Unveiling Novel miRNA–mRNA Interactions and Their Prognostic Roles in Triple-Negative Breast Cancer: Insights into miR-210, miR-183, miR-21, and miR-181b
Abstract
1. Introduction
2. Results
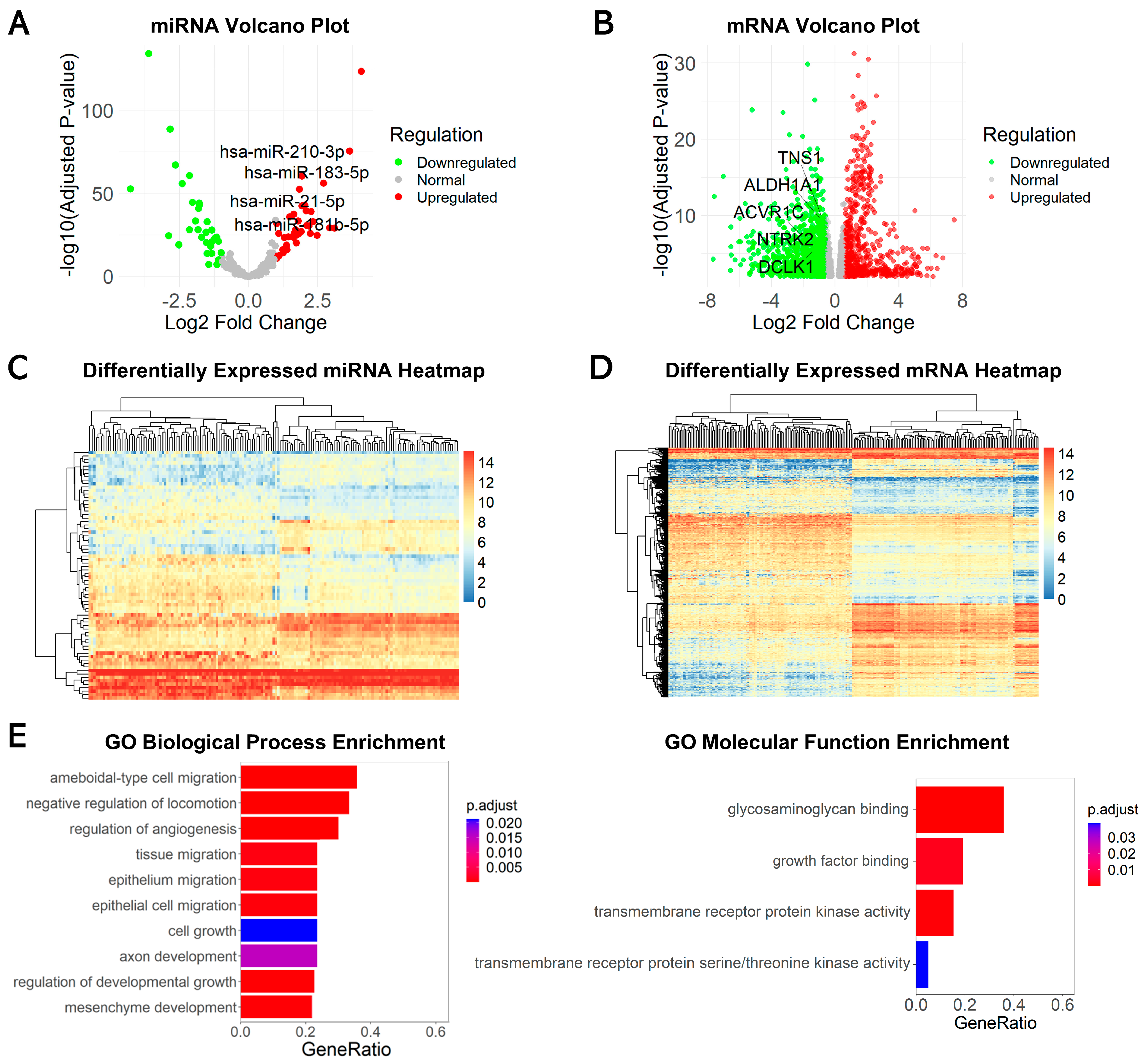
2.1. Identification of Differentially Expressed miRNAs and mRNAs
2.2. Screening of Prognostic-Related Hub miRNAs in TNBC
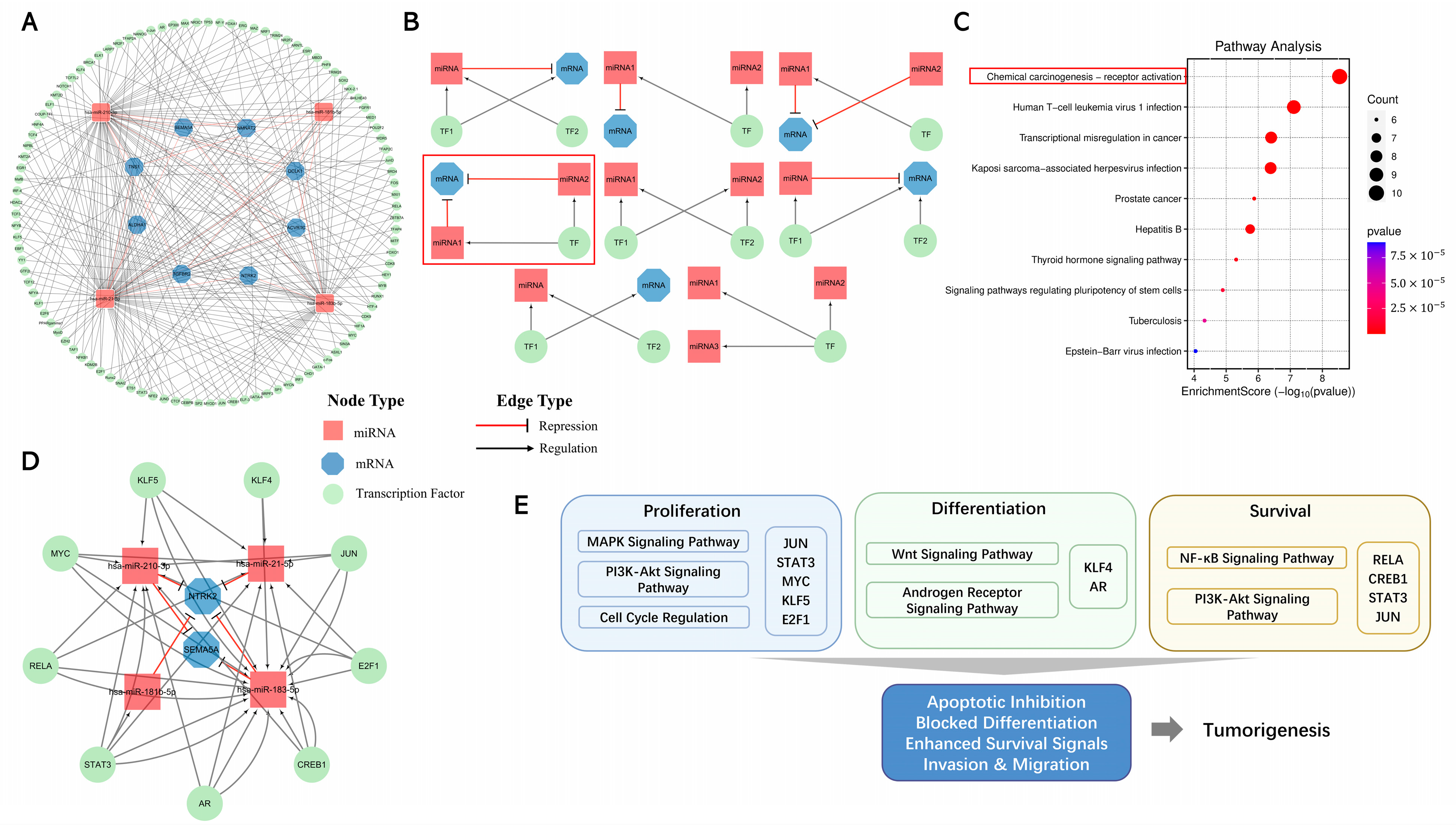
2.3. Identification of Putative miRNA–Target Interaction Network in TNBC
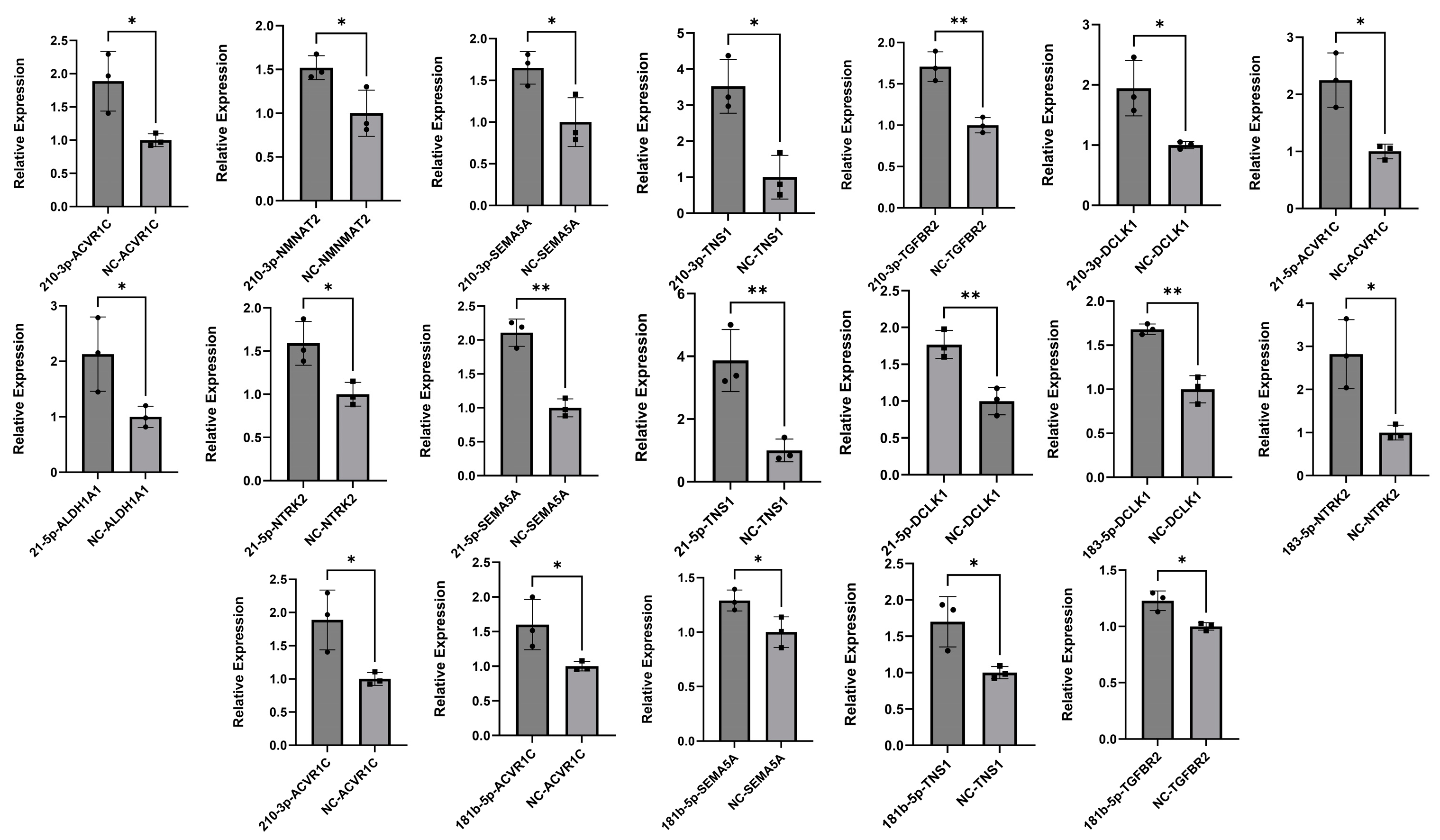
2.4. Validation of MTI Pairs
2.5. miRNA Inhibition Analysis
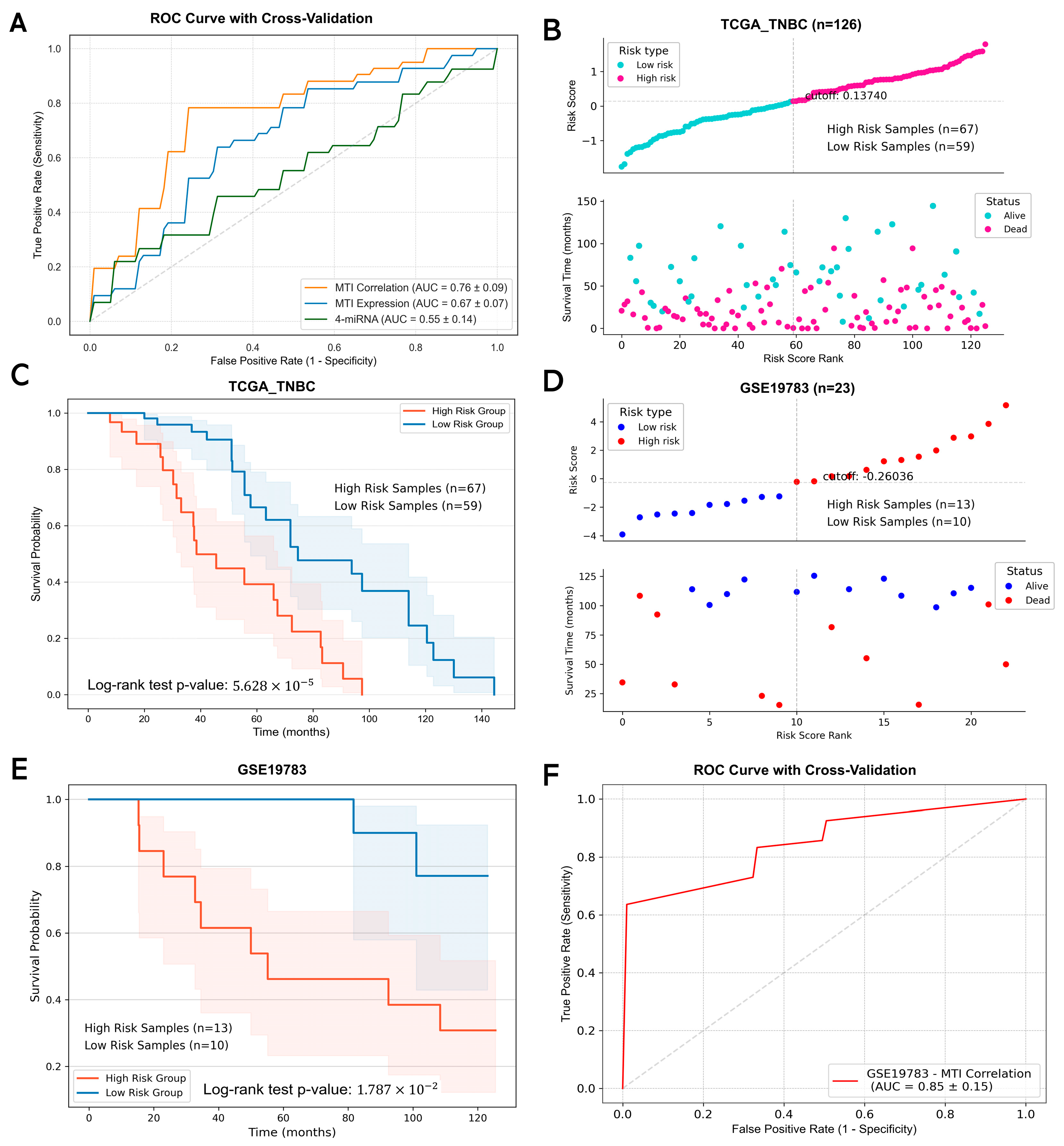
2.6. Identification of Prognosis-Related MTI Risk Model in TNBC
2.7. Interactions of TFs with miRNAs and mRNAs in TNBC Progression
3. Discussion
4. Materials and Methods
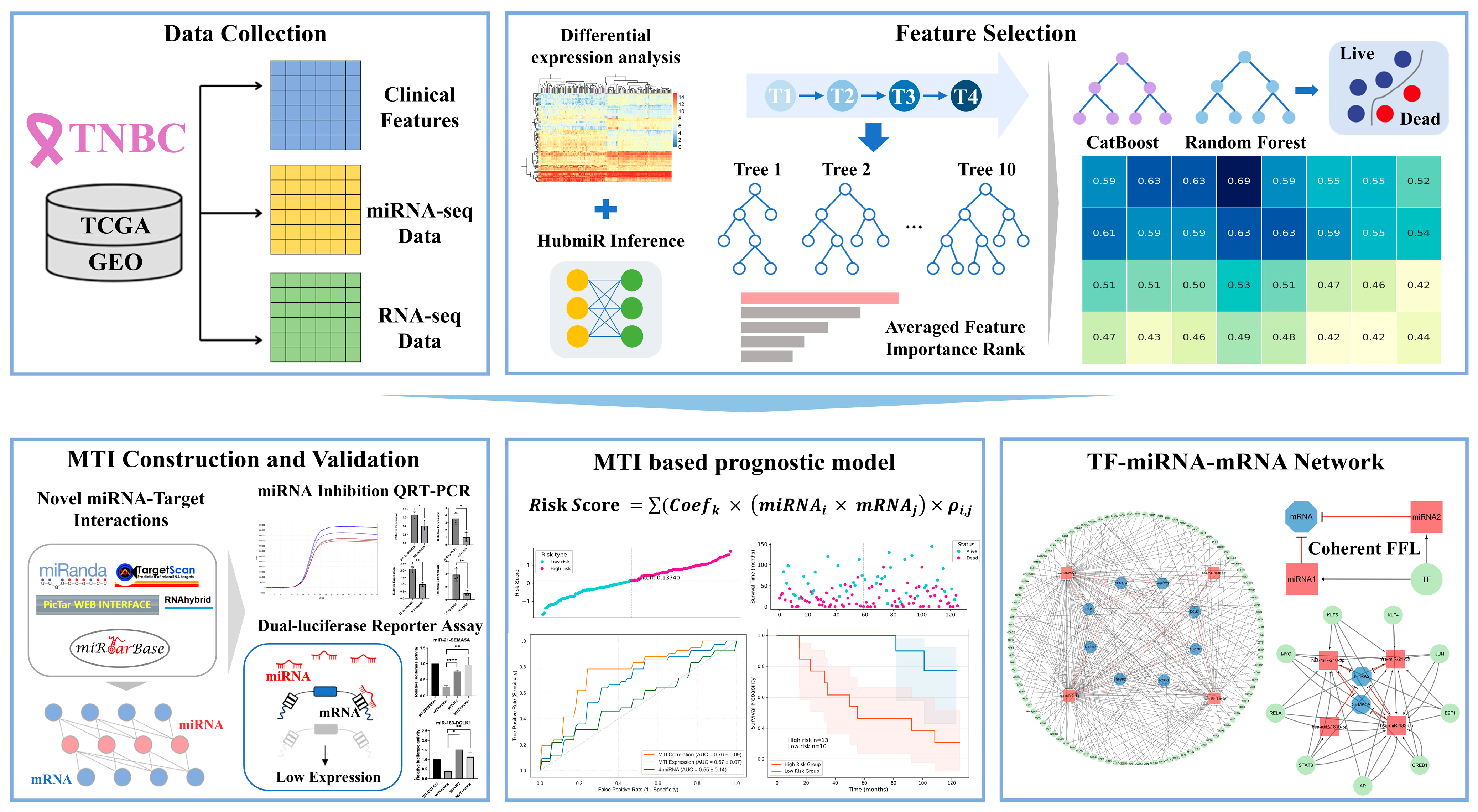
4.1. Data Collection
4.2. Differential Expression Analysis of miRNAs and mRNAs
4.3. Learning miRNA and mRNA Regulatory Correlation via a Neural Network
4.4. Selection of the Prognosis-Related miRNAs
4.5. miRNA Target Gene Identification
4.6. Cell Culture
4.7. Cell Transfection
4.8. Dual-Luciferase Reporter Assay Detection
4.9. Quantitative Real-Time PCR (qRT-PCR)
4.10. Identification of the MTI Prognostic Risk Model
4.11. Upstream TFs Identification
4.12. Detection of Four-Node Regulatory Motifs
4.13. Functional Enrichment Analysis
4.14. Survival Analysis
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zagami, P.; Carey, L.A. Triple negative breast cancer: Pitfalls and progress. NPJ Breast Cancer 2022, 8, 95. [Google Scholar] [CrossRef]
- Liao, L. Inequality in breast cancer: Global statistics from 2022 to 2050. Breast 2024, 79, 103851. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Chang, C.J.; Cheng, J.S. Survival, treatment regimens and medical costs of women newly diagnosed with metastatic triple-negative breast cancer. Sci. Rep. 2022, 12, 729. [Google Scholar] [CrossRef]
- Penault-Llorca, F.; Viale, G. Pathological and molecular diagnosis of triple-negative breast cancer: A clinical perspective. Ann. Oncol. 2012, 23 (Suppl. S6), vi19–vi22. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Wolff, A.C.; Mangu, P.B.; Temin, S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Oncol. Pract. 2010, 6, 195–197. [Google Scholar] [CrossRef]
- Kumar, V.; Gautam, M.; Chaudhary, A.; Chaurasia, B. Impact of three miRNA signature as potential diagnostic marker for triple negative breast cancer patients. Sci. Rep. 2023, 13, 21643. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, D. Role of microRNAs in triple-negative breast cancer and new therapeutic concepts (Review). Oncol. Lett. 2024, 28, 431. [Google Scholar] [CrossRef]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Fuchs Wightman, F.; Giono, L.E.; Fededa, J.P.; de la Mata, M. Target RNAs Strike Back on MicroRNAs. Front. Genet. 2018, 9, 435. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Zhang, F.; Wang, D. Comparative Analysis of microRNA Binding Site Distribution and microRNA-Mediated Gene Expression Repression of Oncogenes and Tumor Suppressor Genes. Genes 2022, 13, 481. [Google Scholar] [CrossRef]
- Lai, X.; Wolkenhauer, O.; Vera, J. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016, 44, 6019–6035. [Google Scholar] [CrossRef]
- Assidicky, R.; Tokat, U.M.; Tarman, I.O.; Saatci, O.; Ersan, P.G.; Raza, U.; Ogul, H.; Riazalhosseini, Y.; Can, T.; Sahin, O. Targeting HIF1-alpha/miR-326/ITGA5 axis potentiates chemotherapy response in triple-negative breast cancer. Breast Cancer Res. Treat. 2022, 193, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Kalecky, K.; Modisette, R.; Pena, S.; Cho, Y.R.; Taube, J. Integrative analysis of breast cancer profiles in TCGA by TNBC subgrouping reveals novel microRNA-specific clusters, including miR-17-92a, distinguishing basal-like 1 and basal-like 2 TNBC subtypes. BMC Cancer 2020, 20, 141. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Perri, P.; Ponzoni, M.; Corrias, M.V.; Ceccherini, I.; Candiani, S.; Bachetti, T. A Focus on Regulatory Networks Linking MicroRNAs, Transcription Factors and Target Genes in Neuroblastoma. Cancers 2021, 13, 5528. [Google Scholar] [CrossRef]
- Jiang, L.; Zhong, M.; Chen, T.; Zhu, X.; Yang, H.; Lv, K. Gene regulation network analysis reveals core genes associated with survival in glioblastoma multiforme. J. Cell Mol. Med. 2020, 24, 10075–10087. [Google Scholar] [CrossRef] [PubMed]
- Beg, A.; Parveen, R.; Fouad, H.; Yahia, M.E.; Hassanein, A.S. Identification of Driver Genes and miRNAs in Ovarian Cancer through an Integrated In-Silico Approach. Biology 2023, 12, 192. [Google Scholar] [CrossRef]
- Cai, X.-X.; Pang, Y.-X.; Cui, S.-D.; Chen, Y.-D.; Lin, Y.-C.-D.; Xu, T.-S.; Chen, Y.-G.; Xu, J.-T.; Wan, J.-T.; Ma, J.-J.; et al. HubmiR: Identifying Hub-miRNAs in Cancers Based on a Neural Network Framework and Applied on Single-cell and Bulk Transcriptome. Nucleic Acids Res. 2025, submitted.
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Conner, S.J.; Guarin, J.R.; Le, T.T.; Fatherree, J.P.; Kelley, C.; Payne, S.L.; Parker, S.R.; Bloomer, H.; Zhang, C.; Salhany, K.; et al. Cell morphology best predicts tumorigenicity and metastasis in vivo across multiple TNBC cell lines of different metastatic potential. Breast Cancer Res. 2024, 26, 43. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Myers, K.A. The role of hypoxia-induced miR-210 in cancer progression. Int. J. Mol. Sci. 2015, 16, 6353–6372. [Google Scholar] [CrossRef] [PubMed]
- Lian, M.; Mortoglou, M.; Uysal-Onganer, P. Impact of Hypoxia-Induced miR-210 on Pancreatic Cancer. Curr. Issues Mol. Biol. 2023, 45, 9778–9792. [Google Scholar] [CrossRef]
- Fang, H.; Xie, J.; Zhang, M.; Zhao, Z.; Wan, Y.; Yao, Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am. J. Transl. Res. 2017, 9, 953. [Google Scholar]
- Neel, J.-C.; Lebrun, J.-J. Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell. Signal. 2013, 25, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Zalles, N.; Distefano, R.; Nigita, G.; Veneziano, D.; Gasparini, P.; Croce, C.M. Synergistic apoptotic effect of miR-183-5p and Polo-Like kinase 1 inhibitor NMS-P937 in breast cancer cells. Cell Death Differ. 2022, 29, 407–419. [Google Scholar] [CrossRef]
- Santana, T.; de Oliveira Passamai, L.; de Miranda, F.S.; Borin, T.F.; Borges, G.F.; Luiz, W.B.; Campos, L.C.G. The Role of miRNAs in the Prognosis of Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Diagnostics 2022, 13, 127. [Google Scholar] [CrossRef]
- Shang, C.; Chen, Q.; Zu, F.; Ren, W. Integrated analysis identified prognostic microRNAs in breast cancer. BMC Cancer 2022, 22, 1170. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qin, S.; Yi, Y.; Gao, H.; Liu, X.; Ma, F.; Guan, M. Delving into the Heterogeneity of Different Breast Cancer Subtypes and the Prognostic Models Utilizing scRNA-Seq and Bulk RNA-Seq. Int. J. Mol. Sci. 2022, 23, 9936. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Li, H.; Zhang, X.; Luo, Z.; Chen, Y.; Zhao, H.; Lv, H.; Zheng, H.; Fu, Z.; Tang, W.; et al. Application of Novel Transcription Factor Machine Learning Model and Targeted Drug Combination Therapy Strategy in Triple Negative Breast Cancer. Int. J. Mol. Sci. 2023, 24, 13497. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Rodriguez-Bautista, R.; Caro-Sanchez, C.H.; Cabrera-Galeana, P.; Alanis-Funes, G.J.; Gutierrez-Millan, E.; Avila-Rios, S.; Matias-Florentino, M.; Reyes-Teran, G.; Diaz-Chavez, J.; Villarreal-Garza, C.; et al. Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior. Cancers 2021, 13, 6256. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, W.; Flamant, S.; Rasko, J.E. Predicting microRNA targets and functions: Traps for the unwary. Nat. Methods 2009, 6, 397–398. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Gennarino, V.A.; Sardiello, M.; Avellino, R.; Meola, N.; Maselli, V.; Anand, S.; Cutillo, L.; Ballabio, A.; Banfi, S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009, 19, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Clément, T.; Salone, V.; Rederstorff, M. Dual luciferase gene reporter assays to study miRNA function. In Small Non-Coding RNAs: Methods and Protocols; Humana Press: New York, NY, USA, 2015; pp. 187–198. [Google Scholar]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K. TRANSFAC® and its module TRANSCompel®: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wernicke, S.; Rasche, F. FANMOD: A tool for fast network motif detection. Bioinformatics 2006, 22, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Survminer: Drawing Survival Curves Using ‘ggplot2’, R Package Version 0.3; 2017; Volume 1. Available online: https://rpkgs.datanovia.com/survminer/index.html (accessed on 12 August 2024).
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable continuous data distribution system for determining survival in Kaplan-Meier estimator. Comput. Methods Programs Biomed. 2019, 177, 133–139. [Google Scholar] [CrossRef] [PubMed]



| Diseases | miRNAs | Target Genes |
|---|---|---|
| Triple-negative breast cancer | hsa-miR-542-3p | BIRC5 |
| hsa-miR-31-5p | SATB2 | |
| hsa-miR-218-5p | SOST, SFRP2 | |
| hsa-miR-498 | BRCA1 | |
| hsa-miR-455-3p | EI24 | |
| hsa-miR-27a-3p | GSK3B | |
| hsa-miR-155-3p | NLRP3 | |
| hsa-miR-496 | Del-1 | |
| hsa-miR-96-3p | BRCA1 | |
| hsa-miR-10b-3p | BRCA1 | |
| hsa-miR-199a-5p | TGFB2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Cai, X.; Huang, J.; Huang, H.-Y.; Wang, Y.-F.; Ji, X.; Huang, Y.; Ni, J.; Zuo, H.; Li, S.; et al. Unveiling Novel miRNA–mRNA Interactions and Their Prognostic Roles in Triple-Negative Breast Cancer: Insights into miR-210, miR-183, miR-21, and miR-181b. Int. J. Mol. Sci. 2025, 26, 1916. https://doi.org/10.3390/ijms26051916
Xu J, Cai X, Huang J, Huang H-Y, Wang Y-F, Ji X, Huang Y, Ni J, Zuo H, Li S, et al. Unveiling Novel miRNA–mRNA Interactions and Their Prognostic Roles in Triple-Negative Breast Cancer: Insights into miR-210, miR-183, miR-21, and miR-181b. International Journal of Molecular Sciences. 2025; 26(5):1916. https://doi.org/10.3390/ijms26051916
Chicago/Turabian StyleXu, Jiatong, Xiaoxuan Cai, Junyang Huang, Hsi-Yuan Huang, Yong-Fei Wang, Xiang Ji, Yuxin Huang, Jie Ni, Huali Zuo, Shangfu Li, and et al. 2025. "Unveiling Novel miRNA–mRNA Interactions and Their Prognostic Roles in Triple-Negative Breast Cancer: Insights into miR-210, miR-183, miR-21, and miR-181b" International Journal of Molecular Sciences 26, no. 5: 1916. https://doi.org/10.3390/ijms26051916
APA StyleXu, J., Cai, X., Huang, J., Huang, H.-Y., Wang, Y.-F., Ji, X., Huang, Y., Ni, J., Zuo, H., Li, S., Lin, Y.-C.-D., & Huang, H.-D. (2025). Unveiling Novel miRNA–mRNA Interactions and Their Prognostic Roles in Triple-Negative Breast Cancer: Insights into miR-210, miR-183, miR-21, and miR-181b. International Journal of Molecular Sciences, 26(5), 1916. https://doi.org/10.3390/ijms26051916






