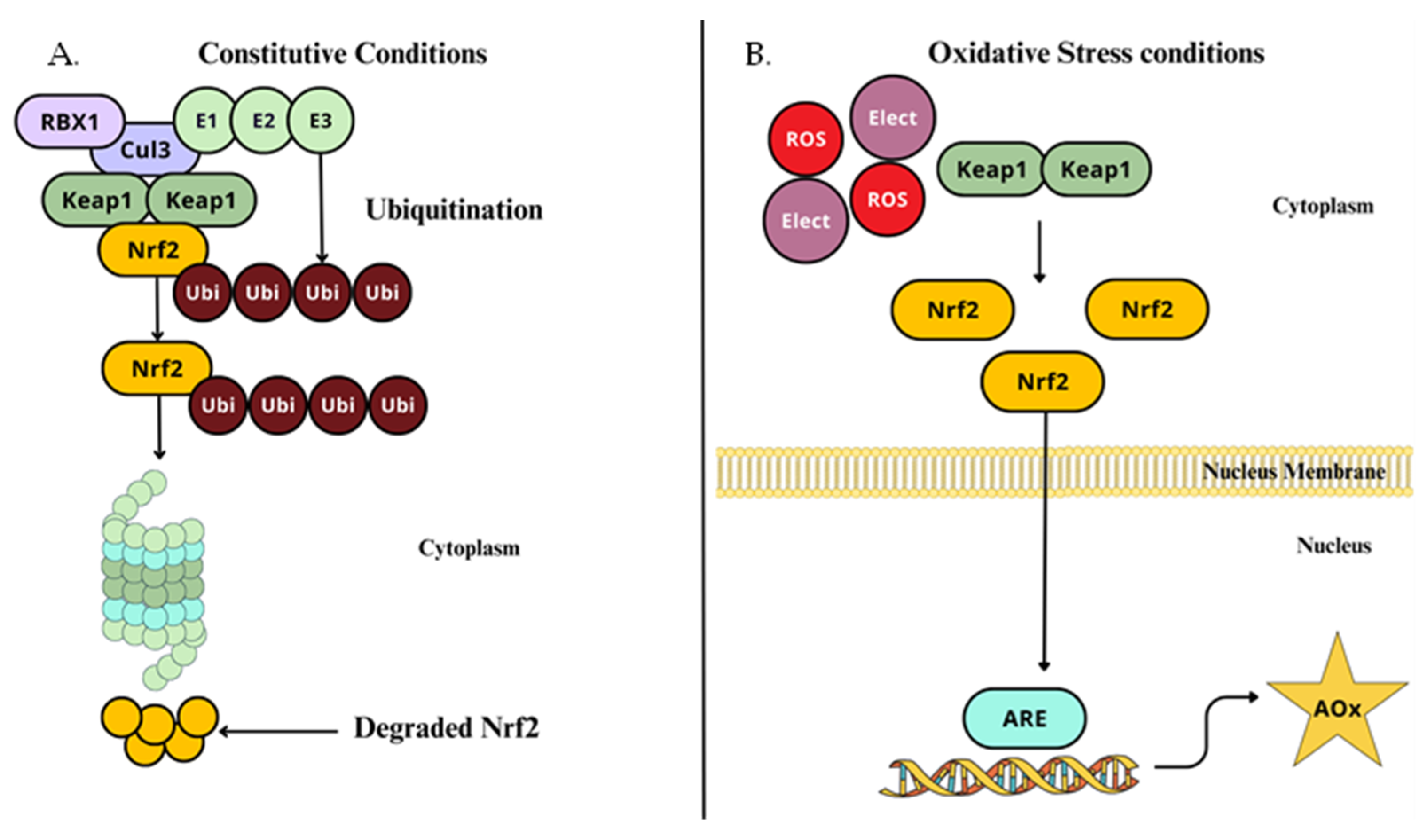
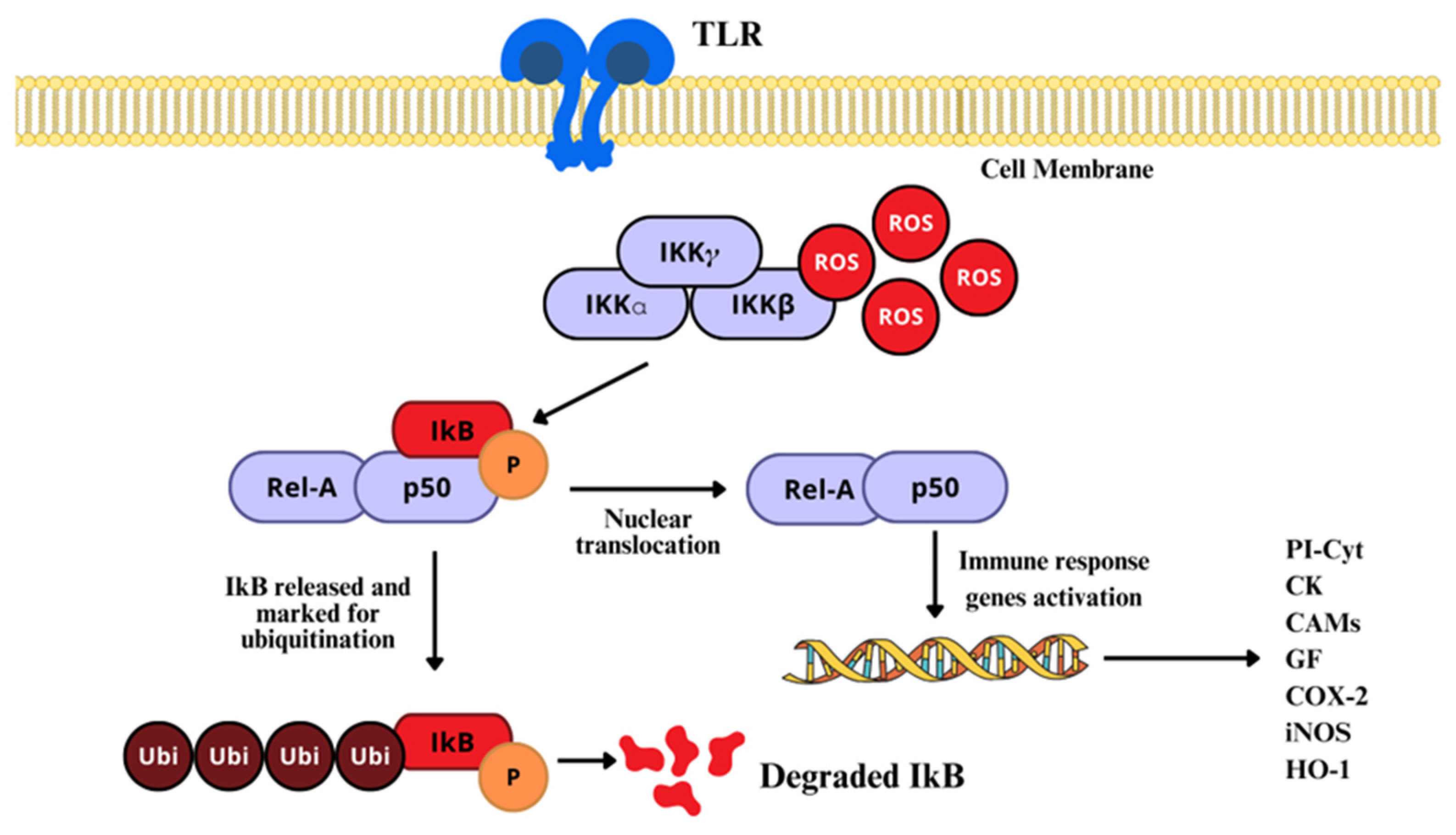

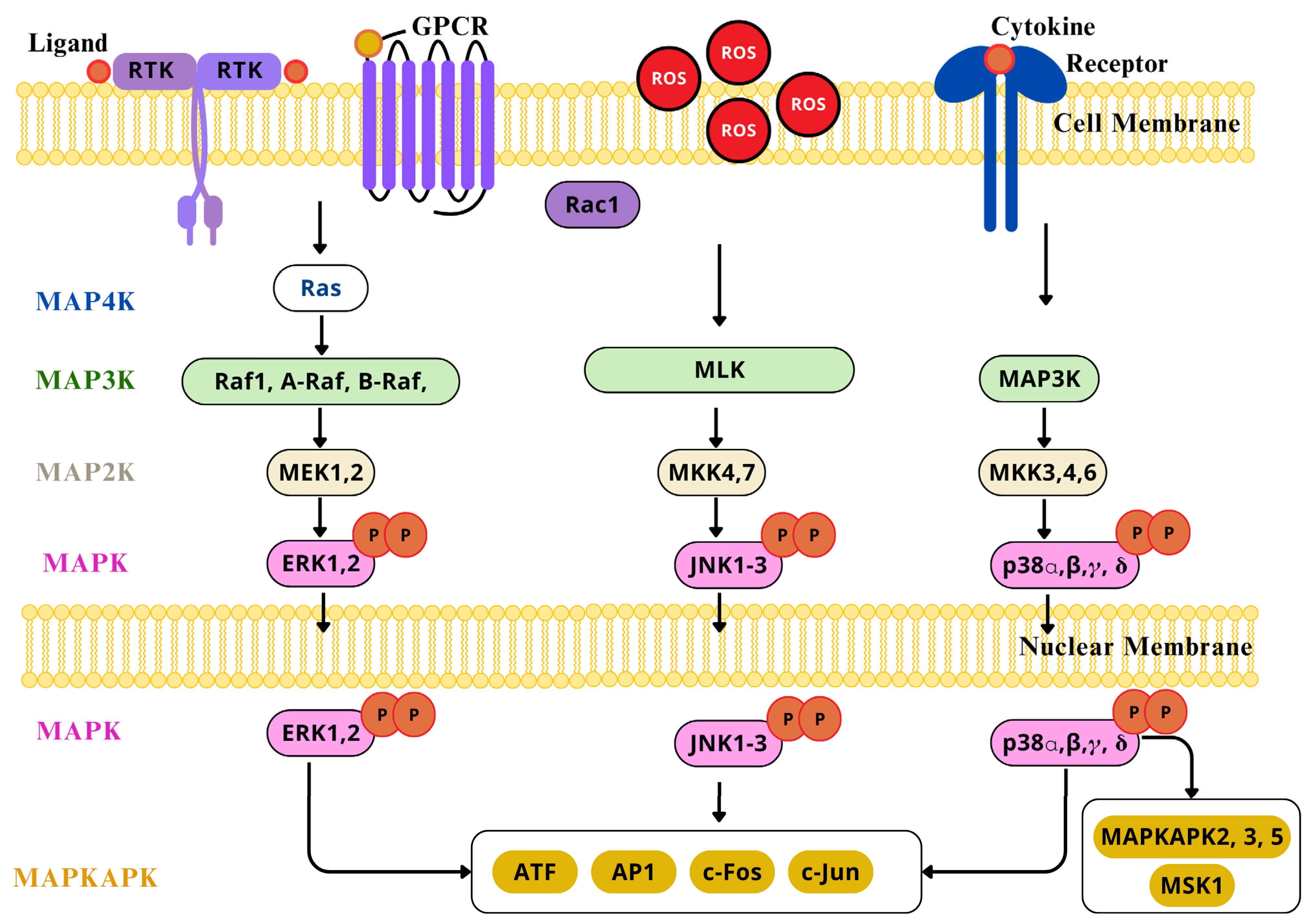
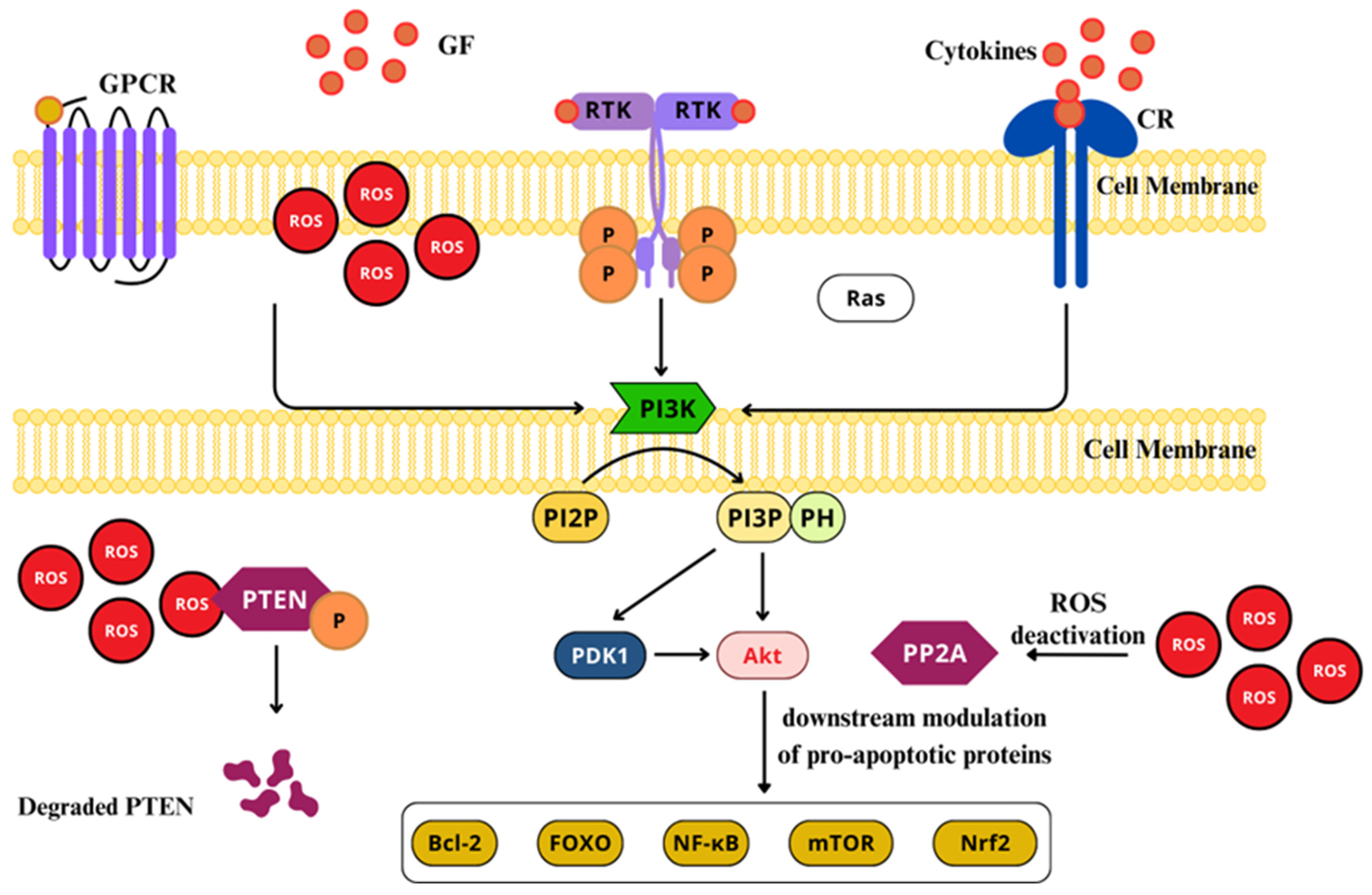
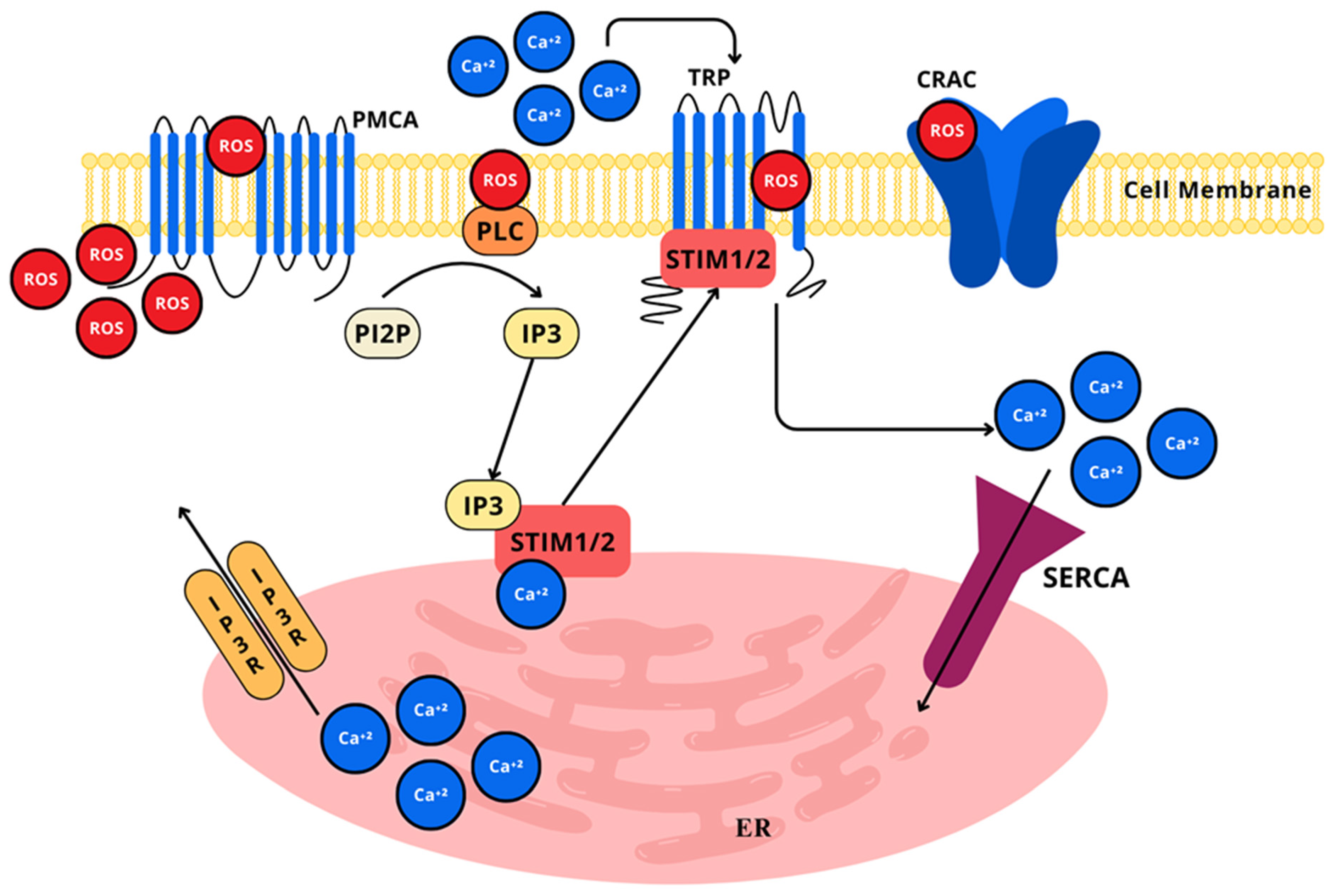
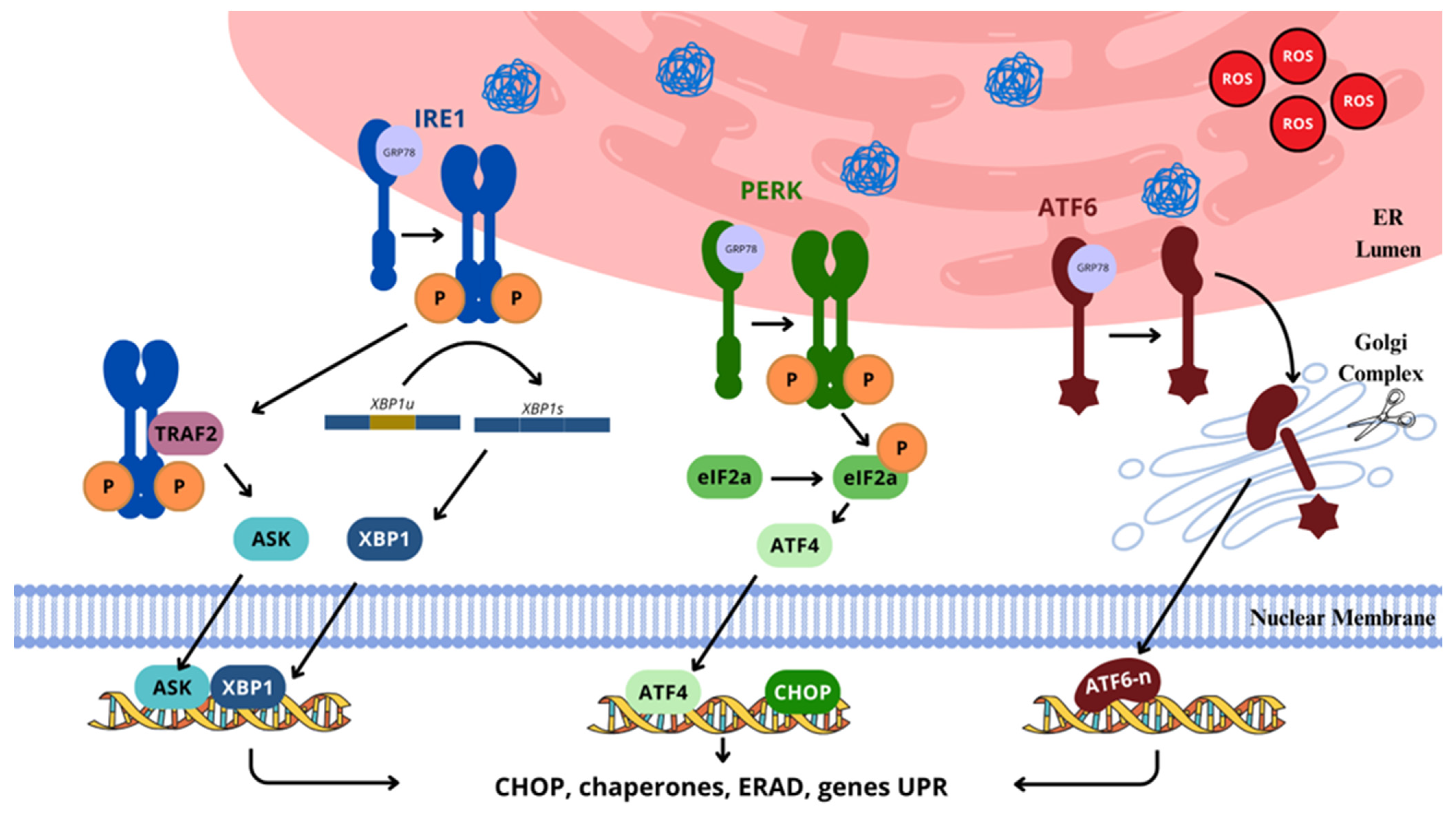
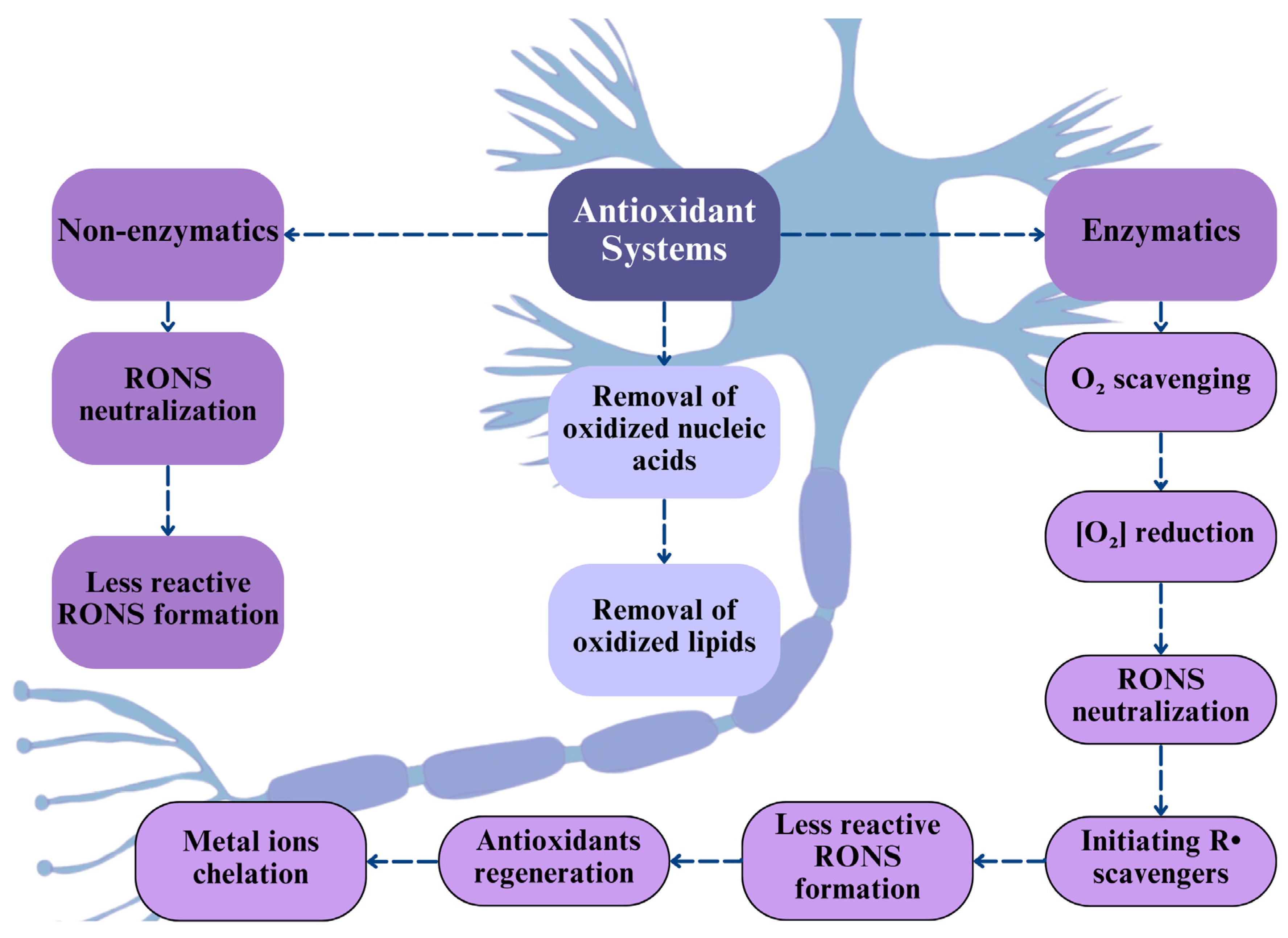
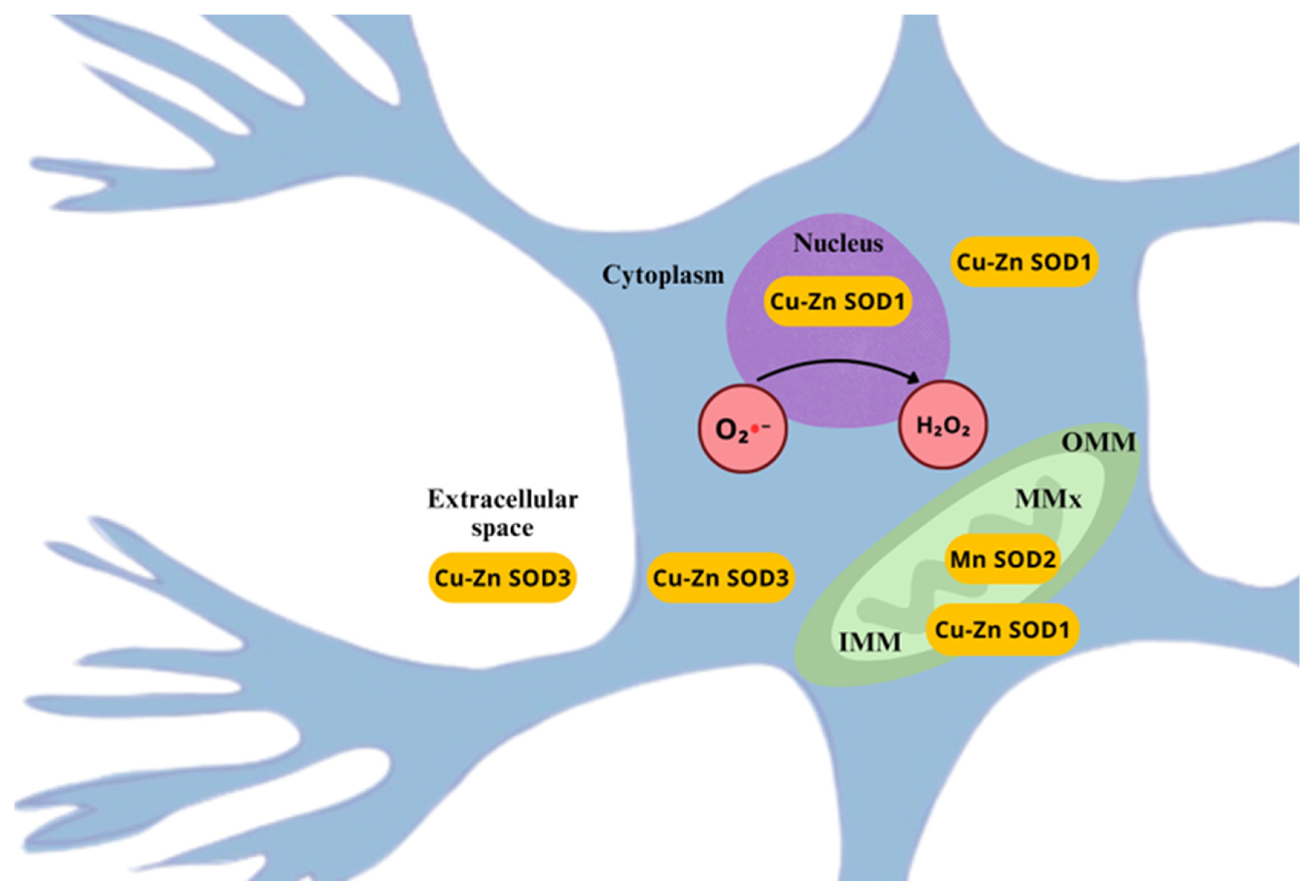
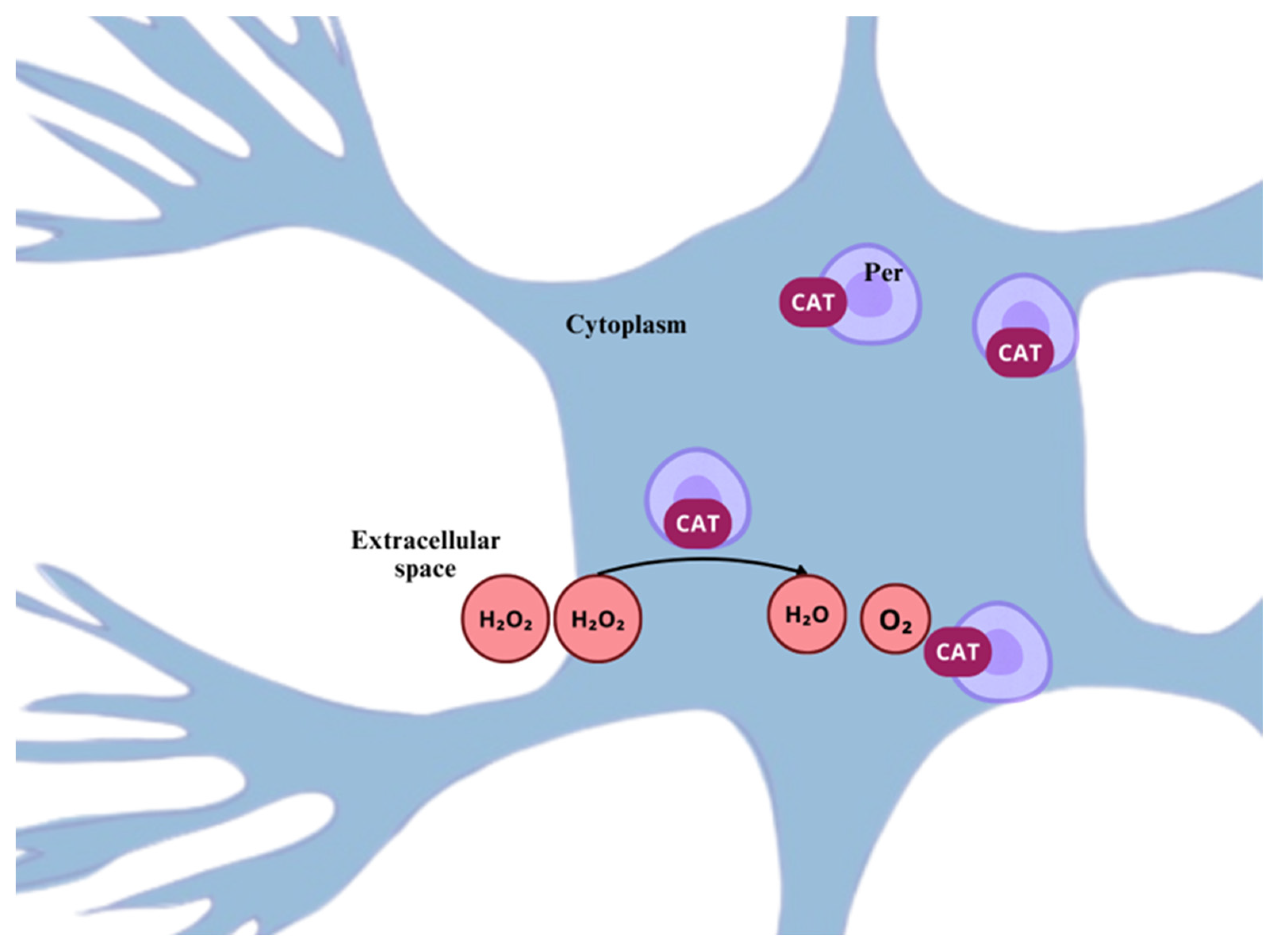
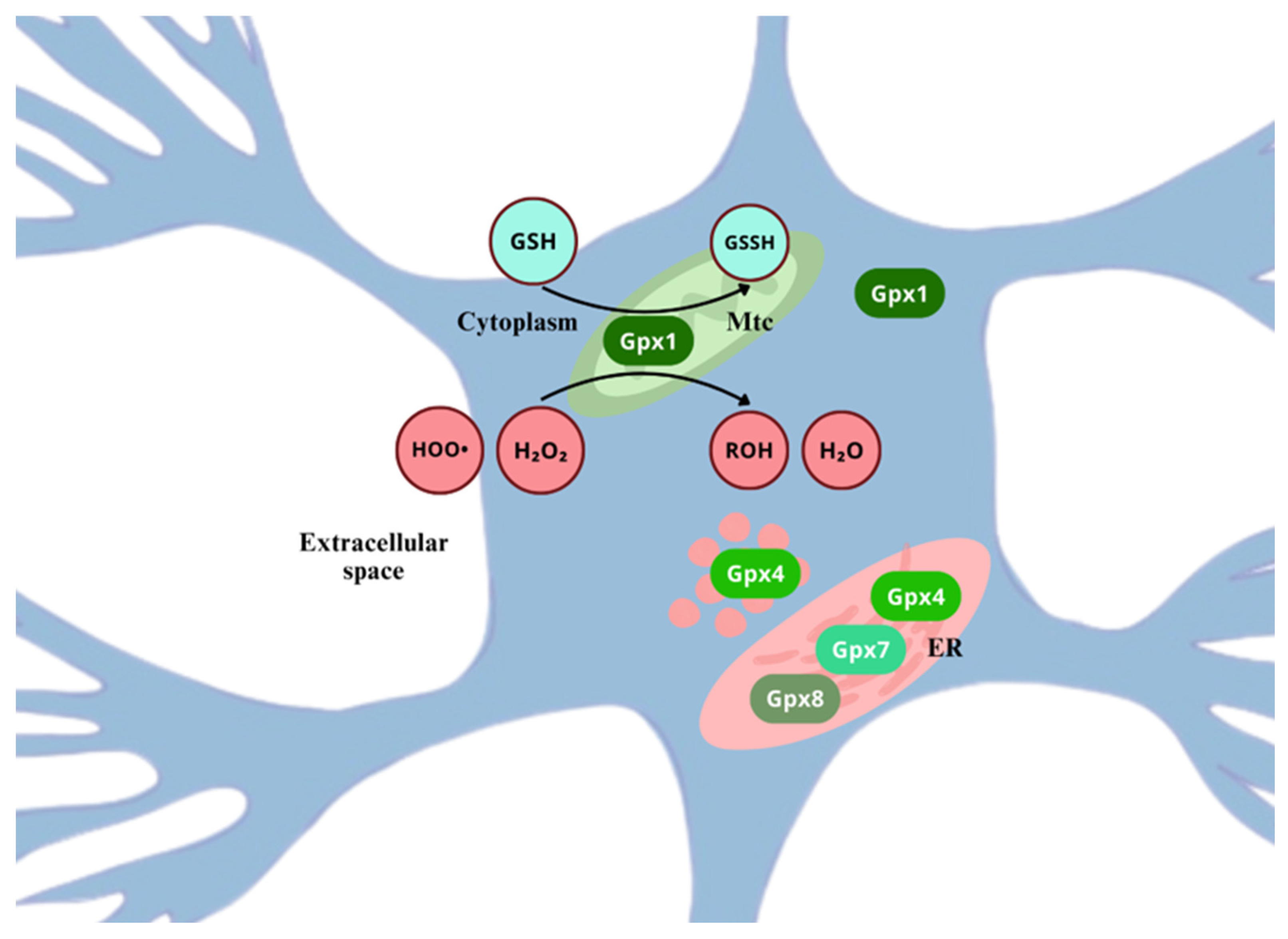
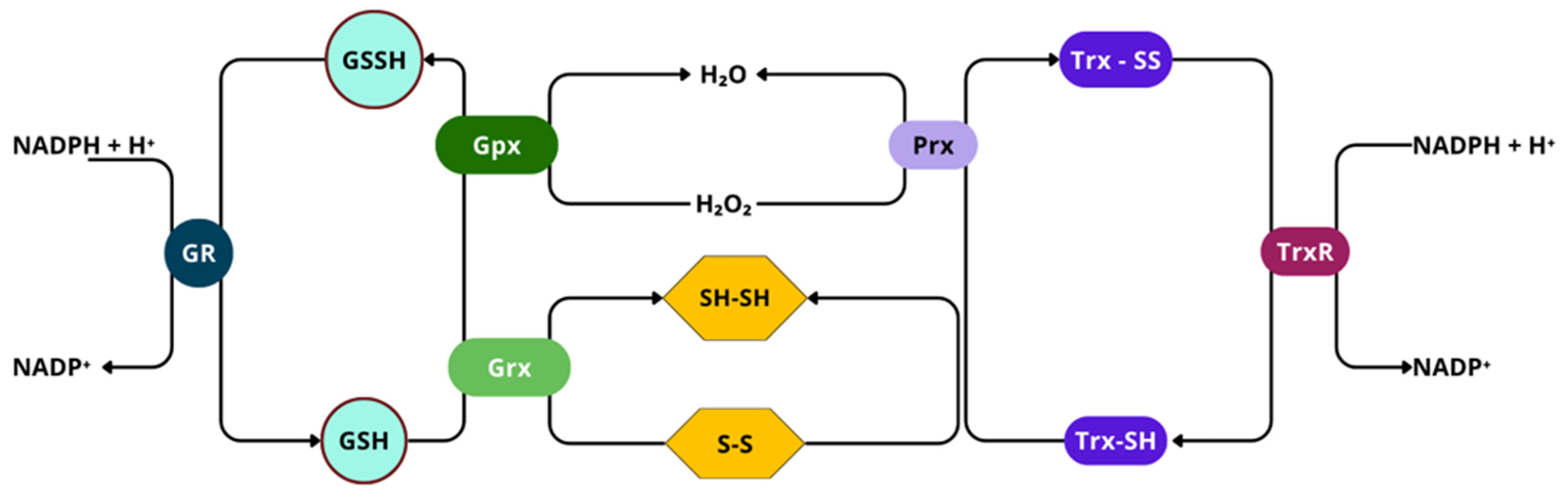


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, Á.J.C.; de Lavor, M.S.L. Nitroxidative Stress, Cell—Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain. Int. J. Mol. Sci. 2025, 26, 2050. https://doi.org/10.3390/ijms26052050
Silva ÁJC, de Lavor MSL. Nitroxidative Stress, Cell—Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain. International Journal of Molecular Sciences. 2025; 26(5):2050. https://doi.org/10.3390/ijms26052050
Chicago/Turabian StyleSilva, Álvaro José Chávez, and Mário Sérgio Lima de Lavor. 2025. "Nitroxidative Stress, Cell—Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain" International Journal of Molecular Sciences 26, no. 5: 2050. https://doi.org/10.3390/ijms26052050
APA StyleSilva, Á. J. C., & de Lavor, M. S. L. (2025). Nitroxidative Stress, Cell—Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain. International Journal of Molecular Sciences, 26(5), 2050. https://doi.org/10.3390/ijms26052050




