An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival
Abstract
1. Introduction
2. Results
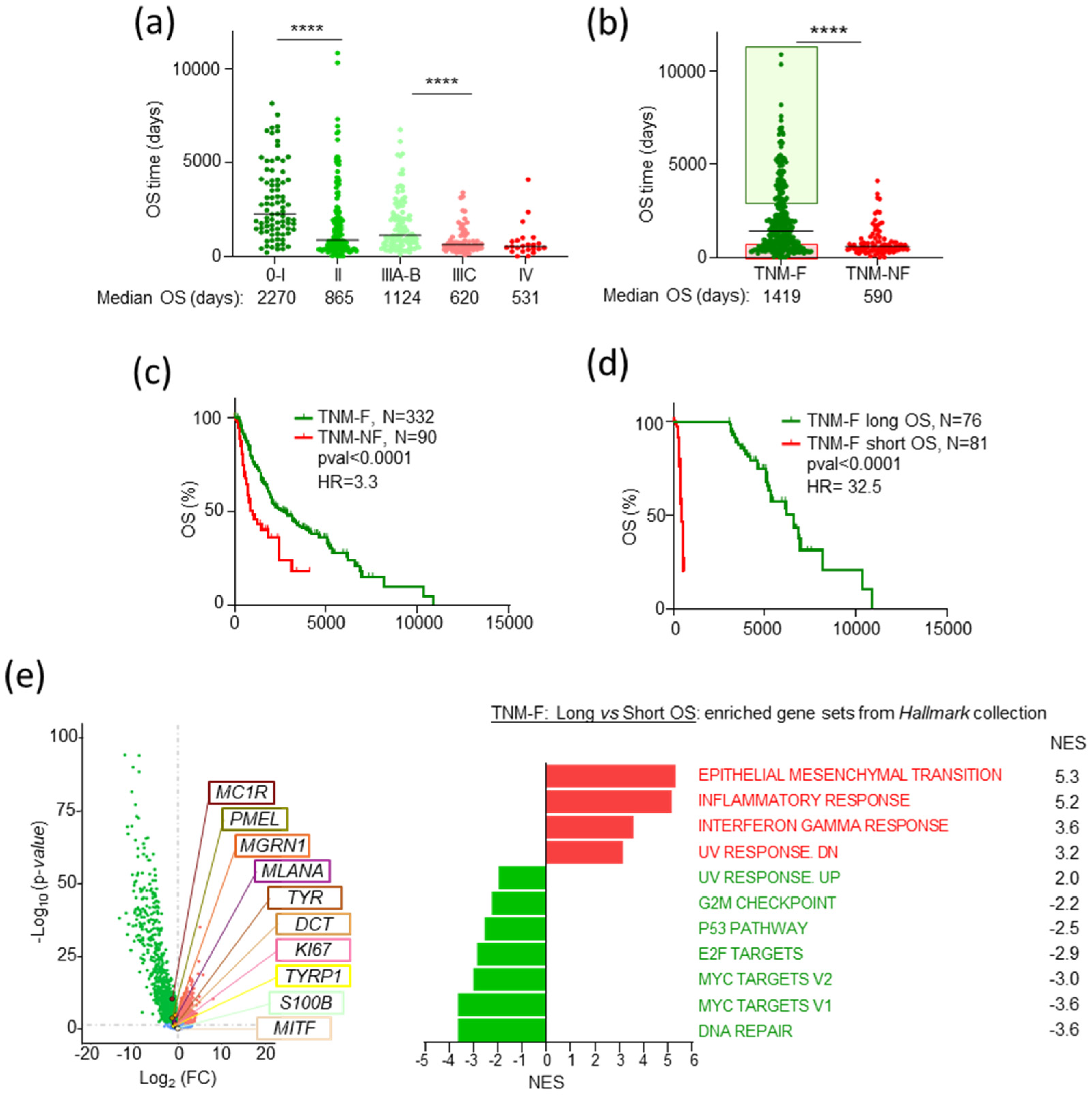
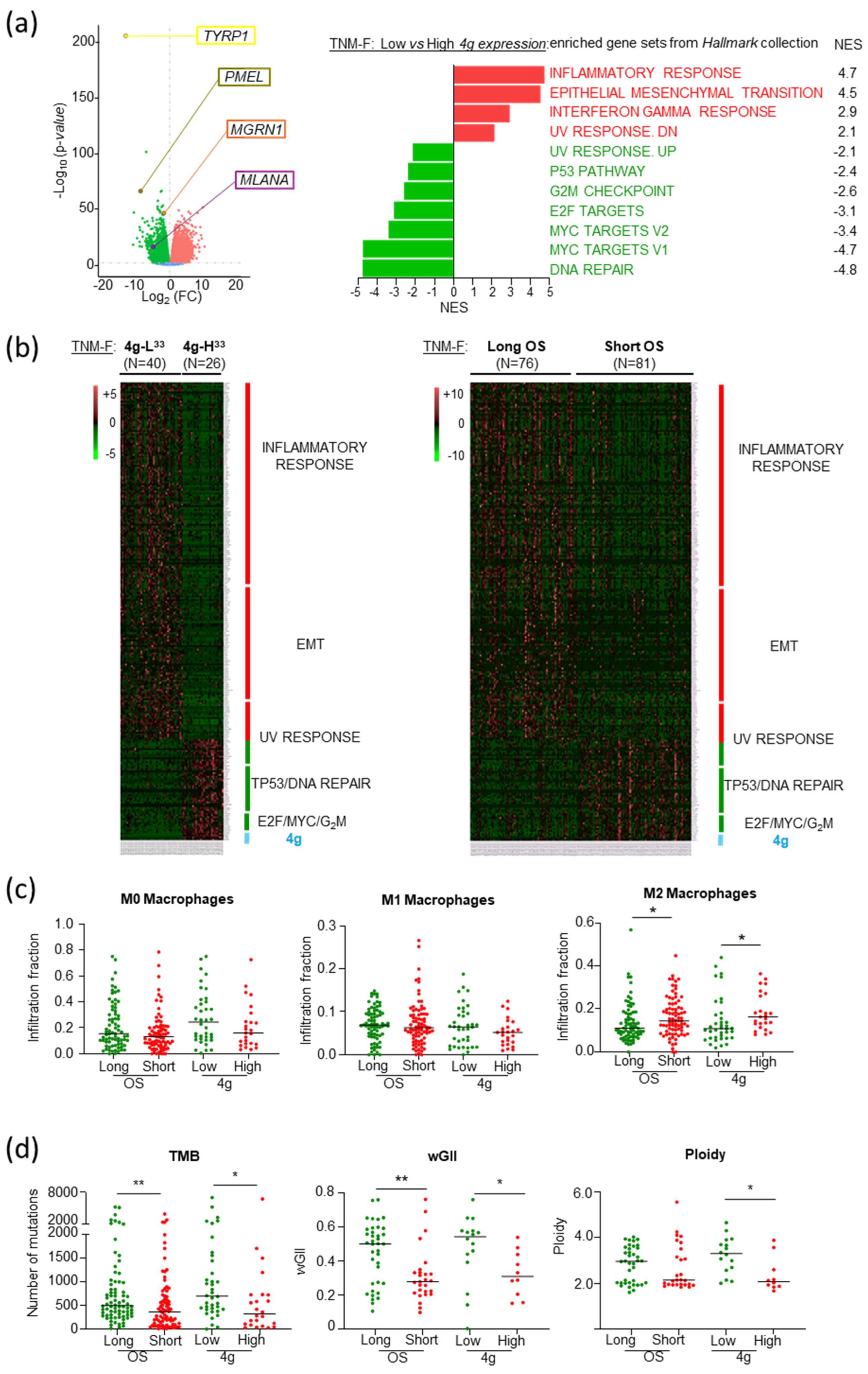
2.1. TNM-F Patients with Short OS Exhibit a Specific Transcriptomic Profile
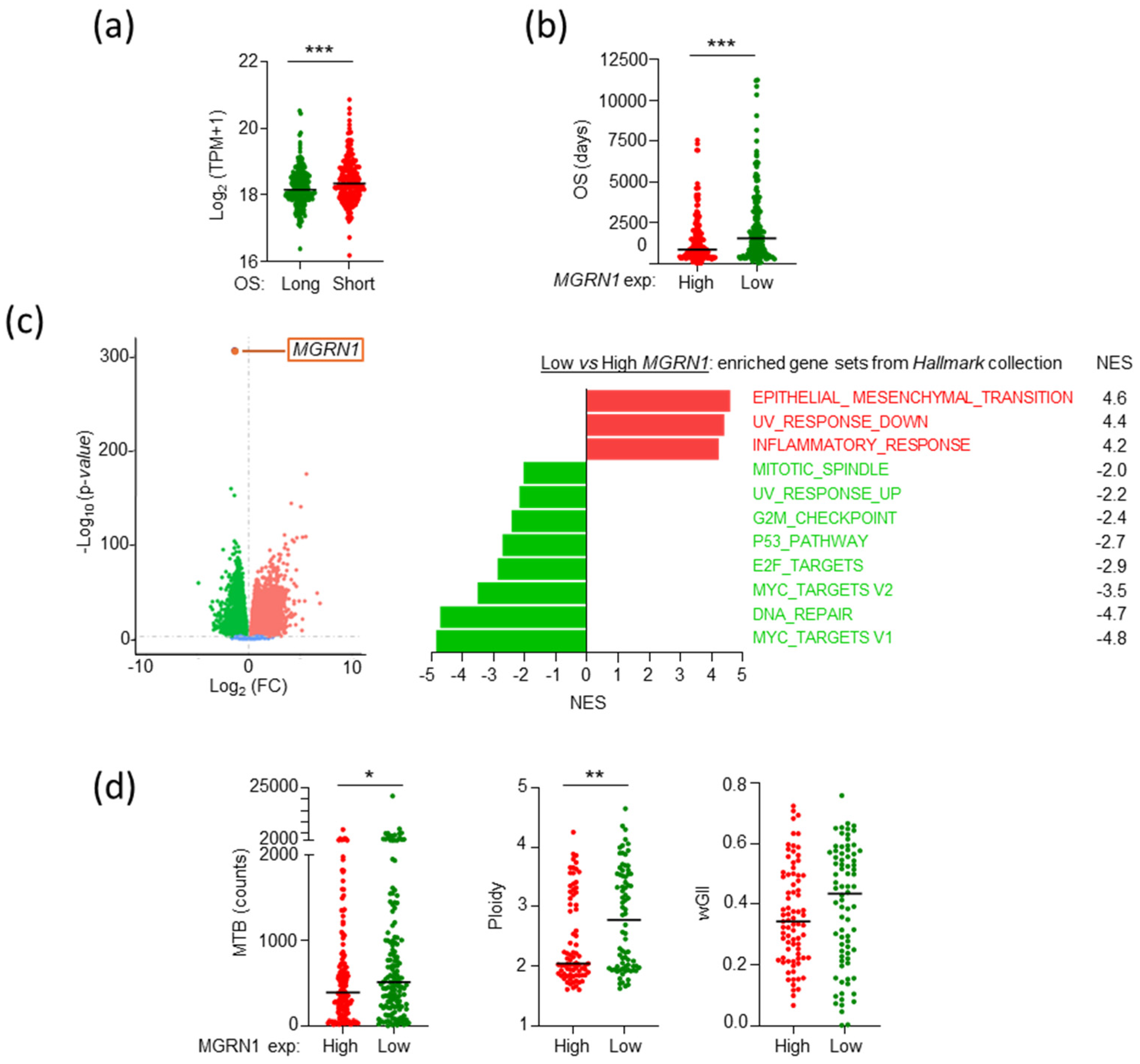
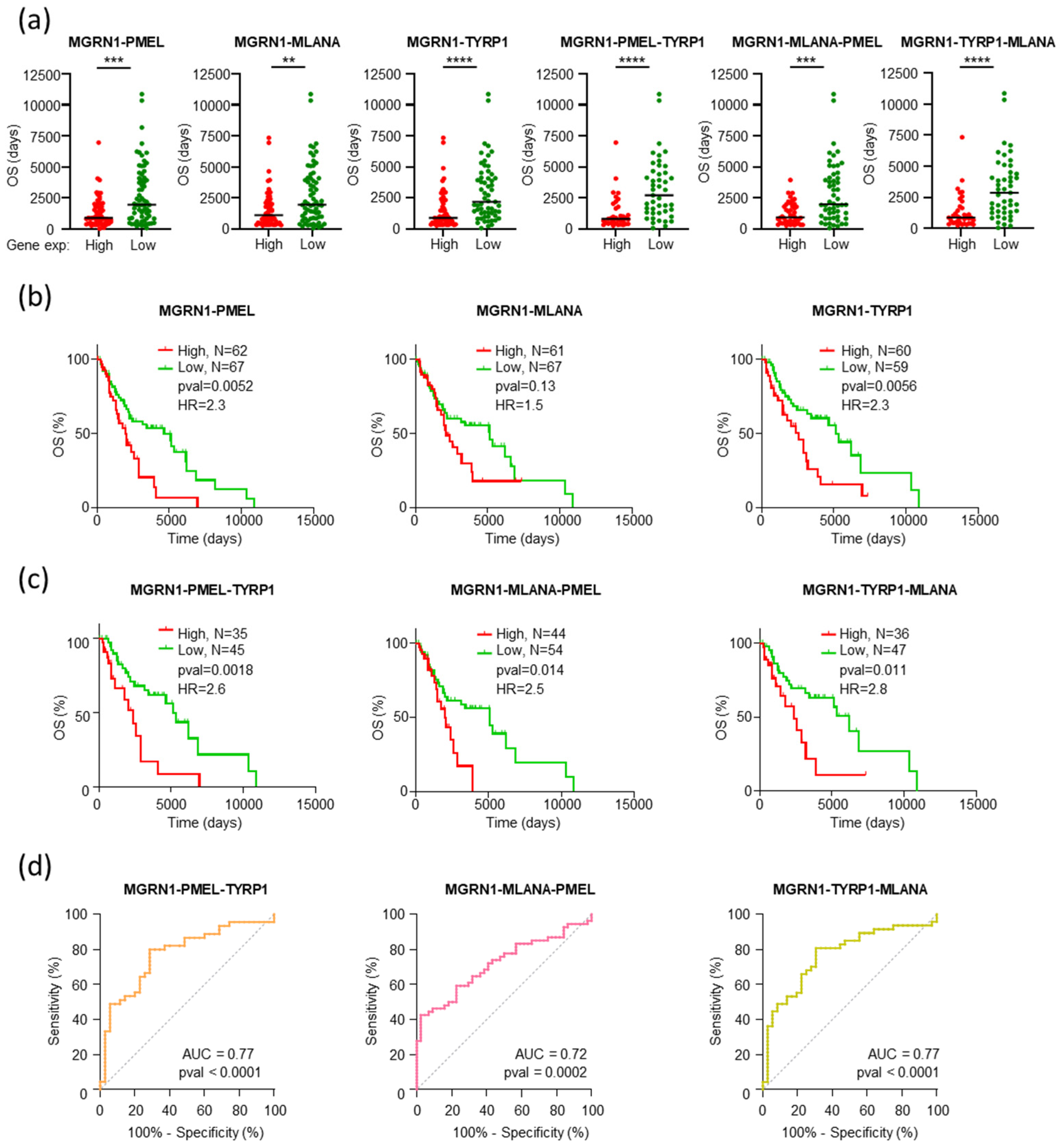
2.2. The Level of Expression of MGRN1 and Genes Involved in Melanocyte Differentiation Predicts Patient Survival Better than Driver Genes
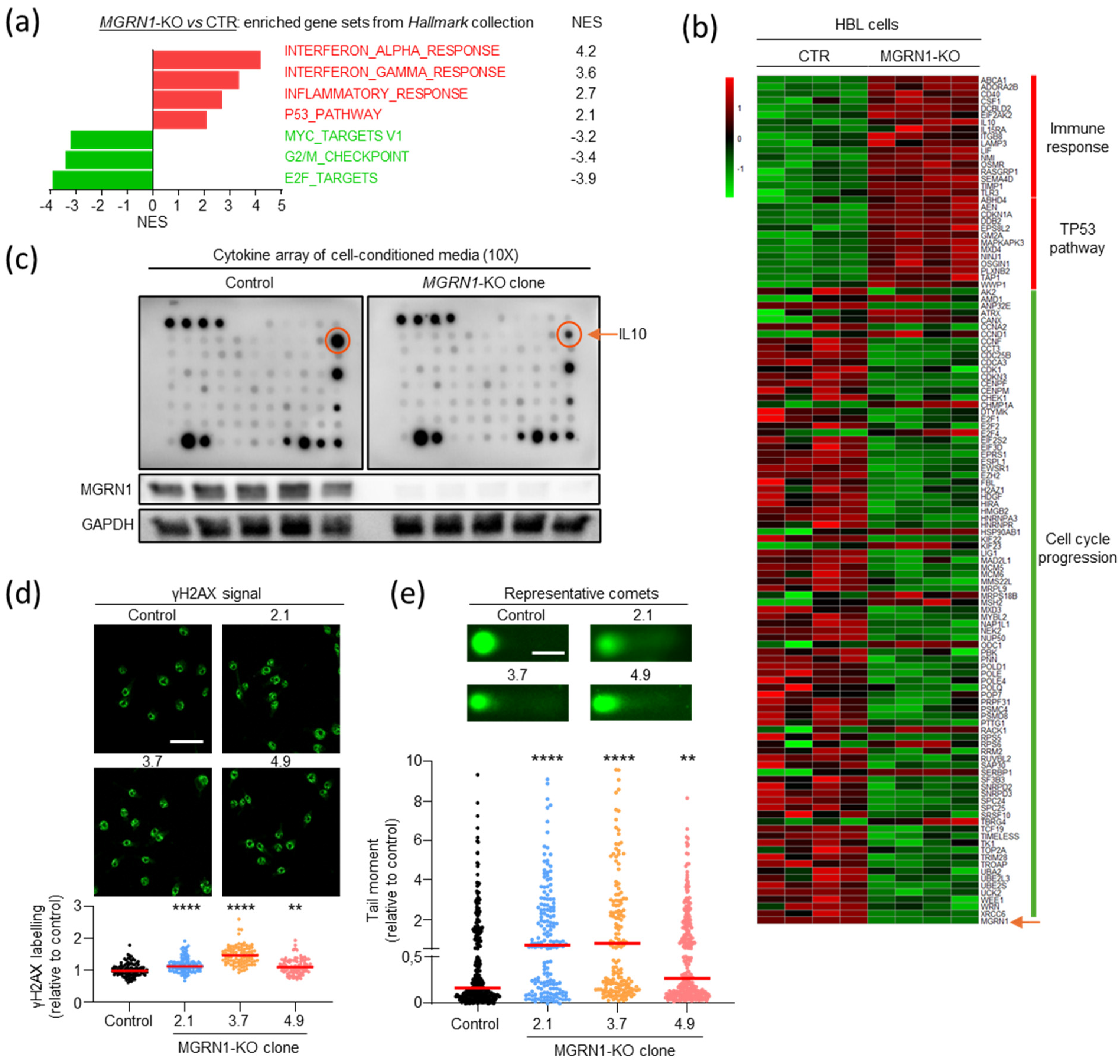
2.3. Comparable Gene Expression Patterns in Patients with Longer OS and Low MGRN1, PMEL, MLANA, or TYRP1 Expression
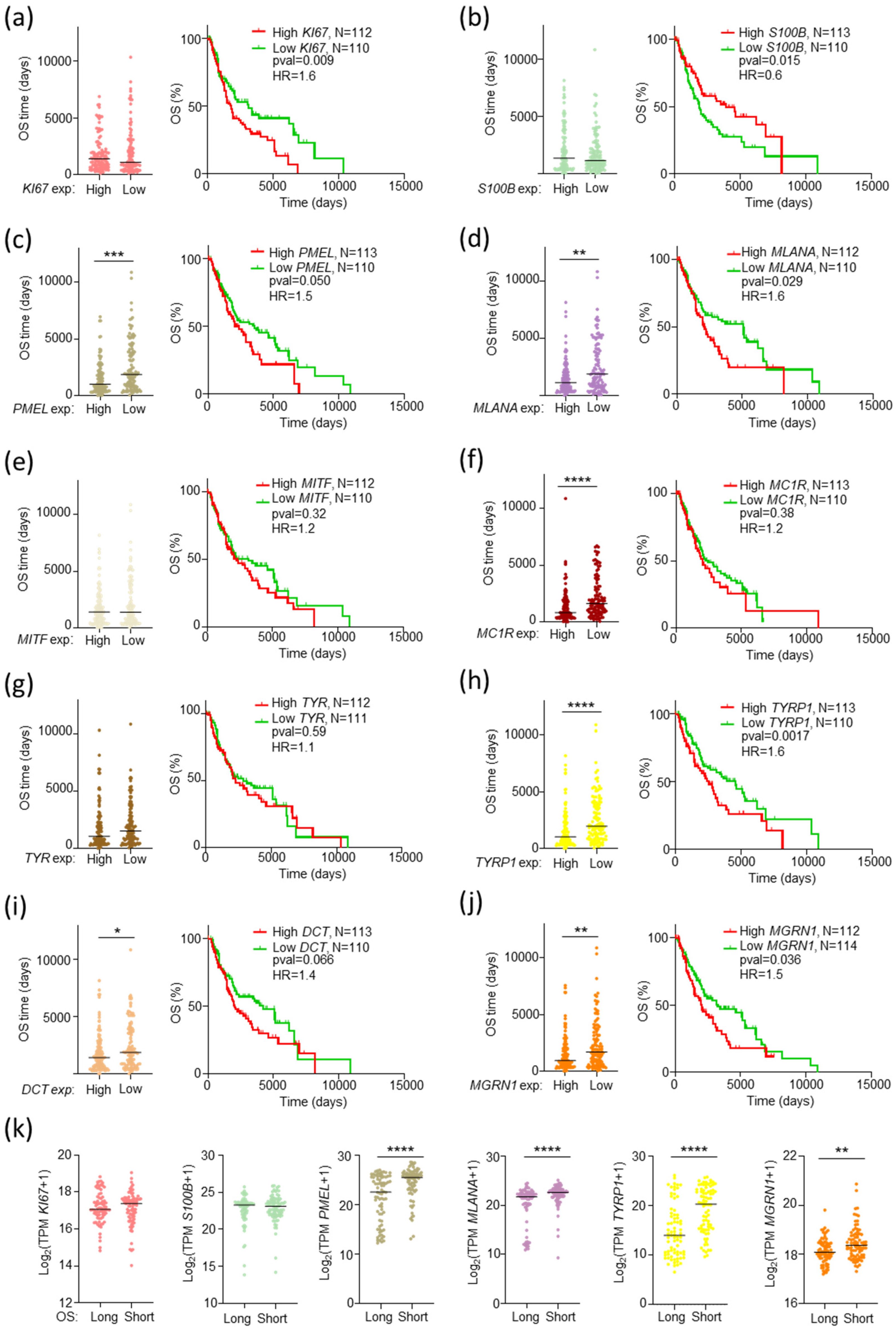
2.4. An MGRN1-Based Expression Panel Complements TNM-Based Prediction of Outcome
2.5. Increased Genomic Stability and Infiltration of M2 Macrophages as Molecular Features Associated with Poor Prognosis in TNM-F Patients
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Generation of MGRN1-KO Cells
4.3. RNAseq Analysis
4.4. Differential Expression and Enrichment Analysis
4.5. Data Processing
4.6. Immunoblotting
4.7. Analysis of Secreted Cytokines in Cell Culture Media
4.8. Immunofluorescence, Confocal Microscopy, and Image Quantification
4.9. Comet Assays
4.10. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| 4g | 4-gene signature |
| AJCC | American Joint Committee on Cancer |
| EMT | Epithelial-Mesenchimal Transition |
| GSEA | Gene Set Enrichment Analysis |
| HR | Hazard Ratio |
| MGRN1 | Mahogunin Ring Finger-1 |
| MM | Cutaneous Melanoma |
| OS | Overall Survival |
| SKCM | Cutaneous Melanoma |
| TCGA | The Cancer Genome Atlas |
| TNM | Tumor-Node-Metastasis |
| TNM-F | Favorable, low-medium-grade TNM |
| TNM-NF | Unfavorable, high-grade TNM |
| WT | Wild type |
References
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous Melanoma. Lancet 2023, 402, 485–502. [Google Scholar] [CrossRef]
- Arnold, M.; de Vries, E.; Whiteman, D.C.; Jemal, A.; Bray, F.; Parkin, D.M.; Soerjomataram, I. Global Burden of Cutaneous Melanoma Attributable to Ultraviolet Radiation in 2012. Int. J. Cancer 2018, 143, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Marquard, A.M.; Eklund, A.C.; Joshi, T.; Krzystanek, M.; Favero, F.; Wang, Z.C.; Richardson, A.L.; Silver, D.P.; Szallasi, Z.; Birkbak, N.J. Pan-Cancer Analysis of Genomic Scar Signatures Associated with Homologous Recombination Deficiency Suggests Novel Indications for Existing Cancer Drugs. Biomark. Res. 2015, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Sosinsky, A.; Ambrose, J.; Cross, W.; Turnbull, C.; Henderson, S.; Jones, L.; Hamblin, A.; Arumugam, P.; Chan, G.; Chubb, D.; et al. Insights for Precision Oncology from the Integration of Genomic and Clinical Data of 13,880 Tumors from the 100,000 Genomes Cancer Programme. Nat. Med. 2024, 30, 279–289. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Giampietri, C.; Scatozza, F.; Crecca, E.; Vigiano Benedetti, V.; Natali, P.G.; Facchiano, A. Analysis of Gene Expression Levels and Their Impact on Survival in 31 Cancer-Types Patients Identifies Novel Prognostic Markers and Suggests Unexplored Immunotherapy Treatment Options in a Wide Range of Malignancies. J. Transl. Med. 2022, 20, 467. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Li, M.; Gao, X.; Wang, X. Identification of Tumor Mutation Burden-Associated Molecular and Clinical Features in Cancer by Analyzing Multi-Omics Data. Front. Immunol. 2023, 14, 1090838. [Google Scholar] [CrossRef] [PubMed]
- Koelblinger, P.; Emberger, M.; Drach, M.; Cheng, P.F.; Lang, R.; Levesque, M.P.; Bauer, J.W.; Dummer, R. Increased Tumour Cell PD-L1 Expression, Macrophage and Dendritic Cell Infiltration Characterise the Tumour Microenvironment of Ulcerated Primary Melanomas. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Geukes Foppen, M.H.; Goldinger, S.M.; et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.B.; Arachchi, H.; Arora, A.; Auman, J.T.; Ayala, B.; et al. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.-M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-Genome Landscapes of Major Melanoma Subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Krauthammer, M.; Kong, Y.; Bacchiocchi, A.; Evans, P.; Pornputtapong, N.; Wu, C.; McCusker, J.P.; Ma, S.; Cheng, E.; Straub, R.; et al. Exome Sequencing Identifies Recurrent Mutations in NF1 and RASopathy Genes in Sun-Exposed Melanomas. Nat. Genet. 2015, 47, 996–1002. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.-P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A Landscape of Driver Mutations in Melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Tímár, J.; Ladányi, A. Molecular Pathology of Skin Melanoma: Epidemiology, Differential Diagnostics, Prognosis and Therapy Prediction. Int. J. Mol. Sci. 2022, 23, 5384. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, D.; Leininger, J.; Hamby, C.; Safai, B. Diagnostic and Prognostic Biomarkers in Melanoma. J. Clin. Aesthet. Dermatol. 2014, 7, 13–24. [Google Scholar]
- Zhang, S.; Chen, K.; Liu, H.; Jing, C.; Zhang, X.; Qu, C.; Yu, S. PMEL as a Prognostic Biomarker and Negatively Associated With Immune Infiltration in Skin Cutaneous Melanoma (SKCM). J. Immunother. 2021, 44, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.C.; Sheltzer, J.M. Genome-Wide Identification and Analysis of Prognostic Features in Human Cancers. Cell Rep. 2022, 38, 110569. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma Staging: Evidence-based Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Pavlick, A.C.; Ariyan, C.E.; Buchbinder, E.I.; Davar, D.; Gibney, G.T.; Hamid, O.; Hieken, T.J.; Izar, B.; Johnson, D.B.; Kulkarni, R.P.; et al. Society for Immunotherapy of Cancer (SITC) Clinical Practice Guideline on Immunotherapy for the Treatment of Melanoma, Version 3.0. J. Immunother. Cancer 2023, 11, e006947. [Google Scholar] [CrossRef]
- Kahlon, N.; Doddi, S.; Yousif, R.; Najib, S.; Sheikh, T.; Abuhelwa, Z.; Burmeister, C.; Hamouda, D.M. Melanoma Treatments and Mortality Rate Trends in the US, 1975 to 2019. JAMA Netw. Open 2022, 5, e2245269. [Google Scholar] [CrossRef] [PubMed]
- Gimotty, P.A.; Elder, D.E.; Fraker, D.L.; Botbyl, J.; Sellers, K.; Elenitsas, R.; Ming, M.E.; Schuchter, L.; Spitz, F.R.; Czerniecki, B.J.; et al. Identification of High-Risk Patients Among Those Diagnosed With Thin Cutaneous Melanomas. J. Clin. Oncol. 2007, 25, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Cao, M.; Puertolas, T.; Manzano, J.L.; Maldonado, C.; Yelamos, O.; Berciano-Guerrero, M.Á.; Cerezuela, P.; Martin-Liberal, J.; Muñoz-Couselo, E.; Espinosa, E.; et al. Access to Melanoma Drugs in Spain: A Cross-Sectional Survey. Clin. Transl. Oncol. 2024, 26, 2572–2583. [Google Scholar] [CrossRef]
- Arenberger, P.; Fialova, A.; Gkalpakiotis, S.; Pavlikova, A.; Puzanov, I.; Arenbergerova, M. Melanoma Antigens Are Biomarkers for Ipilimumab Response. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Váraljai, R.; Elouali, S.; Lueong, S.S.; Wistuba-Hamprecht, K.; Seremet, T.; Siveke, J.T.; Becker, J.C.; Sucker, A.; Paschen, A.; Horn, P.A.; et al. The Predictive and Prognostic Significance of Cell-free DNA Concentration in Melanoma. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 387–395. [Google Scholar] [CrossRef]
- Dixon, A.J.; Steinman, H.K.; Nirenberg, A.; Zouboulis, C.C.; Sladden, M.; Popescu, C.; Anderson, S.; Longo, C.; Thomas, J.M. BAUSSS Biomarker Improves Melanoma Survival Risk Assessment. J. Eur. Acad. Dermatol. Venereol. 2024. [Google Scholar] [CrossRef]
- Asato, M.A.; Neto, F.A.M.; de Toledo Moraes, M.P.; Ocanha-Xavier, J.P.; Takita, L.C.; Fung, M.A.; Marques, M.E.A.; Xavier-Júnior, J.C.C. The Utility of PRAME and Ki-67 as Prognostic Markers for Cutaneous Melanoma. Am. J. Dermatopathol. 2025, 47, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, B.E.G.; Bracken, M.B.; Rimm, D.L. Tissue Biomarkers for Prognosis in Cutaneous Melanoma: A Systematic Review and Meta-Analysis. JNCI J. Natl. Cancer Inst. 2009, 101, 452–474. [Google Scholar] [CrossRef]
- Winnepenninckx, V.; Lazar, V.; Michiels, S.; Dessen, P.; Stas, M.; Alonso, S.R.; Avril, M.-F.; Ortiz Romero, P.L.; Robert, T.; Balacescu, O.; et al. Gene Expression Profiling of Primary Cutaneous Melanoma and Clinical Outcome. JNCI J. Natl. Cancer Inst. 2006, 98, 472–482. [Google Scholar] [CrossRef]
- Garg, M.; Couturier, D.-L.; Nsengimana, J.; Fonseca, N.A.; Wongchenko, M.; Yan, Y.; Lauss, M.; Jönsson, G.B.; Newton-Bishop, J.; Parkinson, C.; et al. Tumour Gene Expression Signature in Primary Melanoma Predicts Long-Term Outcomes. Nat. Commun. 2021, 12, 1137. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Oliva, A.B.; Olivares, C.; Jiménez-Cervantes, C.; García-Borrón, J.C. Mahogunin Ring Finger-1 (MGRN1) E3 Ubiquitin Ligase Inhibits Signaling from Melanocortin Receptor by Competition with Galphas. J. Biol. Chem. 2009, 284, 31714–31725. [Google Scholar] [CrossRef]
- Herraiz, C.; Garcia-Borron, J.C.; Jiménez-Cervantes, C.; Olivares, C. MC1R Signaling. Intracellular Partners and Pathophysiological Implications. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 2448–2461. [Google Scholar] [CrossRef]
- Reissmann, M.; Ludwig, A. Pleiotropic Effects of Coat Colour-Associated Mutations in Humans, Mice and Other Mammals. Semin. Cell Dev. Biol. 2013, 24, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Sirés-Campos, J.; Lambertos, A.; Delevoye, C.; Raposo, G.; Bennett, D.C.; Sviderskaya, E.; Jiménez-Cervantes, C.; Olivares, C.; García-Borrón, J.C. Mahogunin Ring Finger 1 Regulates Pigmentation by Controlling the PH of Melanosomes in Melanocytes and Melanoma Cells. Cell. Mol. Life Sci. 2022, 79, 47. [Google Scholar] [CrossRef] [PubMed]
- Abrisqueta, M.; Cerdido, S.; Sánchez-Beltrán, J.; Martínez-Vicente, I.; Herraiz, C.; Lambertos, A.; Olivares, C.; Sevilla, A.; Alonso, S.; Boyano, M.D.; et al. MGRN1 as a Phenotypic Determinant of Human Melanoma Cells and a Potential Biomarker. Life 2022, 12, 1118. [Google Scholar] [CrossRef]
- Cerdido, S.; Abrisqueta, M.; Sánchez-Beltrán, J.; Lambertos, A.; Castejón-Griñán, M.; Muñoz, C.; Olivares, C.; García-Borrón, J.C.; Jiménez-Cervantes, C.; Herraiz, C. MGRN1 Depletion Promotes Intercellular Adhesion in Melanoma by Upregulation of E-Cadherin and Inhibition of CDC42. Cancer Lett. 2024, 581, 216484. [Google Scholar] [CrossRef]
- Martínez-Vicente, I.; Abrisqueta, M.; Herraiz, C.; Sirés-Campos, J.; Castejón-Griñán, M.; Bennett, D.C.; Olivares, C.; García-Borrón, J.C.; Jiménez-Cervantes, C. Mahogunin Ring Finger 1 Is Required for Genomic Stability and Modulates the Malignant Phenotype of Melanoma Cells. Cancers 2020, 12, 2840. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- El Hajj, P.; Journe, F.; Wiedig, M.; Laios, I.; Salès, F.; Galibert, M.-D.; Van Kempen, L.C.; Spatz, A.; Badran, B.; Larsimont, D.; et al. Tyrosinase-Related Protein 1 MRNA Expression in Lymph Node Metastases Predicts Overall Survival in High-Risk Melanoma Patients. Br. J. Cancer 2013, 108, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Centeno, P.P.; Pavet, V.; Marais, R. The Journey from Melanocytes to Melanoma. Nat. Rev. Cancer 2023, 23, 372–390. [Google Scholar] [CrossRef] [PubMed]
- García-Borrón, J.C.; Abdel-Malek, Z.; Jiménez-Cervantes, C. MC1R, the cAMP Pathway, and the Response to Solar UV: Extending the Horizon beyond Pigmentation. Pigment. Cell Melanoma Res. 2014, 27, 699–720. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Miller, A.J.; Widlund, H.R.; Horstmann, M.A.; Ramaswamy, S.; Fisher, D.E. MLANA/MART1 and SILV/PMEL17/GP100 Are Transcriptionally Regulated by MITF in Melanocytes and Melanoma. Am. J. Pathol. 2003, 163, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, D.; O’Neill, D.W.; Belitskaya-Levy, I.; Vacic, V.; Yu, Y.-L.; Adams, S.; Darvishian, F.; Berman, R.; Shapiro, R.; Pavlick, A.C.; et al. Immune Profile and Mitotic Index of Metastatic Melanoma Lesions Enhance Clinical Staging in Predicting Patient Survival. Proc. Natl. Acad. Sci. USA 2009, 106, 20429–20434. [Google Scholar] [CrossRef]
- Cirenajwis, H.; Ekedahl, H.; Lauss, M.; Harbst, K.; Carneiro, A.; Enoksson, J.; Rosengren, F.; Werner-Hartman, L.; Törngren, T.; Kvist, A.; et al. Molecular Stratification of Metastatic Melanoma Using Gene Expression Profiling: Prediction of Survival Outcome and Benefit from Molecular Targeted Therapy. Oncotarget 2015, 6, 12297–12309. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Pollard, J.W. Tumor-Associated Macrophages. Curr. Biol. 2020, 30, R246–R248. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus Guidelines for the Definition, Detection and Interpretation of Immunogenic Cell Death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Kang, K.; Xie, F.; Mao, J.; Bai, Y.; Wang, X. Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front. Oncol. 2020, 10, 573141. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Alonso, S.R.; Ortiz, P.; Pollán, M.; Pérez-Gómez, B.; Sánchez, L.; Acuña, M.J.; Pajares, R.; Martínez-Tello, F.J.; Hortelano, C.M.; Piris, M.A.; et al. Progression in Cutaneous Malignant Melanoma Is Associated with Distinct Expression Profiles. Am. J. Pathol. 2004, 164, 193–203. [Google Scholar] [CrossRef]
- Kashani-Sabet, M.; Venna, S.; Nosrati, M.; Rangel, J.; Sucker, A.; Egberts, F.; Baehner, F.L.; Simko, J.; Leong, S.P.L.; Haqq, C.; et al. A Multimarker Prognostic Assay for Primary Cutaneous Melanoma. Clin. Cancer Res. 2009, 15, 6987–6992. [Google Scholar] [CrossRef]
- Koh, S.S.; Wei, J.-P.J.; Li, X.; Huang, R.R.; Doan, N.B.; Scolyer, R.A.; Cochran, A.J.; Binder, S.W. Differential Gene Expression Profiling of Primary Cutaneous Melanoma and Sentinel Lymph Node Metastases. Mod. Pathol. 2012, 25, 828–837. [Google Scholar] [CrossRef][Green Version]
- Mandruzzato, S.; Callegaro, A.; Turcatel, G.; Francescato, S.; Montesco, M.C.; Chiarion-Sileni, V.; Mocellin, S.; Rossi, C.R.; Bicciato, S.; Wang, E.; et al. A Gene Expression Signature Associated with Survival in Metastatic Melanoma. J. Transl. Med. 2006, 4, 50. [Google Scholar] [CrossRef]
- Xu, L.; Shen, S.S.; Hoshida, Y.; Subramanian, A.; Ross, K.; Brunet, J.-P.; Wagner, S.N.; Ramaswamy, S.; Mesirov, J.P.; Hynes, R.O. Gene Expression Changes in an Animal Melanoma Model Correlate with Aggressiveness of Human Melanoma Metastases. Mol. Cancer Res. 2008, 6, 760–769. [Google Scholar] [CrossRef]
- Martínez-Jiménez, F.; Movasati, A.; Brunner, S.R.; Nguyen, L.; Priestley, P.; Cuppen, E.; Van Hoeck, A. Pan-Cancer Whole-Genome Comparison of Primary and Metastatic Solid Tumours. Nature 2023, 618, 333–341. [Google Scholar] [CrossRef]
- Aaltonen, L.A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D.J.; Agrawal, N.; Ahn, K.S.; Ahn, S.-M.; Aikata, H.; Akbani, R.; et al. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Mirek, J.; Bal, W.; Olbryt, M. Melanoma Genomics–Will We Go beyond BRAF in Clinics? J Cancer Res. Clin. Oncol. 2024, 150, 433. [Google Scholar] [CrossRef] [PubMed]
- Watt, B.; van Niel, G.; Raposo, G.; Marks, M.S. PMEL: A Pigment Cell-specific Model for Functional Amyloid Formation. Pigment. Cell Melanoma Res. 2013, 26, 300–315. [Google Scholar] [CrossRef]
- Frøsig, T.M.; Lyngaa, R.; Met, Ö.; Larsen, S.K.; Donia, M.; Svane, I.M.; thor Straten, P.; Hadrup, S.R. Broadening the Repertoire of Melanoma-Associated T-Cell Epitopes. Cancer Immunol. Immunother. 2015, 64, 609–620. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. MITF—The First 25 Years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf Regulation of Dia1 Controls Melanoma Proliferation and Invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Marine, J.-C.; Goding, C.R. Melanoma Plasticity and Phenotypic Diversity: Therapeutic Barriers and Opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Wellbrock, C. Phenotype Plasticity as Enabler of Melanoma Progression and Therapy Resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Slominski, R.M.; Sarna, T.; Płonka, P.M.; Raman, C.; Brożyna, A.A.; Slominski, A.T. Melanoma, Melanin, and Melanogenesis: The Yin and Yang Relationship. Front. Oncol. 2022, 12, 842496. [Google Scholar] [CrossRef]
- Journe, F.; Boufker, H.I.; Van Kempen, L.; Galibert, M.-D.; Wiedig, M.; Salès, F.; Theunis, A.; Nonclercq, D.; Frau, A.; Laurent, G.; et al. TYRP1 MRNA Expression in Melanoma Metastases Correlates with Clinical Outcome. Br. J. Cancer 2011, 105, 1726–1732. [Google Scholar] [CrossRef] [PubMed]
- Gilot, D.; Migault, M.; Bachelot, L.; Journé, F.; Rogiers, A.; Donnou-Fournet, E.; Mogha, A.; Mouchet, N.; Pinel-Marie, M.-L.; Mari, B.; et al. A Non-Coding Function of TYRP1 MRNA Promotes Melanoma Growth. Nat. Cell Biol. 2017, 19, 1348–1357. [Google Scholar] [CrossRef]
- Aung, T.N.; Warrell, J.; Martinez-Morilla, S.; Gavrielatou, N.; Vathiotis, I.; Yaghoobi, V.; Kluger, H.M.; Gerstein, M.; Rimm, D.L. Spatially Informed Gene Signatures for Response to Immunotherapy in Melanoma. Clin. Cancer Res. 2024, 30, 3520–3532. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lamberti, G.; Di Federico, A.; Alessi, J.; Ferrara, R.; Sholl, M.L.; Awad, M.M.; Vokes, N.; Ricciuti, B. Tumor Mutational Burden for the Prediction of PD-(L)1 Blockade Efficacy in Cancer: Challenges and Opportunities. Ann. Oncol. 2024, 35, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Furtado, L.V.; Bifulco, C.; Dolderer, D.; Hsiao, S.J.; Kipp, B.R.; Lindeman, N.I.; Ritterhouse, L.L.; Temple-Smolkin, R.L.; Zehir, A.; Nowak, J.A. Recommendations for Tumor Mutational Burden Assay Validation and Reporting. J. Mol. Diagn. 2024, 26, 653–668. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Beltrán, J.; Soler Díaz, J.; Herraiz, C.; Olivares, C.; Cerdido, S.; Cerezuela-Fuentes, P.; García-Borrón, J.C.; Jiménez-Cervantes, C. An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival. Int. J. Mol. Sci. 2025, 26, 1739. https://doi.org/10.3390/ijms26041739
Sánchez-Beltrán J, Soler Díaz J, Herraiz C, Olivares C, Cerdido S, Cerezuela-Fuentes P, García-Borrón JC, Jiménez-Cervantes C. An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival. International Journal of Molecular Sciences. 2025; 26(4):1739. https://doi.org/10.3390/ijms26041739
Chicago/Turabian StyleSánchez-Beltrán, José, Javier Soler Díaz, Cecilia Herraiz, Conchi Olivares, Sonia Cerdido, Pablo Cerezuela-Fuentes, José Carlos García-Borrón, and Celia Jiménez-Cervantes. 2025. "An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival" International Journal of Molecular Sciences 26, no. 4: 1739. https://doi.org/10.3390/ijms26041739
APA StyleSánchez-Beltrán, J., Soler Díaz, J., Herraiz, C., Olivares, C., Cerdido, S., Cerezuela-Fuentes, P., García-Borrón, J. C., & Jiménez-Cervantes, C. (2025). An MGRN1-Based Biomarker Combination Accurately Predicts Melanoma Patient Survival. International Journal of Molecular Sciences, 26(4), 1739. https://doi.org/10.3390/ijms26041739






