Epitope Mapping with Sidewinder: An XL-MS and Structural Modeling Approach
Abstract
1. Introduction
2. Results
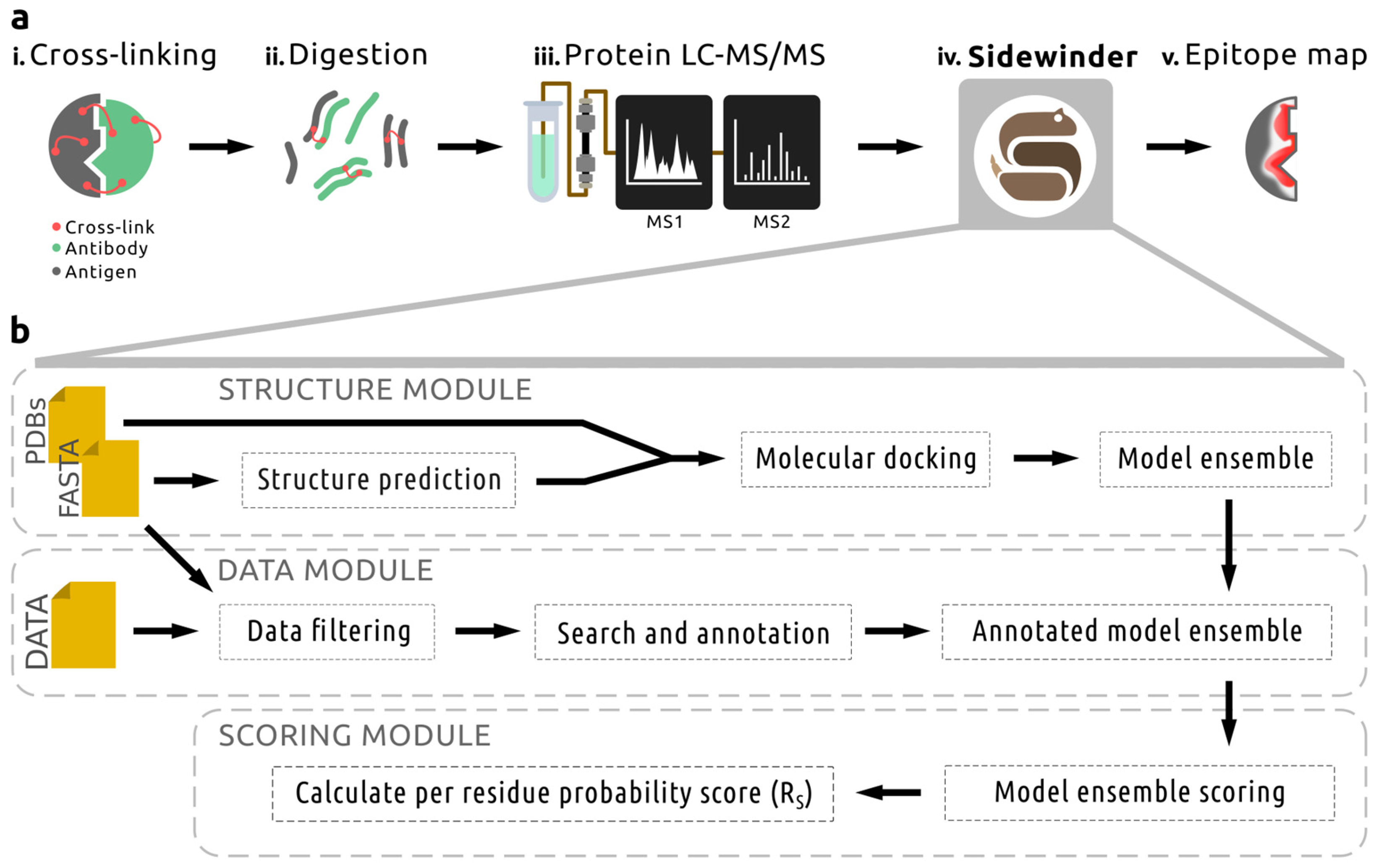
2.1. Sidewinder: A Modular Pipeline for Epitope Mapping
2.2. S. pyogenes Epitope Mapping with Sidewinder
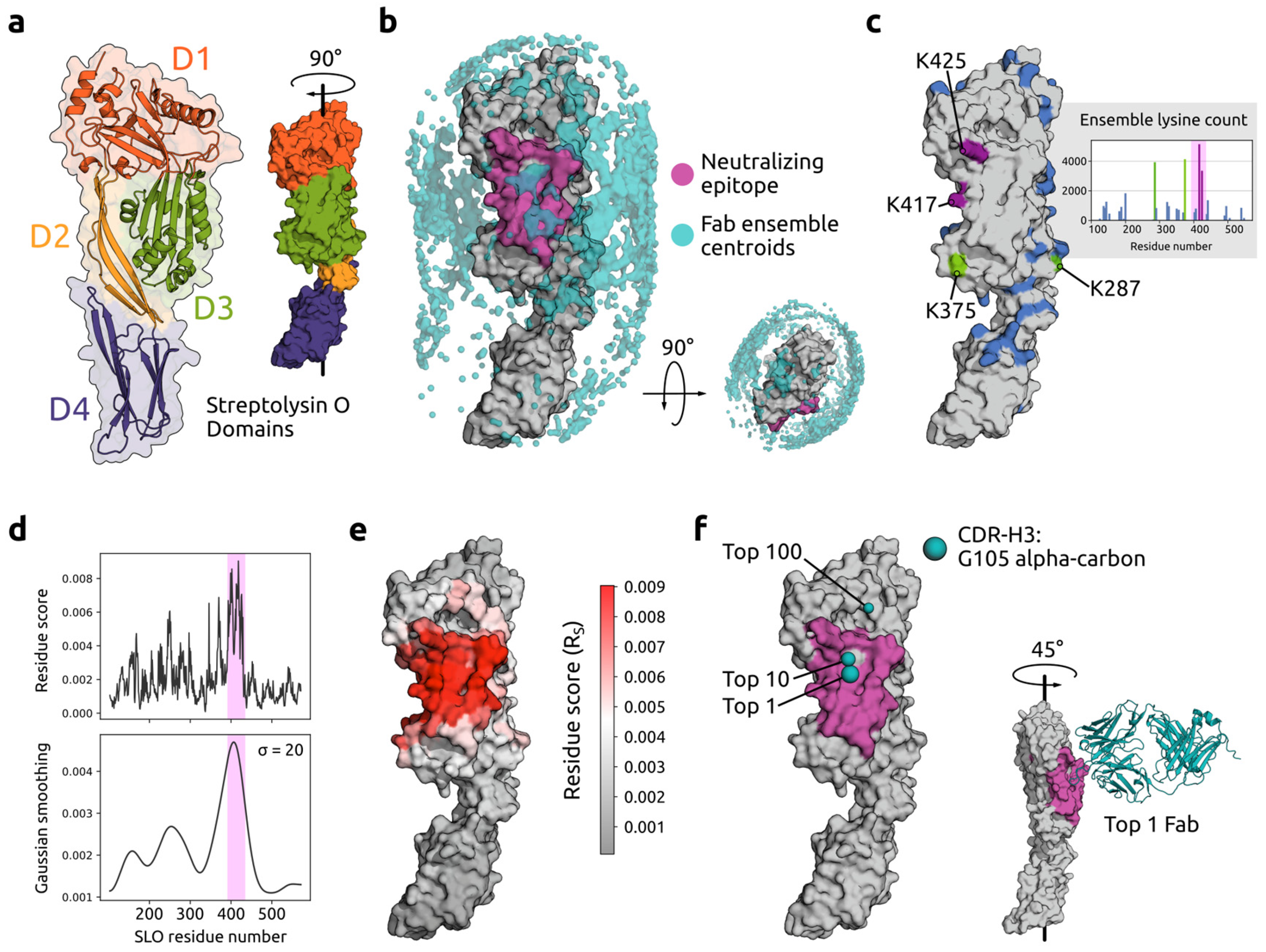
2.2.1. Recapitulating a Protective Epitope of Streptolysin O
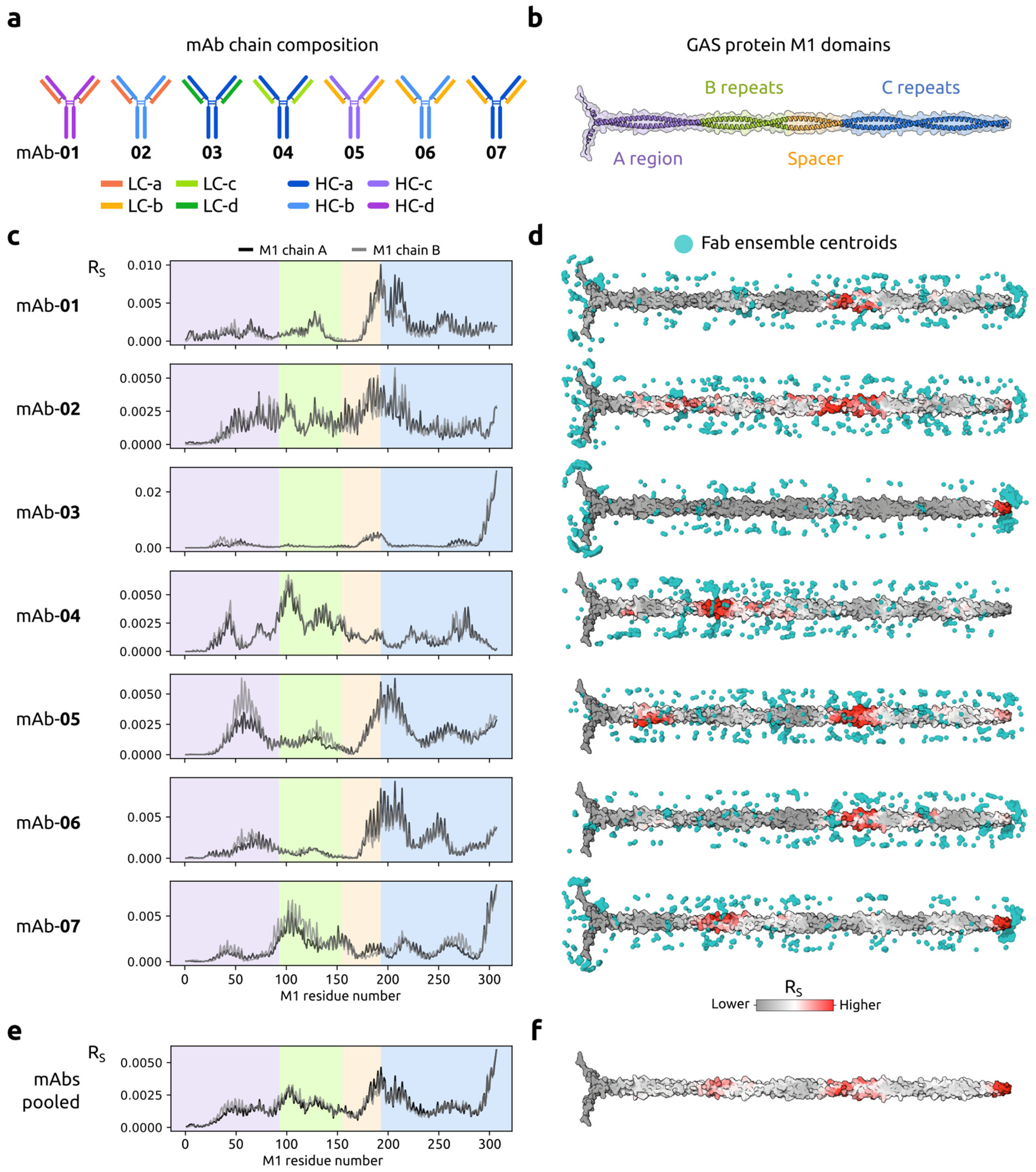
2.2.2. Deconvoluting the Complexity of GAS M Protein Binders
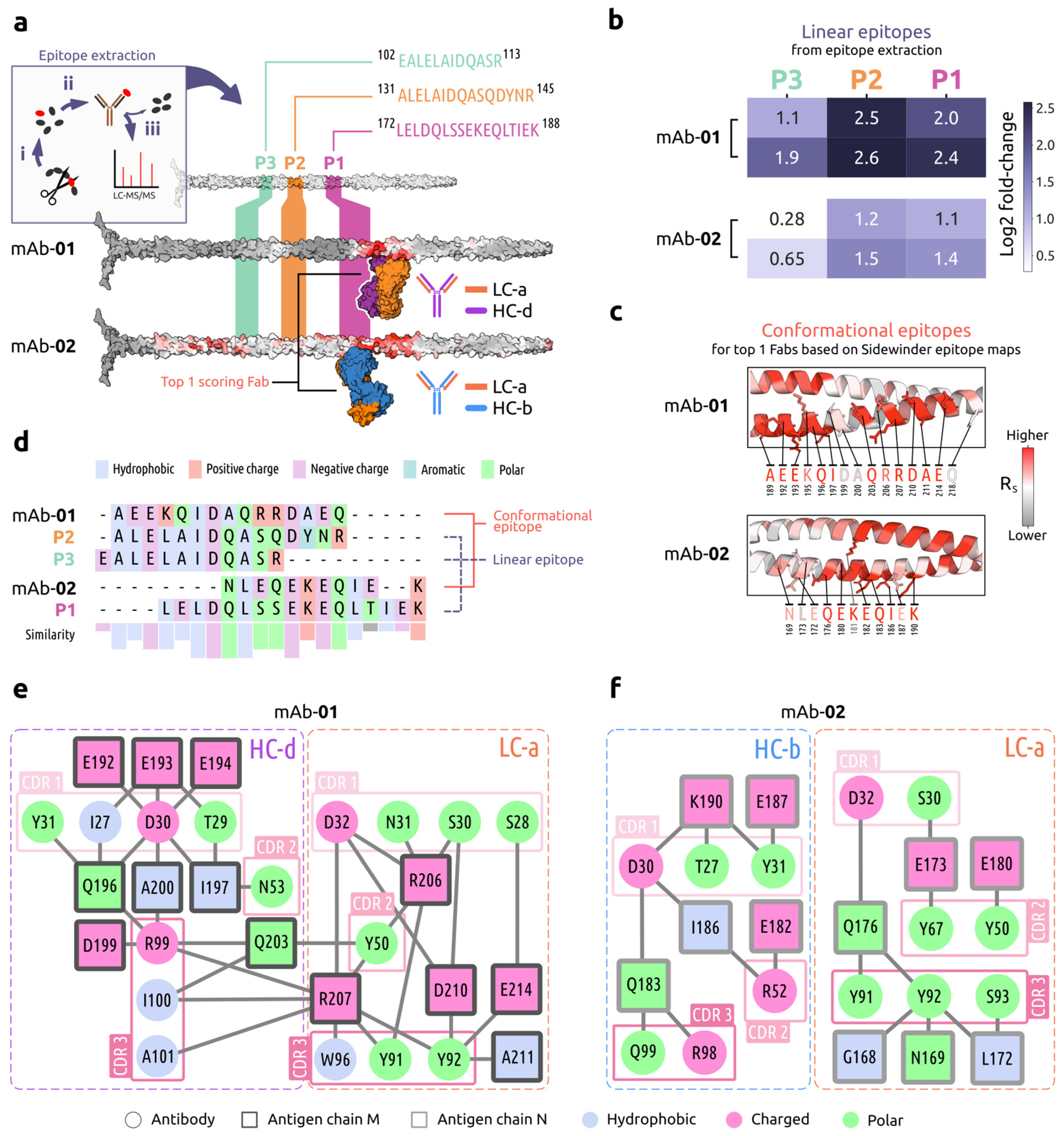
2.2.3. GAS M1 Protein Epitope Validation and Characterization
3. Discussion
4. Materials and Methods
4.1. The Sidewinder Pipeline
4.1.1. Structure Module
4.1.2. Data Module
4.1.3. Scoring Module
- Annotated MS2 ions (): The count of annotated ions in the MS2 spectrum;
- Cross-link ratio (): The ratio between cross-linked peptide length () and ;
- MS2 intensity coverage (): The ratio of the sum of intensities of annotated ions () to the sum of intensities of all ions () in the MS2 spectrum:
- Conforming distance constraints (): The total number of distance constraints (cross-links) satisfied by the interaction model.
4.2. Data Acquisition and Analysis of GAS SLO Neutralizing mAb
4.3. Data Acquisition and Analysis of GAS M1 Protein mAbs
4.3.1. Recombinant Monoclonal Antibody Production
4.3.2. Cross-Linking Mass Spectrometry of M1 mAbs
4.3.3. Analyzing M1 mAbs with Sidewinder
4.3.4. Epitope Extraction of Linear GAS M1 Protein Epitopes
4.3.5. Downstream Structure Refinement and Network Generation
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global Burden of Bacterial Antimicrobial Resistance 1990–2021: A Systematic Analysis with Forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Burton, D.R. Antibodies, Viruses and Vaccines. Nat. Rev. Immunol. 2002, 2, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Bebbington, C.; Yarranton, G. Antibodies for the Treatment of Bacterial Infections: Current Experience and Future Prospects. Curr. Opin. Biotechnol. 2008, 19, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Hitawala, F.N.; Gray, J.J. What Has AlphaFold3 Learned about Antibody and Nanobody Docking, and What Remains Unsolved? bioRxiv 2024. [Google Scholar] [CrossRef]
- Sharon, J.; Rynkiewicz, M.J.; Lu, Z.; Yang, C. Discovery of Protective B-cell Epitopes for Development of Antimicrobial Vaccines and Antibody Therapeutics. Immunology 2014, 142, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-Spectrometric Exploration of Proteome Structure and Function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Zhu, S.; Liuni, P.; Chen, T.; Houy, C.; Wilson, D.J.; James, D.A. Epitope Screening Using Hydrogen/Deuterium Exchange Mass Spectrometry (HDX-MS): An Accelerated Workflow for Evaluation of Lead Monoclonal Antibodies. Biotechnol. J. 2022, 17, e2100358. [Google Scholar] [CrossRef]
- Tang, D.; Gueto-Tettay, C.; Hjortswang, E.; Ströbaek, J.; Ekström, S.; Happonen, L.; Malmström, L.; Malmström, J. Multimodal Mass Spectrometry Identifies a Conserved Protective Epitope in S. pyogenes Streptolysin O. Anal. Chem. 2024, 96, 9060–9068. [Google Scholar] [CrossRef]
- Tran, M.H.; Schoeder, C.T.; Schey, K.L.; Meiler, J. Computational Structure Prediction for Antibody-Antigen Complexes From Hydrogen-Deuterium Exchange Mass Spectrometry: Challenges and Outlook. Front. Immunol. 2022, 13, 859964. [Google Scholar] [CrossRef]
- Herzog, F.; Kahraman, A.; Boehringer, D.; Mak, R.; Bracher, A.; Walzthoeni, T.; Leitner, A.; Beck, M.; Hartl, F.-U.; Ban, N.; et al. Structural Probing of a Protein Phosphatase 2A Network by Chemical Cross-Linking and Mass Spectrometry. Science 2012, 337, 1348–1352. [Google Scholar] [CrossRef]
- Mädler, S.; Seitz, M.; Robinson, J.; Zenobi, R. Does Chemical Cross-Linking with NHS Esters Reflect the Chemical Equilibrium of Protein-Protein Noncovalent Interactions in Solution? J. Am. Soc. Mass Spectrom. 2010, 21, 1775–1783. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Reilly, F.J.; Rappsilber, J. Cross-Linking Mass Spectrometry: Methods and Applications in Structural, Molecular and Systems Biology. Nat. Struct. Mol. Biol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Piersimoni, L.; Kastritis, P.L.; Arlt, C.; Sinz, A. Cross-Linking Mass Spectrometry for Investigating Protein Conformations and Protein–Protein Interactions—A Method for All Seasons. Chem. Rev. 2022, 122, 7500–7531. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; O’Reilly, F.J. Cross-Linking Mass Spectrometry for Mapping Protein Complex Topologies in Situ. Essays Biochem. 2023, 67, 215–228. [Google Scholar] [CrossRef]
- Yu, C.; Huang, L. Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Anal. Chem. 2018, 90, 144–165. [Google Scholar] [CrossRef]
- Khakzad, H.; Happonen, L.; Malmström, J.; Malmström, L. Cheetah-MS: A Web Server to Model Protein Complexes Using Tandem Cross-Linking Mass Spectrometry Data. Bioinformatics 2021, 37, 4871–4872. [Google Scholar] [CrossRef]
- Hauri, S.; Khakzad, H.; Happonen, L.; Teleman, J.; Malmström, J.; Malmström, L. Rapid Determination of Quaternary Protein Structures in Complex Biological Samples. Nat. Commun. 2019, 10, 192. [Google Scholar] [CrossRef]
- Huang, S.-Y. Search Strategies and Evaluation in Protein–Protein Docking: Principles, Advances and Challenges. Drug Discov. Today 2014, 19, 1081–1096. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Yamamori, Y.; Tomii, K. Protein–Protein Interaction Prediction Methods: From Docking-Based to AI-Based Approaches. Biophys. Rev. 2022, 14, 1341–1348. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Akbar, R.; Robert, P.A.; Pavlović, M.; Jeliazkov, J.R.; Snapkov, I.; Slabodkin, A.; Weber, C.R.; Scheffer, L.; Miho, E.; Haff, I.H.; et al. A Compact Vocabulary of Paratope-Epitope Interactions Enables Predictability of Antibody-Antigen Binding. Cell Rep. 2021, 34, 108856. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.J. High-Resolution Protein–Protein Docking. Curr. Opin. Struct. Biol. 2006, 16, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Stahl, K.; Warneke, R.; Demann, L.; Bremenkamp, R.; Hormes, B.; Brock, O.; Stülke, J.; Rappsilber, J. Modelling Protein Complexes with Crosslinking Mass Spectrometry and Deep Learning. Nat. Commun. 2024, 15, 7866. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ruffolo, J.A.; Chu, L.-S.; Mahajan, S.P.; Gray, J.J. Fast, Accurate Antibody Structure Prediction from Deep Learning on Massive Set of Natural Antibodies. Nat. Commun. 2023, 14, 2389. [Google Scholar] [CrossRef]
- Abanades, B.; Wong, W.K.; Boyles, F.; Georges, G.; Bujotzek, A.; Deane, C.M. ImmuneBuilder: Deep-Learning Models for Predicting the Structures of Immune Proteins. Commun. Biol. 2023, 6, 575. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Ambrosetti, F.; Jiménez-García, B.; Roel-Touris, J.; Bonvin, A.M.J.J. Modeling Antibody-Antigen Complexes by Information-Driven Docking. Structure 2020, 28, 119–129.e2. [Google Scholar] [CrossRef]
- Giulini, M.; Schneider, C.; Cutting, D.; Desai, N.; Deane, C.M.; Bonvin, A.M.J.J. Towards the Accurate Modelling of Antibody−antigen Complexes from Sequence Using Machine Learning and Information-Driven Docking. Bioinformatics 2024, 40, btae583. [Google Scholar] [CrossRef]
- Zhu, W.; Shenoy, A.; Kundrotas, P.; Elofsson, A. Evaluation of AlphaFold-Multimer Prediction on Multi-Chain Protein Complexes. Bioinformatics 2023, 39, btad424. [Google Scholar] [CrossRef]
- Yin, R.; Pierce, B.G. Evaluation of AlphaFold Antibody–Antigen Modeling with Implications for Improving Predictive Accuracy. Protein Sci. 2024, 33, e4865. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, G. Subdominance in Antibody Responses: Implications for Vaccine Development. Microbiol. Mol. Biol. Rev. 2020, 85, e00078–20. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, S.; Rivera-Hernandez, T.; Curren, B.F.; Harbison-Price, N.; Oliveira, D.M.P.D.; Jespersen, M.G.; Davies, M.R.; Walker, M.J. Pathogenesis, Epidemiology and Control of Group A Streptococcus Infection. Nat. Rev. Microbiol. 2023, 21, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Massell, B.F.; Honikman, L.H.; Amezcua, J. Rheumatic Fever Following Streptococcal Vaccination: Report of Three Cases. JAMA 1969, 207, 1115–1119. [Google Scholar] [CrossRef]
- Azuar, A.; Jin, W.; Mukaida, S.; Hussein, W.M.; Toth, I.; Skwarczynski, M. Recent Advances in the Development of Peptide Vaccines and Their Delivery Systems against Group A Streptococcus. Vaccines 2019, 7, 58. [Google Scholar] [CrossRef]
- Fan, J.; Toth, I.; Stephenson, R.J. Recent Scientific Advancements towards a Vaccine against Group A Streptococcus. Vaccines 2024, 12, 272. [Google Scholar] [CrossRef]
- Smeesters, P.R.; McMillan, D.J.; Sriprakash, K.S. The Streptococcal M Protein: A Highly Versatile Molecule. Trends Microbiol. 2010, 18, 275–282. [Google Scholar] [CrossRef]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable Data Analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef]
- van Pee, K.; Neuhaus, A.; D’Imprima, E.; Mills, D.J.; Kühlbrandt, W.; Yildiz, Ö. CryoEM Structures of Membrane Pore and Prepore Complex Reveal Cytolytic Mechanism of Pneumolysin. eLife 2017, 6, e23644. [Google Scholar] [CrossRef]
- Chiarot, E.; Faralla, C.; Chiappini, N.; Tuscano, G.; Falugi, F.; Gambellini, G.; Taddei, A.; Capo, S.; Cartocci, E.; Veggi, D.; et al. Targeted Amino Acid Substitutions Impair Streptolysin O Toxicity and Group A Streptococcus Virulence. mBio 2013, 4, e00387–12. [Google Scholar] [CrossRef]
- Feil, S.C.; Ascher, D.B.; Kuiper, M.J.; Tweten, R.K.; Parker, M.W. Structural Studies of Streptococcus Pyogenes Streptolysin O Provide Insights into the Early Steps of Membrane Penetration. J. Mol. Biol. 2014, 426, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Heusel, M.; Malmström, E.; Bratanis, E.; Tang, D.; Hartman, E.; Olofsson, B.; Mohanty, T.; Khakzad, H.; Happonen, L.; Malmström, L.; et al. Time-resolved proteogenomics analysis of the circulating antibody response against the streptococcal M1 protein. 2025; manuscript in preparation. [Google Scholar]
- Collin, M.; Olsén, A. Generation of a Mature Streptococcal Cysteine Proteinase Is Dependent on Cell Wall-anchored M1 Protein. Mol. Microbiol. 2000, 36, 1306–1318. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Toledo, A.G.; Hjortswang, E.; Sorrentino, J.T.; Lewis, N.E.; Bläckberg, A.; Ekström, S.; Izadi, A.; Nordenfelt, P.; Malmström, L.; et al. Dissecting the Properties of Circulating IgG against Group A Streptococcus through a Combined Systems Antigenomics-Serology Workflow. bioRxiv 2023. [Google Scholar] [CrossRef]
- Leman, J.K.; Weitzner, B.D.; Lewis, S.M.; Adolf-Bryfogle, J.; Alam, N.; Alford, R.F.; Aprahamian, M.; Baker, D.; Barlow, K.A.; Barth, P.; et al. Macromolecular Modeling and Design in Rosetta: Recent Methods and Frameworks. Nat. Methods 2020, 17, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A Web Server for Predicting the Binding Affinity of Protein–Protein Complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Iacobucci, C.; Sinz, A. To Be or Not to Be? Five Guidelines to Avoid Misassignments in Cross-Linking/Mass Spectrometry. Anal. Chem. 2017, 89, 7832–7835. [Google Scholar] [CrossRef]
- Wallner, B. AFsample: Improving Multimer Prediction with AlphaFold Using Massive Sampling. Bioinformatics 2023, 39, btad573. [Google Scholar] [CrossRef]
- Raouraoua, N.; Mirabello, C.; Véry, T.; Blanchet, C.; Wallner, B.; Lensink, M.F.; Brysbaert, G. MassiveFold: Unveiling AlphaFold’s Hidden Potential with Optimized and Parallelized Massive Sampling. Nat. Comput. Sci. 2024, 4, 824–828. [Google Scholar] [CrossRef]
- Bahnan, W.; Happonen, L.; Khakzad, H.; Ahnlide, V.K.; de Neergaard, T.; Wrighton, S.; André, O.; Bratanis, E.; Tang, D.; Hellmark, T.; et al. A Human Monoclonal Antibody Bivalently Binding Two Different Epitopes in Streptococcal M Protein Mediates Immune Function. EMBO Mol. Med. 2023, 15, EMMM202216208. [Google Scholar] [CrossRef]
- McNamara, C.; Zinkernagel, A.S.; Macheboeuf, P.; Cunningham, M.W.; Nizet, V.; Ghosh, P. Coiled-Coil Irregularities and Instabilities in Group A Streptococcus M1 Are Required for Virulence. Science 2008, 319, 1405–1408. [Google Scholar] [CrossRef]
- Heath, D.G.; Cleary, P.P. Fc-Receptor and M-Protein Genes of Group A Streptococci Are Products of Gene Duplication. Proc. Natl. Acad. Sci. USA 1989, 86, 4741–4745. [Google Scholar] [CrossRef] [PubMed]
- Nordenfelt, P.; Waldemarson, S.; Linder, A.; Mörgelin, M.; Karlsson, C.; Malmström, J.; Björck, L. Antibody Orientation at Bacterial Surfaces Is Related to Invasive Infection. J. Exp. Med. 2012, 209, 2367–2381. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.-L.S.; Flores, C.M.J.; Reynolds, S.; Wun, C.; Calcutt, A.; Baker, S.B.; Murugappan, S.; Depelsenaire, A.C.I.; Dooley, J.; Fahey, P.V.; et al. M-Protein Based Vaccine Induces Immunogenicity and Protection from Streptococcus Pyogenes When Delivered on a High-Density Microarray Patch (HD-MAP). npj Vaccines 2020, 5, 74. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, S.; Pandey, M.; Dooley, J.; Calcutt, A.; Batzloff, M.; Ozberk, V.; Mills, J.-L.; Good, M. Preclinical Safety and Immunogenicity of Streptococcus Pyogenes (Strep A) Peptide Vaccines. Sci. Rep. 2021, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Ohue, M.; Matsuzaki, Y.; Uchikoga, N.; Ishida, T.; Akiyama, Y. MEGADOCK: An All-to-All Protein-Protein Interaction Prediction System Using Tertiary Structure Data. Protein Pept. Lett. 2014, 21, 766–778. [Google Scholar] [CrossRef]
- Ohue, M.; Shimoda, T.; Suzuki, S.; Matsuzaki, Y.; Ishida, T.; Akiyama, Y. MEGADOCK 4.0: An Ultra–High-Performance Protein–Protein Docking Software for Heterogeneous Supercomputers. Bioinformatics 2014, 30, 3281–3283. [Google Scholar] [CrossRef]
- Chothia, C.; Lesk, A.M.; Tramontano, A.; Levitt, M.; Smith-Gill, S.J.; Air, G.; Sheriff, S.; Padlan, E.A.; Davies, D.; Tulip, W.R.; et al. Conformations of Immunoglobulin Hypervariable Regions. Nature 1989, 342, 877–883. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Chaudhury, S.; Lyskov, S.; Gray, J.J. PyRosetta: A Script-Based Interface for Implementing Molecular Modeling Algorithms Using Rosetta. Bioinformatics 2010, 26, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ströbaek, J.; Tang, D.; Gueto-Tettay, C.; Gomez Toledo, A.; Olofsson, B.; Hartman, E.; Heusel, M.; Malmström, J.; Malmström, L. Epitope Mapping with Sidewinder: An XL-MS and Structural Modeling Approach. Int. J. Mol. Sci. 2025, 26, 1488. https://doi.org/10.3390/ijms26041488
Ströbaek J, Tang D, Gueto-Tettay C, Gomez Toledo A, Olofsson B, Hartman E, Heusel M, Malmström J, Malmström L. Epitope Mapping with Sidewinder: An XL-MS and Structural Modeling Approach. International Journal of Molecular Sciences. 2025; 26(4):1488. https://doi.org/10.3390/ijms26041488
Chicago/Turabian StyleStröbaek, Joel, Di Tang, Carlos Gueto-Tettay, Alejandro Gomez Toledo, Berit Olofsson, Erik Hartman, Moritz Heusel, Johan Malmström, and Lars Malmström. 2025. "Epitope Mapping with Sidewinder: An XL-MS and Structural Modeling Approach" International Journal of Molecular Sciences 26, no. 4: 1488. https://doi.org/10.3390/ijms26041488
APA StyleStröbaek, J., Tang, D., Gueto-Tettay, C., Gomez Toledo, A., Olofsson, B., Hartman, E., Heusel, M., Malmström, J., & Malmström, L. (2025). Epitope Mapping with Sidewinder: An XL-MS and Structural Modeling Approach. International Journal of Molecular Sciences, 26(4), 1488. https://doi.org/10.3390/ijms26041488





