Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis
Abstract
1. Introduction
2. Results
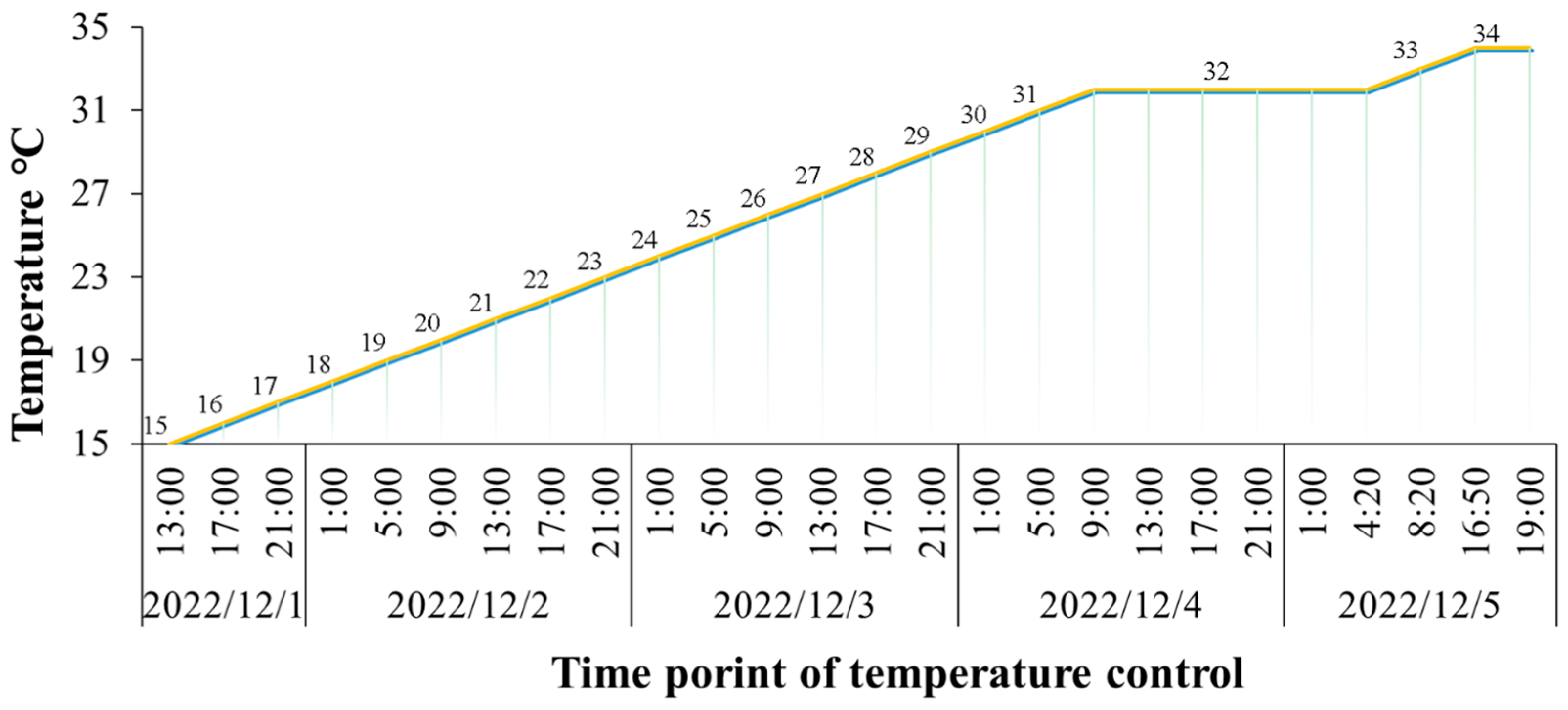
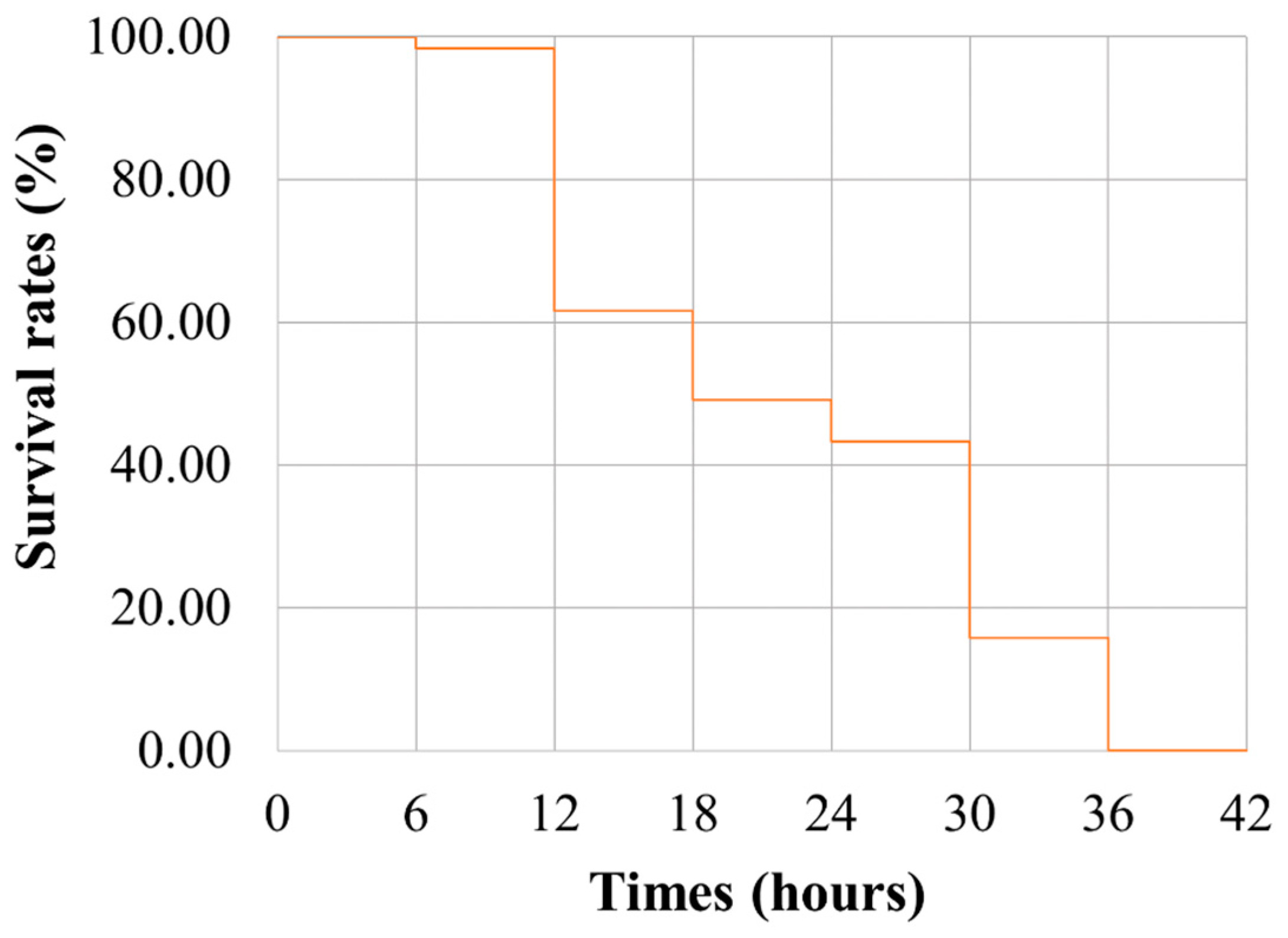
2.1. Statistical Analysis of Heat-Tolerant Phenotypes
2.2. Sequencing and Genotyping
2.3. Construction of Linkage Maps
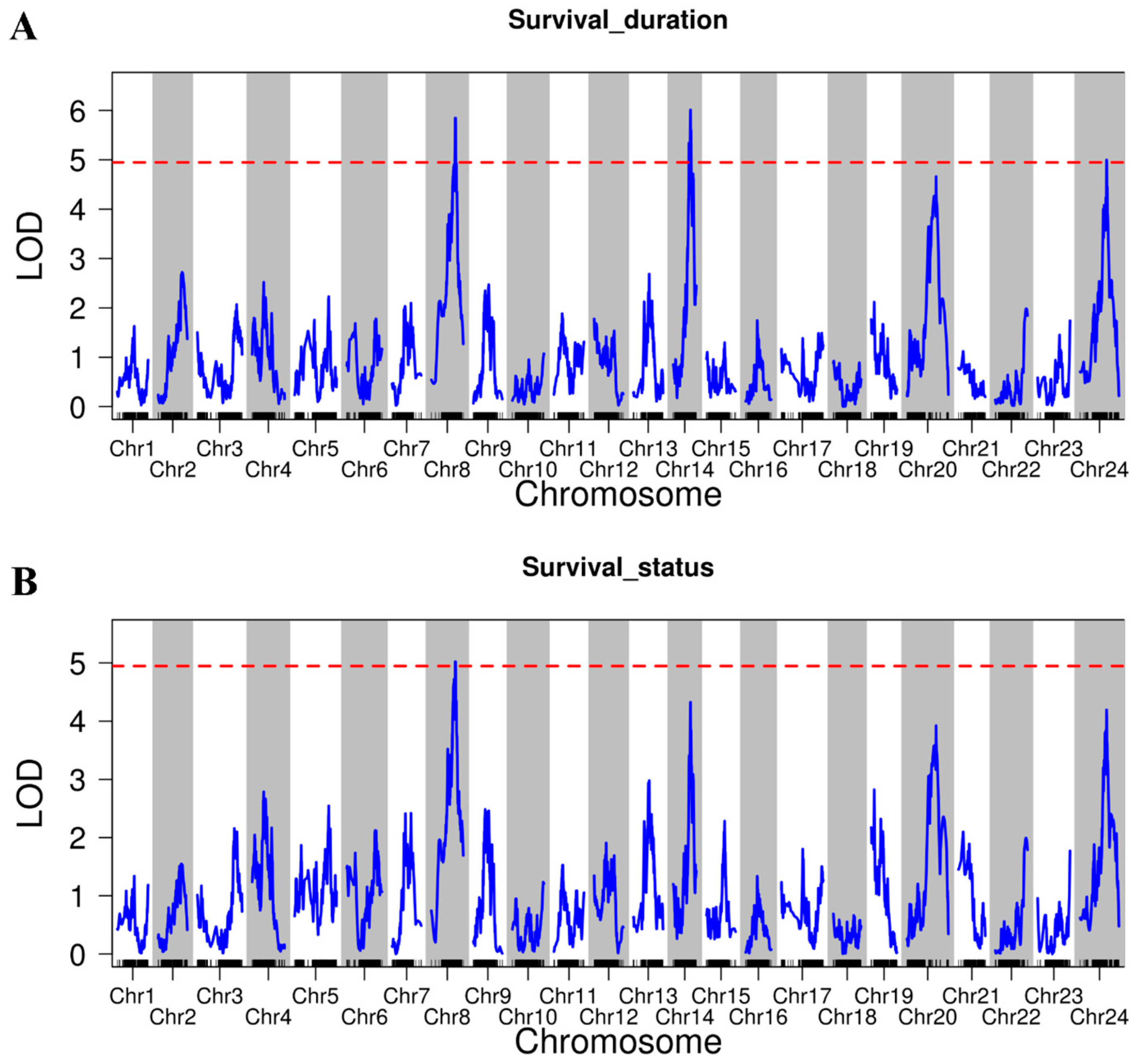
2.4. QTL Mapping and Identification of Heat Tolerance Candidate Genes
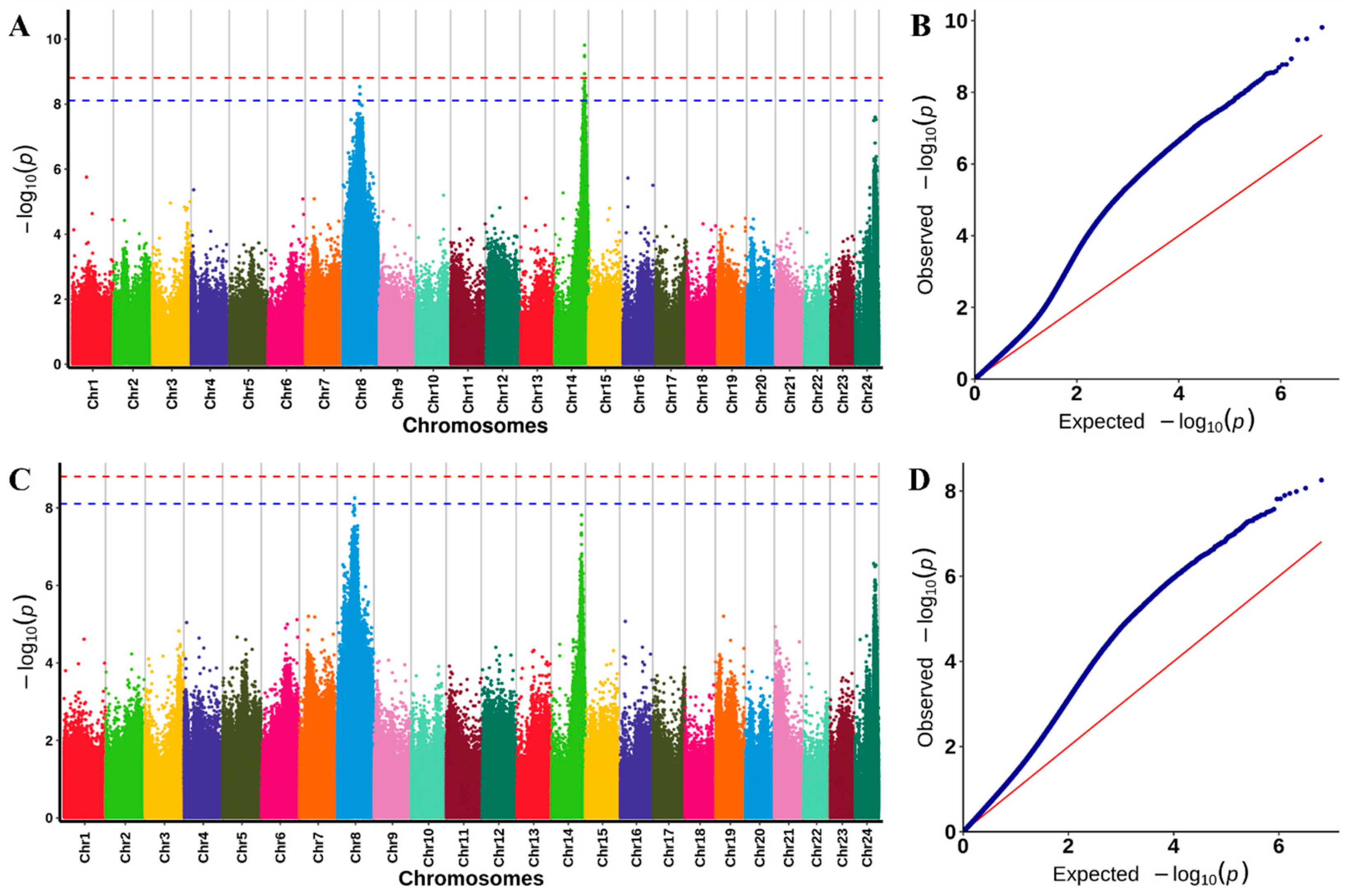
2.5. GWAS Analysis of Heat Tolerance
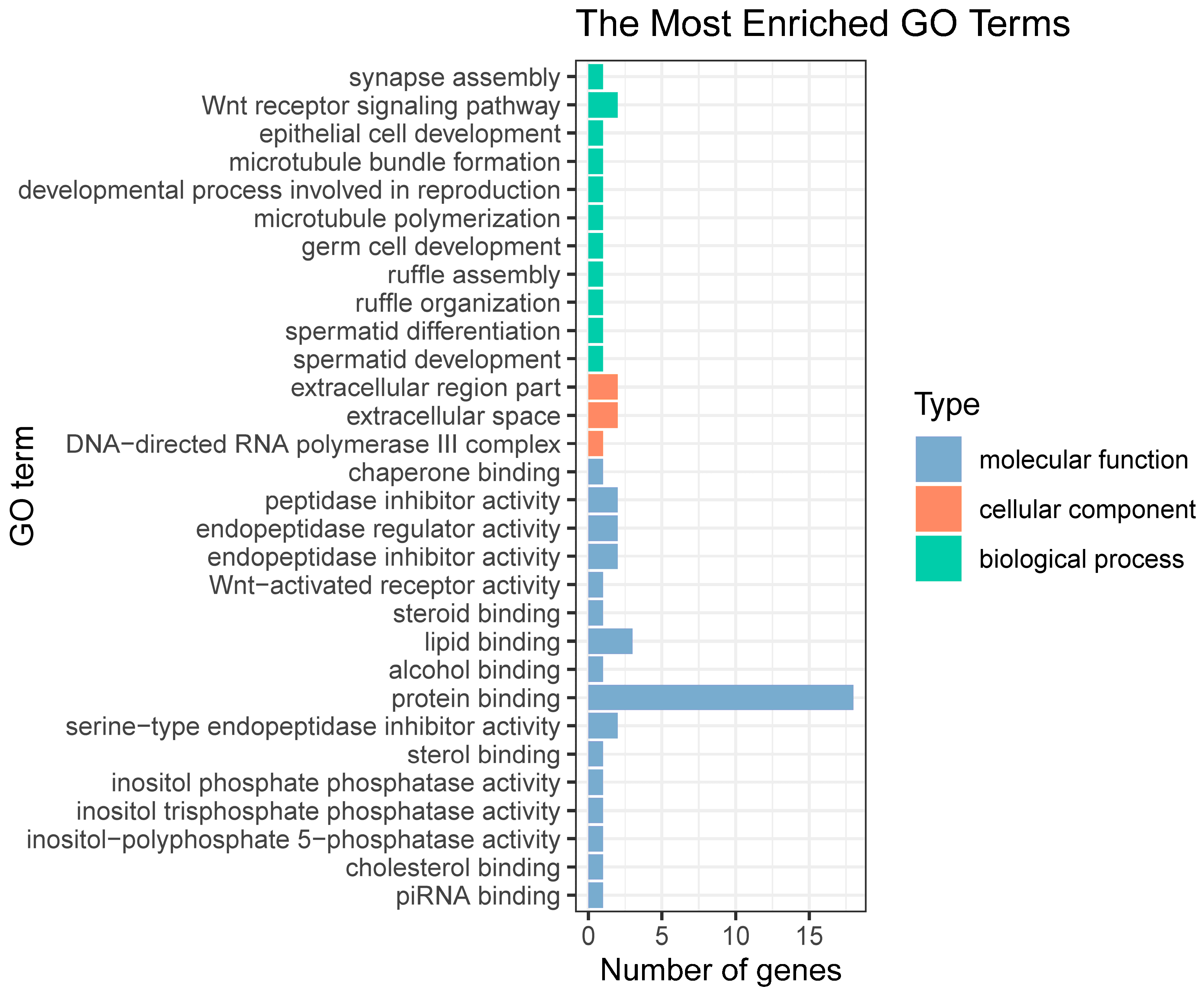
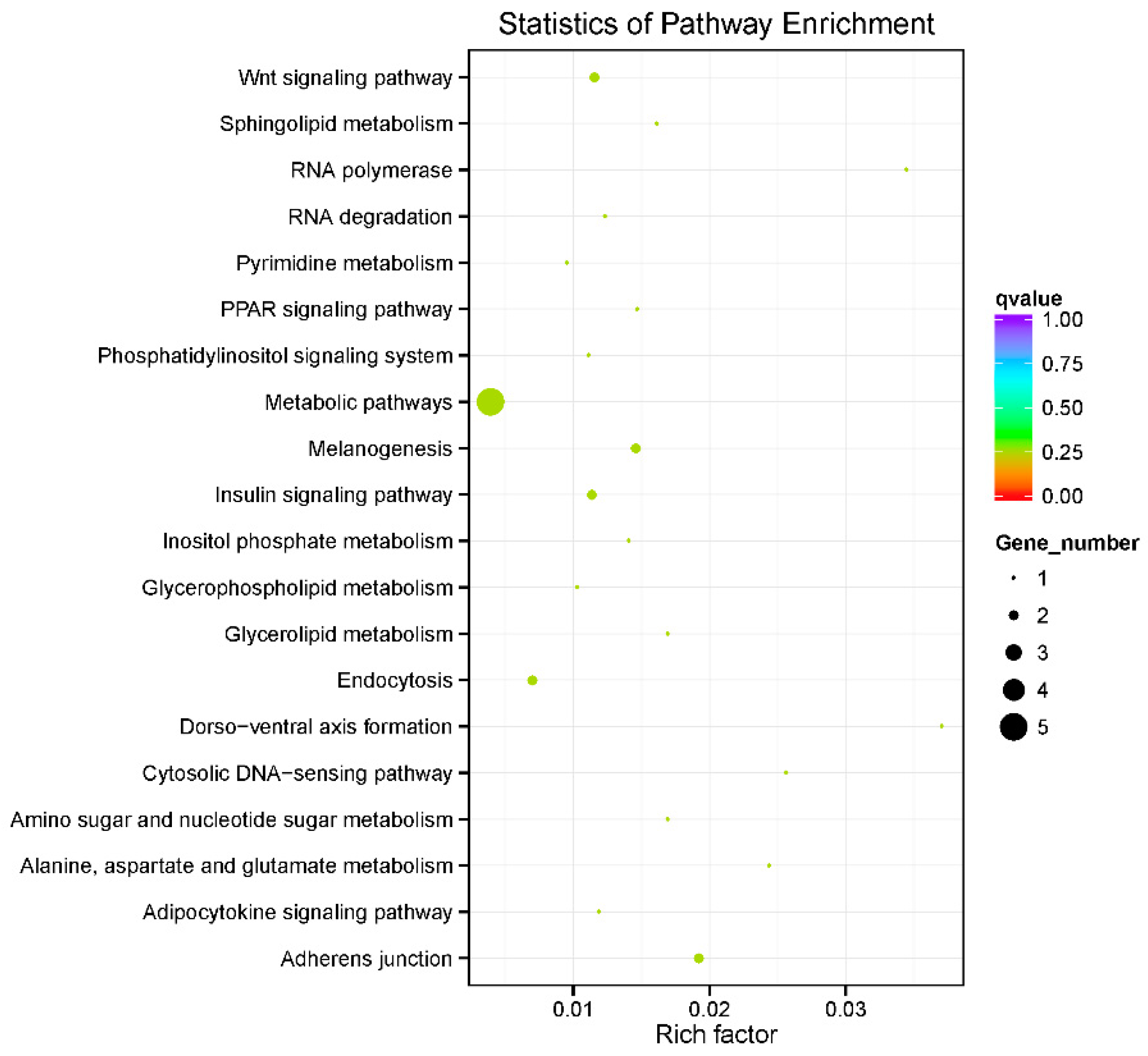
2.6. Functional Enrichment Analysis of Candidate Genes
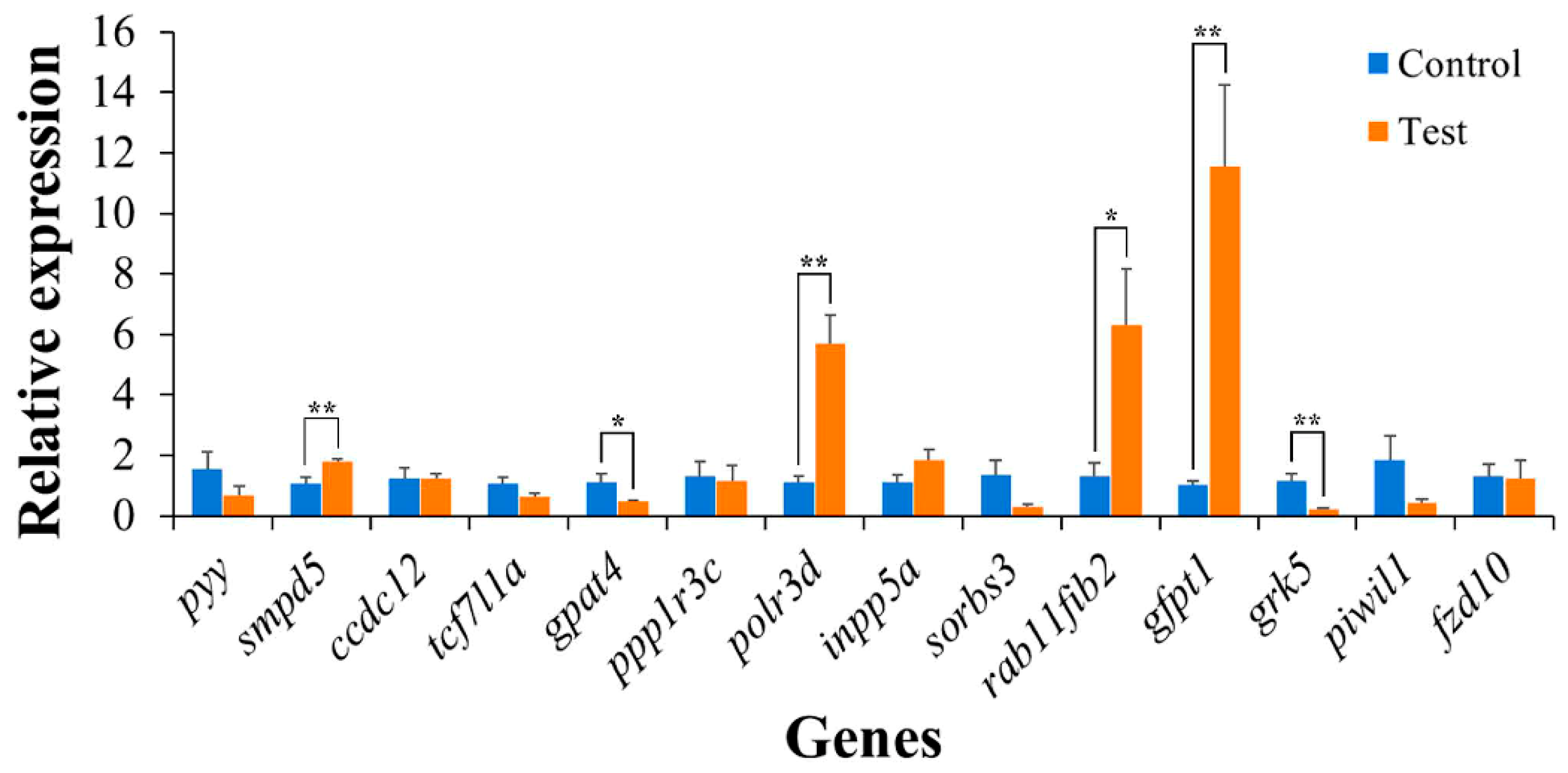
2.7. Identification of High-Temperature Response Gene Candidates
3. Discussion
4. Materials and Methods
4.1. Fish Full-Sib Family Production and Sample Collection
4.2. Genotyping-by-Sequencing Library Construction and Sequencing
4.3. Single-Nucleotide Polymorphism Genotyping
4.4. Construction and Evaluation of the Genetic Linkage Map
4.5. Fine-Mapping of Quantitative Trait Loci for Heat Tolerance Traits
4.6. Genome-Wide Association Studies
4.7. Gene Annotation and Enrichment Analysis
4.8. Identification of Candidate High-Temperature Response Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonfils, C.J.W.; Santer, B.D.; Fyfe, J.C.; Marvel, K.; Phillips, T.J.; Zimmerman, S.R.H. Human influence on joint changes in temperature, rainfall and continental aridity. Nat. Clim. Chang. 2020, 10, 726–731. [Google Scholar] [CrossRef]
- Brander, K.M. Global fish production and climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19709–19714. [Google Scholar] [CrossRef]
- Pan, L.; Zhang, L.; Zhang, Y.; Xie, S.; Qin, Q. Effects of acute high temperature stress on the growth, immune response, and antioxidant capacity of juvenile grass carp (Ctenopharyngodon idella). Aquac. Res. 2018, 49, 144–153. [Google Scholar]
- Li, M.; Chen, L.; Zeng, C.; Qin, J.G.; Chen, H. Effects of high temperature stress on oxidative stress, immune response and heat shock protein gene expression in fish: A review. Aquac. Res. 2017, 48, 753–770. [Google Scholar] [CrossRef]
- Xiong, Y.; Yang, J.; Jiang, T.; Liu, H.B.; Zhong, X.M.; Tang, J.H. Early life history of the small yellow croaker (Larimichthys polyactis) in sandy ridges of the South Yellow Sea. Mar. Biol. Res. 2017, 13, 993–1002. [Google Scholar] [CrossRef]
- Kim, S.; Lim, J.; Lee, K.; Park, S. Effect of twine thickness on size-selectivity of driftnet for the yellow croaker Larimichthys polyactis in southwestern Sea of Korea. Chin. J. Oceanol. Limnol. 2016, 34, 1199–1208. [Google Scholar] [CrossRef]
- Liu, F.; Liu, Y.; Chu, T.; Luo, B.; Zhan, W.; Chen, R. Interspecific hybridization and genetic characterization of Larimichthys polyactis (♀) and L. crocea (♂). Aquac. Int. 2019, 27, 663–674. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Cho, Y.; Hossen, S.; Yang, S.W.; Hwang, N.Y.; Lee, W.K.; Kho, K.H. Functional characterization of three GnRH isoforms in small yellow croaker Larimichthys polyactis maintained in captivity: Special emphasis on reproductive dysfunction. Biology 2022, 11, 1200. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, J.; Li, J.; Chen, Z.; Wang, N.; Li, M.; Wang, L.; Si, Y.; Lu, S.; Cui, Z.; et al. Decoding the fish genome opens a new era in important trait research and molecular breeding in China. Sci. China Life Sci. 2024, 67, 2064–2083. [Google Scholar] [CrossRef]
- Everett, M.V.; Seeb, J.E. Detection and mapping of QTL for temperature tolerance and body size in Chinook salmon (Oncorhynchus tshawytscha) using genotyping by sequencing. Evol. Appl. 2014, 7, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Huang, Z.; Wang, X.A.; Xu, Y.; Guo, X. Identification of quantitative trait loci associated with upper temperature tolerance in turbot, Scophthalmus maximus. Sci. Rep. 2021, 11, 21920. [Google Scholar] [CrossRef] [PubMed]
- Lagarde, H.; Lallias, D.; Patrice, P.; Dehaullon, A.; Prchal, M.; François, Y.; D’Ambrosio, J.; Segret, E.; Acin-Perez, A.; Cachelou, F.; et al. Genetic architecture of acute hyperthermia resistance in juvenile rainbow trout (Oncorhynchus mykiss) and genetic correlations with production traits. Genet. Sel. Evol. 2023, 55, 39. [Google Scholar] [CrossRef] [PubMed]
- Seeb, J.E.; Carvalho, G.; Hauser, L.; Naish, K.; Roberts, S.; Seeb, L.W. Single nucleotide polymorphism (SNP) discovery and applications of SNP genotyping in nonmodel organisms. Mol. Ecol. Resour. 2011, 11, 1–8. [Google Scholar] [CrossRef]
- Lee, B.Y.; Kim, M.S.; Choi, B.S.; Nagano, A.J.; Au, D.W.T.; Wu, R.S.S.; Takehana, Y.; Lee, J.S. Construction of high-resolution RAD-seq based linkage map, anchoring reference genome, and QTL mapping of the sex chromosome in the marine Medaka Oryzias melastigma. G3 2019, 9, 3537–3545. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, Y.; Yu, X.; Wang, J.; Luo, W.; Pang, M.; Tong, J. Genome-wide association study reveals SNPs and candidate genes related to growth and body shape in Bighead Carp (Hypophthalmichthys nobilis). Mar. Biotechnol. 2022, 24, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, X.; Wang, Z.; Han, Y.; Jia, Z.; Chen, D.; Xu, Y.; Qiao, Z.; Jiang, X.; Wang, L.; et al. Genome-wide association analysis study on host resistance against the Aeromonas veronii of largemouth bass Micropterus salmoides. Fish Shellfish Immunol. 2025, 157, 110093. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Z.; Chen, L. Genome-wide association study identifies novel loci associated with heat tolerance in Nile tilapia (Oreochromis niloticus). Aquaculture 2023, 560, 739361. [Google Scholar] [CrossRef]
- Liu, T.D.; Huang, D.D.; Chang, L.Y.; Qiao, T.F.; Xia, J.H. Identification of a novel QTL on LG16 associated with acute salt tolerance in Red Tilapia (Oreochromis spp.) using GWAS. Mar. Biotechnol. 2025, 27, 39. [Google Scholar] [CrossRef]
- Fu, S.; Liu, J. Genome-wide association study identified genes associated with ammonia nitrogen tolerance in Litopenaeus vannamei. Front Genet. 2022, 13, 961009. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Shi, R.H.; Li, C.Y.; Qi, H.G.; Liu, S.; Wang, W.; Li, L.; Zhang, G.F. Construction of a high-resolution genetic map of Crassostrea gigas: QTL mapping and GWAS applications revealed candidate genes controlling nutritional traits. Aquaculture 2020, 527, 735427. [Google Scholar] [CrossRef]
- Sallam, A.; Eltaher, S.; Alqudah, A.M.; Belamkar, V.; Baenziger, P.S. Combined GWAS and QTL mapping revealed candidate genes and SNP network controlling recovery and tolerance traits associated with drought tolerance in seedling winter wheat. Genomics 2022, 114, 110358. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, Z.; Luo, B.; Zhong, H.; Ma, P.; Zhang, H.; Hu, H.; Wang, Y.; Zhang, H.; Liu, D. Genetic architecture of maize yield traits dissected by QTL mapping and GWAS in maize. Crop J. 2022, 10, 436–446. [Google Scholar] [CrossRef]
- Ma, F.; Zhao, L.; Ma, R.; Wang, J.; Du, L. FoxO signaling and mitochondria-related apoptosis pathways mediate tsinling lenok trout (Brachymystax lenok tsinlingensis) liver injury under high temperature stress. Int. J. Biol. Macromol. 2023, 251, 126404. [Google Scholar] [CrossRef] [PubMed]
- Charo-Karisa, H.; Rezk, M.A.; Bovenhuis, H.; Komen, H. Heritability of cold tolerance in Nile tilapia, Oreochromis niloticus, juveniles. Aquaculture 2005, 249, 115–123. [Google Scholar] [CrossRef]
- Baer, C.F.; Travis, J. Direct and correlated responses to artificial selection on acute thermal stress tolerance in a live bearing fish. Evolution 2000, 54, 238. [Google Scholar]
- Perry, G.M.L.; Danzmann, R.G.; Ferguson, M.M.; Gibson, J.P. Quantitative trait loci for upper thermal tolerance in outbred strains of rainbow trout (Oncorhynchus mykiss). Heredity 2001, 86, 333–341. [Google Scholar] [CrossRef]
- Sun, X.W.; Liang, L.Q. A genetic linkage map of common carp (Cyprinus carpio L.) and mapping of a locus associated with cold tolerance. Aquaculture 2004, 238, 165–172. [Google Scholar] [CrossRef]
- Quinn, N.L.; McGowan, C.R.; Cooper, G.A.; Koop, B.F.; Davidson, W.S. Identification of genes associated with heat tolerance in Arctic charr exposed to acute thermal stress. Physiol. Genom. 2011, 43, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.Q.; Liu, F.; Qin, G.; Zhan, W.; Wang, M.J.; Lou, B. Transcriptome analysis of the Larimichthys polyactis under heat and cold stress. Cryobiology 2020, 96, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, T.; He, Y.; Zhan, W.; Xie, Q.; Lou, B. Integration of transcriptome and proteome analyses reveals the regulation mechanisms of Larimichthys polyactis liver exposed to heat stress. Fish Shellfish Immunol. 2023, 135, 108704. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.X.; He, P.P.; Wei, P.Y.; Zhang, B.; Zhao, Y.Z.; Li, Q.Y.; Chen, X.L.; Peng, M.; Zeng, D.G.; Yang, C.L.; et al. Proteomic responses under cold stress reveal unique cold tolerance mechanisms in the Pacific White Shrimp (Litopenaeus vannamei). Front. Physiol. 2018, 9, 1399. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Hao, C.; Zhang, Y.; Cheng, J.; Zhang, Z.; Liu, J.; Yi, X.; Cheng, X.; Sun, D.; Xu, Y.; et al. A combined association mapping and linkage analysis of kernel number per spike in common wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 1412. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Zhang, L.; Niyitanga, S.; Afzal, M.Z.; Qi, J. Principles and approaches of association mapping in plant breeding. Trop. Plant Biol. 2020, 13, 212–224. [Google Scholar] [CrossRef]
- Liu, F.; Zhan, W.; Xie, Q.P.; Chen, H.L.; Lou, B.; Xu, W.T. A first genetic linage map construction and QTL mapping for growth traits in Larimichthys polyactis. Sci. Rep. 2020, 10, 11621. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Q.; Jensen, J. Association Studies and Genomic Prediction for Genetic Improvements in Agriculture. Front. Plant Sci. 2022, 13, 904230. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, T.; Geng, X.; Liu, S.; Chen, A.; Yao, J.; Jiang, C.; Tan, S.; Su, B.; Liu, Z. A genome-wide association study of heat stress-associated SNPs in catfish. Anim. Genet. 2017, 48, 233–236. [Google Scholar] [CrossRef]
- Yoshida, G.M.; Yáñez, J.M. Increased accuracy of genomic predictions for growth under chronic thermal stress in rainbow trout by prioritizing variants from GWAS using imputed sequence data. Evol. Appl. 2021, 15, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Z.; Wang, S.; Zhao, A.; Wang, H.; Liu, Z.; Sui, M.; Bao, L.; Zeng, Q.; Hu, J.; et al. Genome-wide association analysis reveals the genetic basis of thermal tolerance in dwarf surf clam Mulinia lateralis. Genomics 2024, 116, 110904. [Google Scholar] [CrossRef] [PubMed]
- San, L.Z.; Wang, G.X.; He, Z.W.; Liu, Y.F.; Cao, W.; Zhang, Y.T.; Yang, Y.C.; Han, T.; Qin, Y.W.; Yang, T.L.; et al. Genome-wide association study for high-temperature tolerance in the Japanese flounder. Animal 2024, 18, 101273. [Google Scholar] [CrossRef]
- Khan, A.; Ahmad, M.; Shani, M.Y.; Khan, M.K.R.; Rahimi, M.; Tan, D.K.Y. Identifying the physiological traits associated with DNA marker using genome wide association in wheat under heat stress. Sci. Rep. 2024, 14, 20134. [Google Scholar] [CrossRef]
- Rizzo, M.A.; Kolesnick, R.N. Sphingomyelinase and ceramide in cell signaling. J. Biol. Chem. 2000, 275, 26361–26364. [Google Scholar] [CrossRef]
- Yabu, T.; Shimuzu, A.; Yamashita, M. A novel mitochondrial sphingomyelinase in zebrafish cells. J. Biol. Chem. 2009, 284, 20349–20363. [Google Scholar] [CrossRef] [PubMed]
- Levade, T.; Malagarie-Cazenave, S.; Gouazé, V.; Ségui, B.; Tardy, C.; Betito, S.; Andrieu-Abadie, N.; Cuvillier, O. Ceramide in apoptosis: A revisited role. Neurochem. Res. 2002, 27, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Luberto, C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000, 10, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Hales, C.M.; Griner, R.; Hobdy-Henderson, K.C.; Dorn, M.C.; Hardy, D.; Kumar, R.; Navarre, J.; Chan, E.K.; Lapierre, L.A.; Goldenring, J.R. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001, 276, 39067–39075. [Google Scholar] [CrossRef] [PubMed]
- Prekeris, R.; Davies, J.M.; Scheller, R.H. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001, 276, 38966–38970. [Google Scholar] [CrossRef]
- Schafer, J.C.; Baetz, N.W.; Lapierre, L.A.; McRae, R.E.; Roland, J.T.; Goldenring, J.R. Rab11-FIP2 interaction with MYO5B regulates movement of Rab11a-containing recycling vesicles. Traffic 2014, 15, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef]
- Finger, F.P.; White, J.G. Fusion and fission: Membrane trafficking in animal cytokinesis. Cell 2002, 108, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Hales, C.M.; Vaerman, J.P.; Goldenring, J.R. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 2002, 277, 50415–50421. [Google Scholar] [CrossRef]
- Naslavsky, N.; Rahajeng, J.; Sharma, M.; Jovic, M.; Caplan, S. Interactions between EHD proteins and Rab11-FIP2: A role for EHD3 in early endosomal transport. Mol. Biol. Cell 2006, 17, 163–177. [Google Scholar] [CrossRef]
- Girbig, M.; Misiaszek, A.D.; Vorländer, M.K.; Lafita, A.; Grötsch, H.; Baudin, F.; Bateman, A.; Müller, C.W. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nat. Struct. Mol. Biol. 2021, 28, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Bernard, G.; Chouery, E.; Putorti, M.L.; Tétreault, M.; Takanohashi, A.; Carosso, G.; Clément, I.; Boespflug-Tanguy, O.; Rodriguez, D.; Delague, V.; et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011, 89, 415–423. [Google Scholar] [CrossRef]
- Choquet, K.; Pinard, M.; Yang, S.; Moir, R.D.; Poitras, C.; Dicaire, M.J.; Sgarioto, N.; Larivière, R.; Kleinman, C.L.; Willis, I.M.; et al. The leukodystrophy mutation Polr3b R103H causes homozygote mouse embryonic lethality and impairs RNA polymerase III biogenesis. Mol. Brain 2019, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Watt, K.E.; Macintosh, J.; Bernard, G.; Trainor, P.A. RNA polymerases I and III in development and disease. Semin. Cell Dev. Biol. 2022, 136, 49–63. [Google Scholar] [CrossRef]
- Farshadyeganeh, P.; Nazim, M.; Zhang, R.; Ohkawara, B.; Nakajima, K.; Rahman, M.A.; Nasrin, F.; Ito, M.; Takeda, J.I.; Ohe, K.; et al. Splicing regulation of GFPT1 muscle-specific isoform and its roles in glucose metabolisms and neuromuscular junction. iScience 2023, 26, 107746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Farshadyeganeh, P.; Ohkawara, B.; Nakajima, K.; Takeda, J.I.; Ito, M.; Zhang, S.; Miyasaka, Y.; Ohno, T.; Mori-Yoshimura, M.; et al. Muscle-specific lack of Gfpt1 triggers ER stress to alleviate misfolded protein accumulation. Dis. Models Mech. 2024, 17, dmm050768. [Google Scholar] [CrossRef]
- Wang, Z.V.; Deng, Y.; Gao, N.; Pedrozo, Z.; Li, D.L.; Morales, C.R.; Criollo, A.; Luo, X.; Tan, W.; Jiang, N.; et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 2014, 156, 1179–1192. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Li, C.; Liu, E.; Zhu, H.; Ling, Q. Heat stress-induced endoplasmic reticulum stress promotes liver apoptosis in largemouth bass (Micropterus salmoides). Aquaculture 2022, 546, 737401. [Google Scholar] [CrossRef]
- Wilfling, F.; Wang, H.; Haas, J.T.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.X.; Graham, M.; Christiano, R.; Fröhlich, F.; et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 2013, 24, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Konrad, R.J.; Li, S.D.; Hammond, C. Glycerolipid acyltransferases in triglyceride metabolism and energy homeostasis-potential as drug targets. Endocr. Metab. Immune Disord.-Drug Targets 2012, 12, 197–206. [Google Scholar] [CrossRef]
- Shan, D.; Li, J.L.; Wu, L.; Li, D.; Hurov, J.; Tobin, J.F.; Gimeno, R.E.; Cao, J. GPAT3 and GPAT4 are regulated by insulin-stimulated phosphorylation and play distinct roles in adipogenesis. J. Lipid Res. 2010, 51, 1971–1981. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Shen, Y.; Zhang, Z.; Jia, Q.; Xu, M.; Zhang, T.; Fang, H.; Yu, X.; Li, L.; Liu, D.; et al. A GPAT1 Mutation in Arabidopsis Enhances Plant Height but Impairs Seed Oil Biosynthesis. Int. J. Mol. Sci. 2021, 22, 785. [Google Scholar] [CrossRef]
- Sok, V.; Jacinto, A.Z.; Peng, N.; Eldemerdash, M.; Le, L.; Tran, P.D.; Feng, L.F.; Patel, J.R.; Gi, M.; Ammon, J.C.; et al. G protein coupled receptor kinase 5 modifies the nucleolar stress response activated by actinomycin D. Biochem. Cell Biol. 2021, 99, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, J.; Gresham, K.; Koch, W.J. G protein-coupled receptor kinases as therapeutic targets in the heart. Nat. Rev. Cardiol. 2019, 16, 612–622. [Google Scholar] [CrossRef] [PubMed]
- de Lucia, C.; Eguchi, A.; Koch, W.J. New insights in cardiac β-Adrenergic signaling during heart failure and aging. Front. Pharmacol. 2018, 9, 904. [Google Scholar] [CrossRef] [PubMed]
- Sato, P.Y.; Chuprun, J.K.; Schwartz, M.; Koch, W.J. The evolving impact of G protein-coupled receptor kinases in cardiac health and disease. Physiol. Rev. 2015, 95, 377–404. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, P.; Ilavenil, S.; Hwang, I.H.; Kim, D.; Choi, K.C. Ferulic acid stimulates adipocyte-specific secretory proteins to regulate adipose homeostasis in 3T3-L1 adipocytes. Molecules 2021, 26, 1984. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Shen, M.; Ma, L. GRK5 deficiency decreases diet-induced obesity and adipogenesis. Biochem. Biophys. Res. Commun. 2012, 421, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhan, W.; Xie, Q.; Lou, B.; Han, M.; Xu, W.; Tao, S.S. First genetic evaluation of growth traits in Larimichthys polyactis to guide the formulation of selective breeding programs. Aquaculture 2022, 554, 738141. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Rastas, P. Lep-MAP3: Robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 2017, 33, 3726–3732. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tian, Y.; Li, Z.; Liu, Y.; Wang, L.; Li, L.; Pang, Z.; Yang, C.; Wang, Q.; Shao, G. Construction of a high-density genetic linkage map and qtl mapping for growth traits in hybrid Epinephelus fuscoguttatus (♀) and Epinephelus tukula (♂) progeny. Aquaculture 2022, 551, 737921. [Google Scholar] [CrossRef]
- Arends, D.; Prins, P.; Jansen, R.C.; Broman, K.W. R/qtl: High-throughput multiple QTL mapping. Bioinformatics 2010, 26, 2990–2992. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
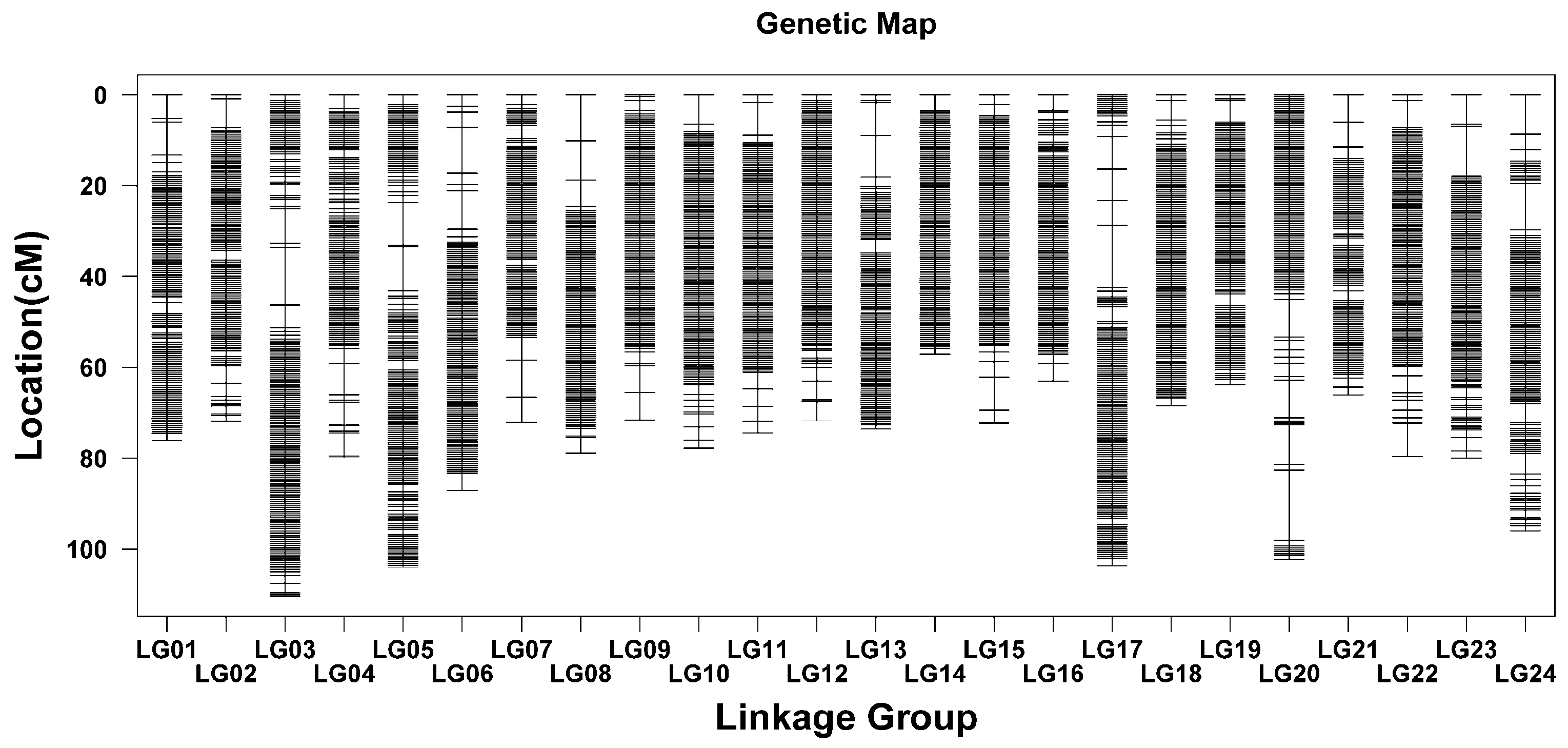
| Linkage Group | Sex-Averaged Linkage Map | Sex-Averaged Bin Map | ||||
|---|---|---|---|---|---|---|
| Marker Number | Genetic Length (cM) | Average Interval (cM) | Marker Number | Genetic Length (cM) | Average Interval (cM) | |
| LG01 | 191,319 | 76.13 | 0.0004 | 134 | 76.13 | 0.57 |
| LG02 | 183,971 | 71.79 | 0.0004 | 130 | 71.79 | 0.55 |
| LG03 | 207,831 | 110.40 | 0.0005 | 179 | 110.40 | 0.62 |
| LG04 | 186,542 | 79.89 | 0.0004 | 123 | 79.89 | 0.65 |
| LG05 | 189,061 | 103.93 | 0.0005 | 171 | 103.93 | 0.61 |
| LG06 | 165,987 | 87.01 | 0.0005 | 133 | 87.01 | 0.65 |
| LG07 | 189,038 | 72.12 | 0.0004 | 119 | 72.12 | 0.61 |
| LG08 | 187,869 | 78.80 | 0.0004 | 123 | 78.80 | 0.64 |
| LG09 | 137,628 | 71.49 | 0.0005 | 134 | 71.49 | 0.53 |
| LG10 | 166,695 | 77.68 | 0.0005 | 145 | 77.68 | 0.54 |
| LG11 | 197,732 | 74.37 | 0.0004 | 129 | 74.37 | 0.58 |
| LG12 | 167,506 | 71.77 | 0.0004 | 142 | 71.77 | 0.51 |
| LG13 | 155,010 | 73.53 | 0.0005 | 125 | 73.53 | 0.59 |
| LG14 | 119,345 | 57.10 | 0.0005 | 129 | 57.10 | 0.44 |
| LG15 | 214,226 | 72.26 | 0.0003 | 129 | 72.26 | 0.56 |
| LG16 | 123,888 | 62.96 | 0.0005 | 125 | 62.96 | 0.50 |
| LG17 | 167,223 | 103.65 | 0.0006 | 151 | 103.65 | 0.69 |
| LG18 | 156,600 | 68.39 | 0.0004 | 142 | 68.39 | 0.48 |
| LG19 | 183,256 | 63.82 | 0.0003 | 134 | 63.82 | 0.48 |
| LG20 | 144,087 | 102.22 | 0.0007 | 127 | 102.22 | 0.81 |
| LG21 | 167,864 | 66.05 | 0.0004 | 110 | 66.05 | 0.60 |
| LG22 | 151,660 | 79.52 | 0.0005 | 137 | 79.52 | 0.58 |
| LG23 | 129,791 | 80.01 | 0.0006 | 130 | 80.01 | 0.62 |
| LG24 | 74,200 | 95.98 | 0.0013 | 136 | 95.98 | 0.71 |
| Total | 3,958,329 | 1900.84 | 0.0005 | 3237 | 1900.84 | 0.59 |
| Traits | Chr | Location (cM) | Number of SNP | Peak Marker | Peak.LOD | PVE (%) | Genes |
|---|---|---|---|---|---|---|---|
| Survival duration | Chr8 | 52.13–62.55 | 6 | Chr8_16044431 | 5.85 | 10.13 | 343 |
| Chr14 | 40.43–45.85 | 10 | Chr14_24682001 | 6.01 | 13.08 | 84 | |
| Chr24 | 57.96–72.17 | 2 | Chr24_16888520 | 5.00 | 6.98 | 189 | |
| Survival status | Chr8 | 50.88–63.80 | 2 | Chr8_16044431 | 5.02 | 17.52 | 375 |
| Trait | Threshold p Value | Chr | SNP Number | Gene Number |
|---|---|---|---|---|
| Survival duration | 7.77 × 10−9 | Chr8 | 2 | 19 |
| Chr14 | 25 | 47 | ||
| Survival status | Chr8 | 1 | 10 |
| Markers | Genes | Description |
|---|---|---|
| Chr8_14197694 | pyy | peptide YY-like |
| smpd5 | Sphingomyelin phosphodiesterase 5 | |
| ccdc12 | coiled-coil domain-containing protein 12 | |
| Chr14_24682001 | tcf7l1a | transcription factor 7-like 1-A isoform X2 |
| gpat4 | glycerol-3-phosphate acyltransferase 4 | |
| ppp1r3c | protein phosphatase 1 regulatory subunit 3C-like | |
| polr3d | DNA-directed RNA polymerase III subunit RPC4 | |
| Chr14_24831514 | INPP5l | inositol polyphosphate 5-phosphatase A |
| sorbs3 | sorbin and SH3 domain-containing protein 2 isoform X5 | |
| Chr14_24852088 | rab11fip2 | rab11 family-interacting protein 2 |
| gfpt1 | glutamine-fructose-6-phosphate aminotransferase | |
| Chr14_25191367 | grk5 | G protein-coupled receptor kinase 5 like |
| Chr14_25303065 | piwil1 | Piwi-like protein 1 |
| fzd10 | frizzled-10 |
| Genes | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| ppp1r3c | TCTGCAGGATTTGGGAAGCA | ATGGCCTGTTCGTTGACACT |
| pyy | AACGGCAAGAAAACAGACGA | GGCTTGGCTGGATATGCGT |
| smpd5 | ATAAACAGCCCGACGAGGAC | TACCAATGACCCACGGCTTC |
| gpat4 | GTATCCTGCTCGGCATCTCC | CATGTAAAGACGCCGGATGC |
| try3 | CTGAATGCCCCCATCCTGAG | CTGATTGTTGCACACCACGG |
| sorbs3 | GGTGTTGGACTACGGGGAAG | CGATCACCTCGCCTTTACGA |
| tcf7l1a | CACCACCACTTCTCCCTAGC | GATTGGCCGGGTGAGGATAG |
| fzd10 | TGGGCTACCTCATCCGACTT | AGCCAGAAACCATGTGAGGG |
| inpp5a | CTGCAACTCCAGTCCTTCCA | ACCAGACTTTGCGTGTCCAG |
| ccdc12 | AGGCAGCTAATCCAGAACCC | TCTCCAGTTTCTTCGCCACA |
| rab11fib2 | ACAGAGCCGTTTGTACGGAG | TATTCTGAGCACTGACCGGC |
| grk5 | TCAAGAGACTGGAGGCTGGA | TGGGATGGAAACACTGCCTG |
| gfpt1 | GAACACTCCCGTCTTCCGAG | AGCGCTCCTCTCTCCTTACA |
| piwil1 | CCCAGAAGATCCGAGCTGAC | TGAATCTGTTGACGCCTCCC |
| hspa8 | GGACGAGTACGATCACCAGC | ATACCTCCTGGCATACCCCC |
| polr3d | CCAGTGAAAACGGAGGTCCA | ACCAGCATCTTTCCCACGAG |
| β-actin | CTCTGTCTGGATCGGAGGCT | GCTGAAGTTGTTGGGTGTTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Liu, H.; Zhang, T.; Guo, D.; Zhan, W.; Ye, T.; Lou, B. Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis. Int. J. Mol. Sci. 2025, 26, 1638. https://doi.org/10.3390/ijms26041638
Liu F, Liu H, Zhang T, Guo D, Zhan W, Ye T, Lou B. Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis. International Journal of Molecular Sciences. 2025; 26(4):1638. https://doi.org/10.3390/ijms26041638
Chicago/Turabian StyleLiu, Feng, Haowen Liu, Tianle Zhang, Dandan Guo, Wei Zhan, Ting Ye, and Bao Lou. 2025. "Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis" International Journal of Molecular Sciences 26, no. 4: 1638. https://doi.org/10.3390/ijms26041638
APA StyleLiu, F., Liu, H., Zhang, T., Guo, D., Zhan, W., Ye, T., & Lou, B. (2025). Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis. International Journal of Molecular Sciences, 26(4), 1638. https://doi.org/10.3390/ijms26041638







