Identification of Salivary Exosome-Derived miRNAs as Potential Biomarkers of Bone Remodeling During Orthodontic Tooth Movement
Abstract
1. Introduction
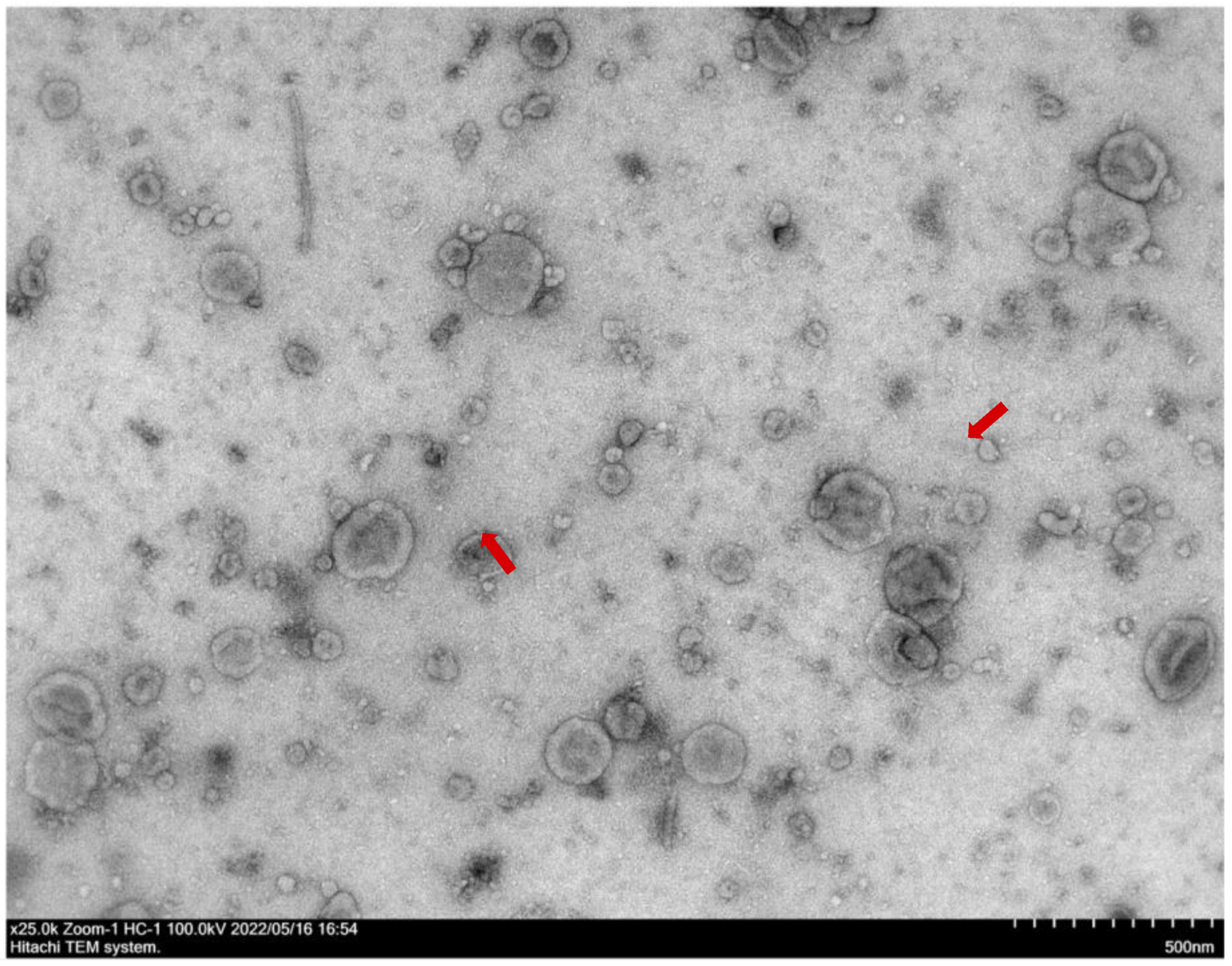
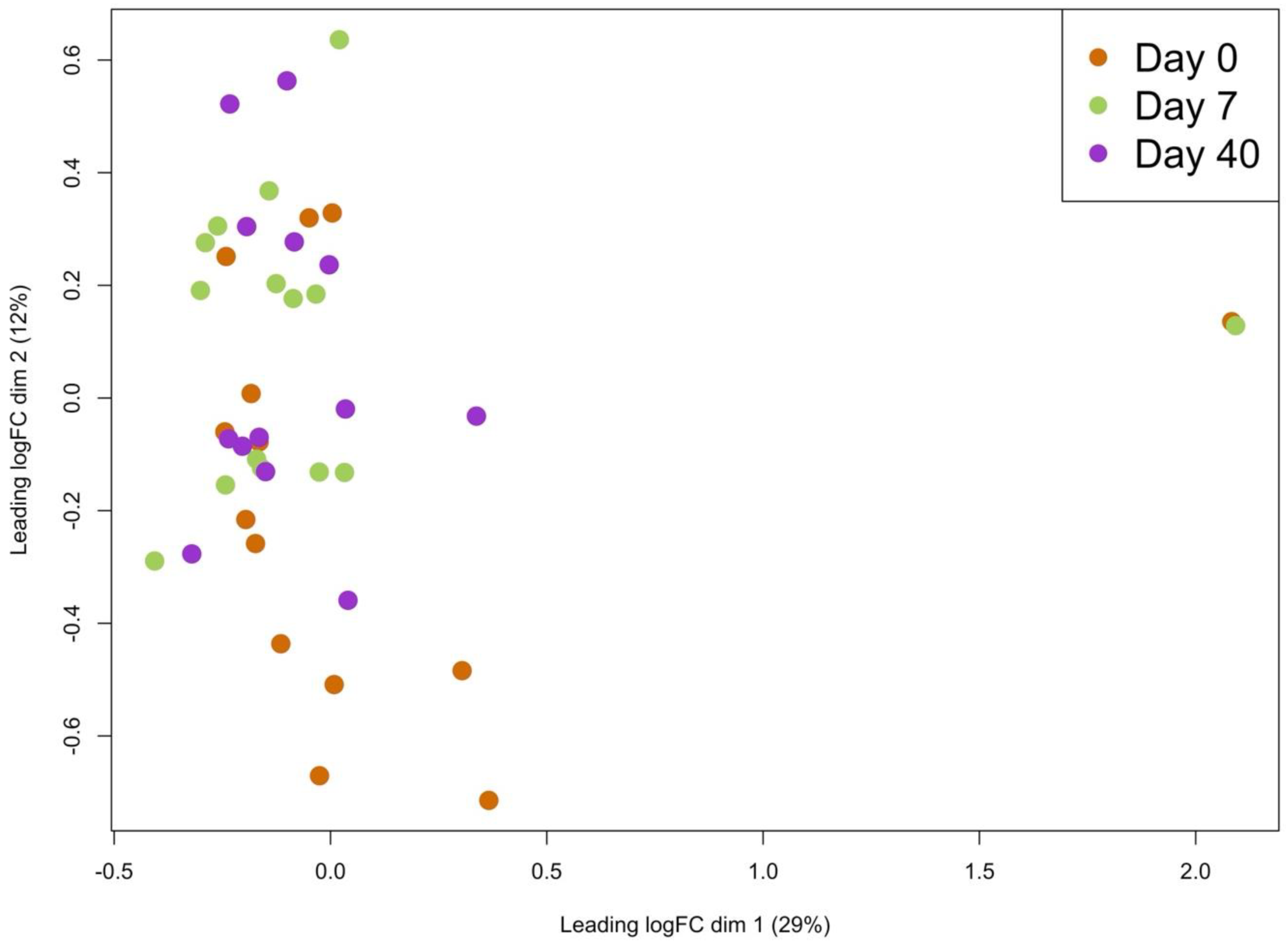
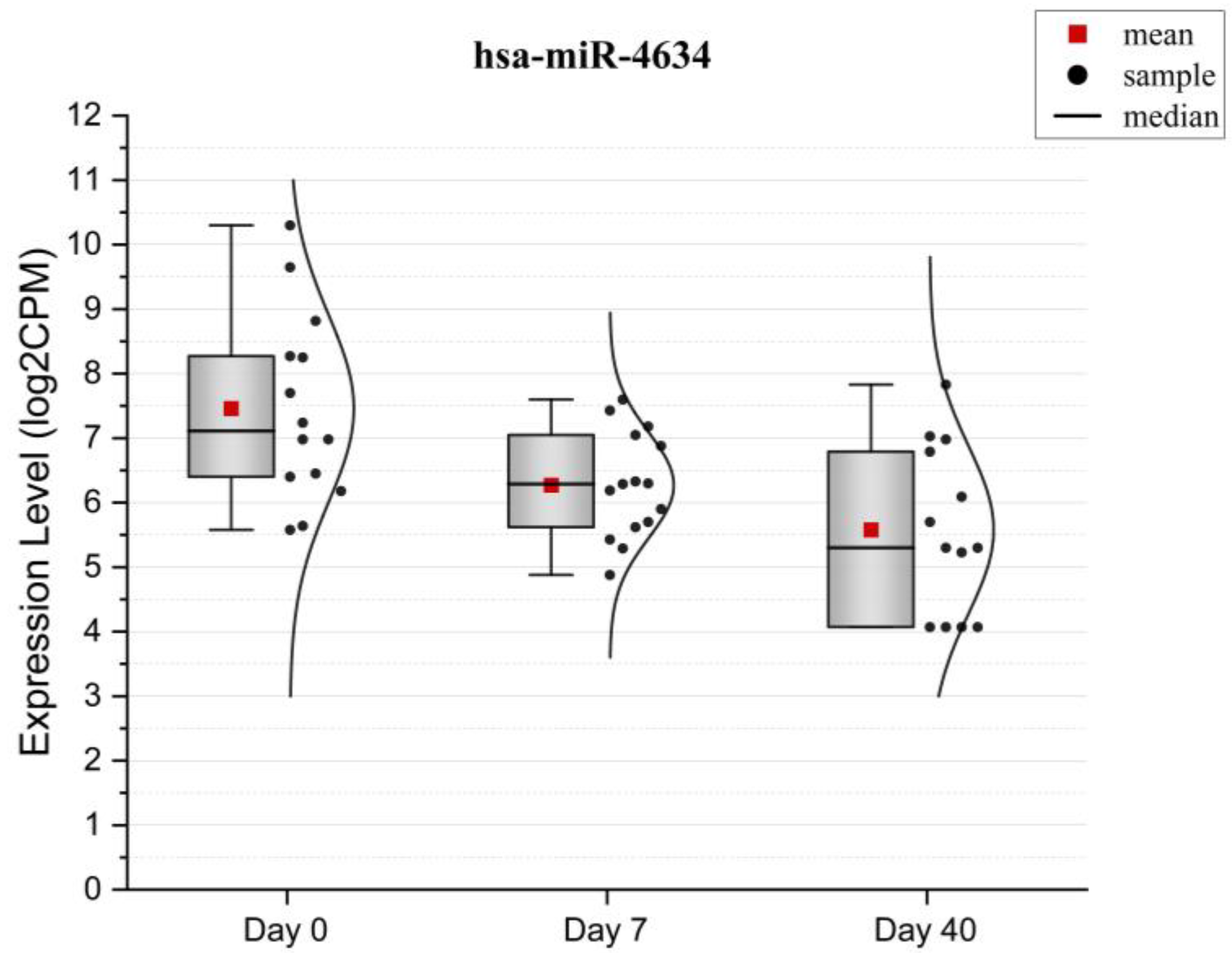
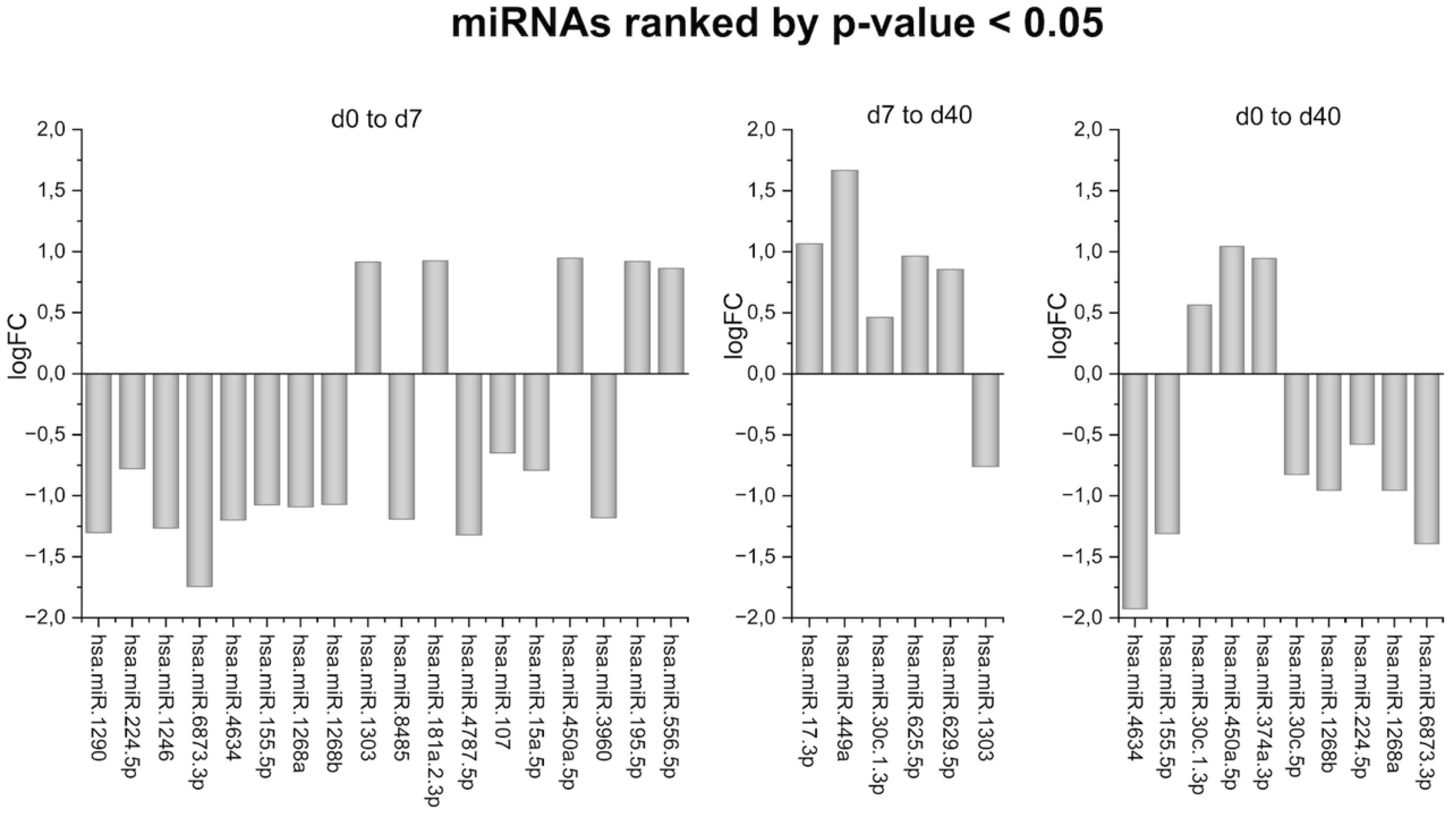
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Size Calculation
4.3. Saliva Collection
4.4. RNA Isolation
4.5. RNA Quantification
4.6. Library Preparation
4.7. Library Quality Control
4.8. Sequencing
4.9. Statistical and Bioinformatical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OTM | orthodontic tooth movement |
| EVs | extracellular vesicles |
| GO | gene ontology |
| NTA TEM | nanoparticle tracking analysis transmission electron microscopy |
References
- Kapoor, P.; Chowdhry, A.; Bagga, D.K.; Bhargava, D.; Aishwarya, S. MicroRNAs in Oral Fluids (Saliva and Gingival Crevicular Fluid) as Biomarkers in Orthodontics: Systematic Review and Integrated Bioinformatic Analysis. Prog. Orthod. 2021, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Atsawasuwan, P.; Lazari, P.; Chen, Y.; Zhou, X.; Viana, G.; Evans, C.A. Secretory MicroRNA-29 Expression in Gingival Crevicular Fluid during Orthodontic Tooth Movement. PLoS ONE 2018, 13, e0194238. [Google Scholar] [CrossRef] [PubMed]
- Großhans, H. Regulation of MicroRNAs; Springer Science & Business Media: Berlin, Germany, 2010. [Google Scholar]
- Zhang, B.; Yang, L.; Zheng, W.; Lin, T. MicroRNA-34 Expression in Gingival Crevicular Fluid Correlated with Orthodontic Tooth Movement. Angle Orthod. 2020, 90, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Pi, C.; Li, Y.P.; Zhou, X.; Gao, B. The Expression and Function of MicroRNAs in Bone Homeostasis. Differentiation 2015, 16, 17. [Google Scholar]
- Wang, C.; Liao, H.; Cao, Z. Role of Osterix and MicroRNAs in Bone Formation and Tooth Development. Med. Sci. Monit. 2016, 22, 2934–2942. [Google Scholar] [CrossRef]
- Park, M.G.; Kim, J.S.; Park, S.Y.; Lee, S.A.; Kim, H.J.; Kim, C.S.; Kim, J.S.; Chun, H.S.; Park, J.C.; Kim, D.K. MicroRNA-27 Promotes the Differentiation of Odontoblastic Cell by Targeting APC and Activating Wnt/β-Catenin Signaling. Gene 2014, 538, 266–272. [Google Scholar] [CrossRef]
- Wang, N. Mechanotransduction Accros the Cell Surface and Through the Cytoskeleton. Science 1993, 260, 1124–1127. [Google Scholar] [CrossRef]
- Shyy, J.Y.-J.; Chient, S. Role of Integrins in Cellular Responses to Mechanical Stress and Adhesion. Curr. Opin. Cell Biol. 1997, 9, 707–713. [Google Scholar] [CrossRef]
- Meyer, C.J.; Alenghat, F.J.; Rim, P.; Hwai, J.; Fong, J.; Fabry, B.; Ingber, D.E. Mechanical Control of Cyclic AMP Signalling and Gene Transcription through Integrins. Nat. Cell Biol. 2000, 2, 666–668. [Google Scholar] [CrossRef]
- Wilson, E.; Sudhir, K.; Ives, H.E. Mechanical Strain of Rat Vascular Smooth Muscle Cells Is Sensed by Specific Extracellular Matrix/Integrin Interactions. J. Clin. Investig. 1995, 96, 2364–2372. [Google Scholar] [CrossRef]
- Clark, E.A.; Brugget, J.S. A Integrins and Signal Transduction Pathways: The Road Taken. Science 1995, 268, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, D.R.; Nedwin, G.E.; Bringman, T.S.; Smith, D.D.; Mundy, G.R. Stimulation of Bone Resorption and inhibition of Bone formation in Vitro by Human Tumour Necrosis Factors. Nature 1986, 319, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin Ligand Is a Cytokine That Regulates Osteoclast Differentiation and Activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, H.; Shima, N.; Nakagawa, N.; Yamaguchi, K.; Kinosaki, M.; Mochizuki, S.-I.; Tomoyasu, A.; Yano, K.; Goto, M.; Murakami, A.; et al. Osteoclast Differentiation Factor Is a Ligand for Osteoprotegerin Osteoclastogenesis-Inhibitory Factor and Is Identical to TRANCERANKL. Proc. Natl. Acad. Sci. USA 1998, 95, 3597–3602. [Google Scholar] [CrossRef] [PubMed]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R. Osteoprotegerin: A Novel Secreted Protein Involved in the Regulation of Bone Density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef]
- Kapoor, P.; Monga, N.; Kharbanda, O.P.; Kapila, S.; Miglani, R.; Moganty, R. Effect of Orthodontic Forces on Levels of Enzymes in Gingival Crevicular Fluid (GCF): A Systematic Review. Dent. Press J. Orthod. 2019, 24, e1–e40. [Google Scholar] [CrossRef]
- De Aguiar, M.C.S.; Perinetti, G.; Capelli, J. The Gingival Crevicular Fluid as a Source of Biomarkers to Enhance Efficiency of Orthodontic and Functional Treatment of Growing Patients. BioMed Res. Int. 2017, 2017, 3257235. [Google Scholar] [CrossRef]
- Almehmadi, A.H.; Alghamdi, F. Biomarkers of Alveolar Bone Resorption in Gingival Crevicular Fluid: A Systematic Review. Arch. Oral Biol. 2018, 93, 12–21. [Google Scholar] [CrossRef]
- Kapoor, P.; Kharbanda, O.P.; Monga, N.; Miglani, R.; Kapila, S. Effect of Orthodontic Forces on Cytokine and Receptor Levels in Gingival Crevicular Fluid: A Systematic Review. Prog. Orthod. 2014, 15, 65. [Google Scholar] [CrossRef]
- Frazão, D.R.; Né, Y.G.d.S.; Ferreira, M.K.M.; Fagundes, N.C.F.; Marañón-Vásquez, G.; Maia, L.C.; Pithon, M.M.; Lima, R.R. Changes in Biomarkers Levels from Gingival Crevicular Fluid in Pre- and Postmenopausal Women Undergoing Orthodontic Treatment: A Systematic Review. J. Orofac. Orthop. 2024, 85, 223–232. [Google Scholar] [CrossRef]
- Prasad, S.; Tyagi, A.K.; Aggarwal, B.B. Detection of Inflammatory Biomarkers in Saliva and Urine: Potential in Diagnosis, Prevention, and Treatment for Chronic Diseases. Exp. Biol. Med. 2016, 241, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Holliday, L.S.; Truzman, E.; Zuo, J.; Han, G.; Torres-Medina, R.; Rody, W.J. Extracellular Vesicle Identification in Tooth Movement Models. Orthod. Craniofac. Res. 2019, 22, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Bartold, P.M.; Ivanovski, S. The Emerging Role of Small Extracellular Vesicles in Saliva and Gingival Crevicular Fluid as Diagnostics for Periodontitis. J. Periodontal. Res. 2022, 57, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2015; Volume 1295, pp. 179–209. [Google Scholar]
- Simpson, R.J.; Lim, J.W.E.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Balmayor, E.R.; Van Griensven, M. MiRNAs Related to Skeletal Diseases. Stem Cells Dev. 2016, 25, 1261–1281. [Google Scholar] [CrossRef]
- Kelch, S.; Balmayor, E.R.; Seeliger, C.; Vester, H.; Kirschke, J.S.; Van Griensven, M. MiRNAs in Bone Tissue Correlate to Bone Mineral Density and Circulating MiRNAs Are Gender Independent in Osteoporotic Patients. Sci. Rep. 2017, 7, 15861. [Google Scholar] [CrossRef]
- Cheshmi, B.; Cheshomi, H. Salivary Exosomes: Properties, Medical Applications, and Isolation Methods. Mol. Biol. Rep. 2020, 47, 6295–6307. [Google Scholar] [CrossRef]
- Nik Mohamed Kamal, N.N.S.B.; Shahidan, W.N.S. Non-Exosomal and Exosomal Circulatory MicroRNAs: Which Are More Valid as Biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef]
- Skoufos, G.; Kakoulidis, P.; Tastsoglou, S.; Zacharopoulou, E.; Kotsira, V.; Miliotis, M.; Mavromati, G.; Grigoriadis, D.; Zioga, M.; Velli, A.; et al. TarBase-v9.0 Extends Experimentally Supported miRNA-gene Interactions to Cell-Types and Virally Encoded miRNAs. Nucleic Acids Res. 2024, 52, D304–D310. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kosior-Jarecka, E.; Czop, M.; Gasińska, K.; Wróbel-Dudzińska, D.; Zalewski, D.P.; Bogucka-Kocka, A.; Kocki, J.; Żarnowski, T. MicroRNAs in the Aqueous Humor of Patients with Different Types of Glaucoma. Graefe’s Arch. Clin. Exp. Ophthalmol. 2021, 259, 2337–2349. [Google Scholar] [CrossRef] [PubMed]
- Faccio, R.; Teitelbaum, S.L.; Fujikawa, K.; Chappel, J.; Zallone, A.; Tybulewicz, V.L.; Ross, F.P.; Swat, W. Vav3 Regulates Osteoclast Function and Bone Mass. Nat. Med. 2005, 11, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Liao, L.; Su, X. Role of Mechano-Sensitive Non-Coding RNAs in Bone Remodeling of Orthodontic Tooth Movement: Recent Advances. Prog. Orthod. 2022, 23, 55. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Davidovitch, Z. Cellular, Molecular, and Tissue-Level Reactions to Orthodontic Force. Am. J. Orthod. Dentofac. Orthop. 2006, 129, e1–e469. [Google Scholar] [CrossRef]
- Bhavna, S. Current Concepts and Applications in Orthodontic Practice Biology of Orthodontic Tooth Movement; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Holland, R.; Bain, C.; Utreja, A. Osteoblast Differentiation during Orthodontic Tooth Movement. Orthod. Craniofac. Res. 2019, 22, 177–182. [Google Scholar] [CrossRef]
- Yoshino, H.; Morita, I.; Murota, S.I.; Ishikawa, I. Mechanical Stress Induces Production of Angiogenic Regulators in Cultured Human Gingival and Periodontal Ligament Fibroblasts. J. Periodontal Res. 2003, 38, 405–410. [Google Scholar] [CrossRef]
- Moin, S.; Kalajzic, Z.; Utreja, A.; Nihara, J.; Wadhwa, S.; Uribe, F.; Nanda, R. Osteocyte Death during Orthodontic Tooth Movement in Mice. Angle Orthod. 2014, 84, 1086–1092. [Google Scholar] [CrossRef]
- Papachristou, D.J.; Papachroni, K.K.; Basdra, E.K.; Papavassiliou, A.G. Signaling Networks and Transcription Factors Regulating Mechanotransduction in Bone. BioEssays 2009, 31, 794–804. [Google Scholar] [CrossRef]
- Reseco, L.; Molina-Crespo, A.; Atienza, M.; Gonzalez, E.; Falcon-Perez, J.M.; Cantero, J.L. Characterization of Extracellular Vesicles from Human Saliva: Effects of Age and Isolation Techniques. Cells 2024, 13, 95. [Google Scholar] [CrossRef]
- Yu, W.; Li, S.; Zhang, G.; Xu, H.H.K.; Zhang, K.; Bai, Y. New Frontiers of Oral Sciences: Focus on the Source and Biomedical Application of Extracellular Vesicles. Front. Bioeng. Biotechnol. 2022, 10, 1023700. [Google Scholar] [CrossRef]
- Li, Y.; Zhan, Q.; Bao, M.; Yi, J.; Li, Y. Biomechanical and Biological Responses of Periodontium in Orthodontic Tooth Movement: Up-Date in a New Decade. Int. J. Oral Sci. 2021, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhao, N.; Peng, L.; Li, Z.; Liu, C.; You, Q.; Fang, B. Biological Characteristics of MicroRNAs Secreted by Exosomes of Periodontal Ligament Stem Cells Due to Mechanical Force. Eur. J. Orthod. 2023, 45, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, T.; Zhang, Z.; Su, J.; Wu, X.; Chen, L.; Ge, X.; Wang, X.; Jiang, N. Periodontal Ligament Cells Derived Small Extracellular Vesicles Are Involved in Orthodontic Tooth Movement. Eur. J. Orthod. 2022, 44, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Justino, A.B.; Teixeira, R.R.; Peixoto, L.G.; Jaramillo, O.L.B.; Espindola, F.S. Effect of Saliva Collection Methods and Oral Hygiene on Salivary Biomarkers. Scand. J. Clin. Lab. Investig. 2017, 77, 415–422. [Google Scholar] [CrossRef]
- Sánchez-Pérez, L.; Irigoyen-Camacho, E.; Sáenz-Martínez, L.; Zepeda Zepeda, M.; Acosta-Gío, E.; Méndez-Ramírez, I. Stability of Unstimulated and Stimulated Whole Saliva Flow Rates in Children. Int. J. Paediatr. Dent. 2016, 26, 346–350. [Google Scholar] [CrossRef]
- Muhlemann, H.R.; Son, S. Gingival Sulcus Bleeding—A Leading Symptom in Initial Gingivitis. Helv. Odontol. Acta 1971, 15, 107–113. [Google Scholar]
- Lange, D.E.; Plagmann, H.C.; Eenboom, A.; Promesberger, A. Clinical Methods for the Objective Evaluation of Oral Hygiene. Deutsche Zahnarztliche Z. 1977, 32, 7–44. [Google Scholar]
- Meyle, J.; Jepsen, S. The Periodontal Screening-Index (PSI). Parodontologie 2000, 11, 17–21. [Google Scholar]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Routledge: Oxfordshire, UK, 2013; ISBN 9781134742707. [Google Scholar]
- Janz, N.K. Exploratory Analyses and Their Role in the Design and Interpretation of Clinical Studies. J. Clin. Epidemiol. 2016, 72, 24–31. [Google Scholar]
- Wassall, R.R.; Preshaw, P.M. Clinical and Technical Considerations in the Analysis of Gingival Crevicular Fluid. Periodontol. 2000 2015, 70, 65–79. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal Information for Studies of Extracellular Vesicles (MISEV2023): From Basic to Advanced Approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The Nf-Core Framework for Community-Curated Bioinformatics Pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow Enables Reproducible Computational Workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef] [PubMed]


| miRNAs | Osteoblast Differentiation | Positive Regulation of Osteoblast Differentiation | Positive Regulation of Bone Mineralization |
|---|---|---|---|
| hsa-miR-107 | ADAP2, ALS2, ALS2CL, ARHGAP12, BVES, CRK, DAB2IP, EFNA5, EPHA1, EPHA3, EPHA4, EPHA5, EPHB3, FGD3, FGD4, ITGB1BP1, NF1, NTRK2, PLXNA1, PLXNA2, PLXNA3, PLXNA4, PLXNB2, PLXNB3, PLXND1, PRKG1, PROM2, PTK2, RAB3GAP1, RAB3GAP2, RABGAP1, RABGAP1L, RALBP1, RAP1GAP, RAPGEF6, RDX, SIPA1L1, SOD1, STMN3, TBC1D10B, VAV2, VAV3 | ACVR1, ACVR2A, ACVR2B, ATRAID, BMP2, BMPR1A, BMPR1B, BMPR2, CCN1, CEBPB, CTHRC1, CTNNB1, CTNNBIP1, FAM20C, FBN2, FBXO5, FERMT2, FGF2, HGF, IFITM1, IGF1, IL6, IL6ST, ILK, JAG1, JUND, LRP5, PPP3CA, SMAD1, SMAD5, SOX11, SUCO, TENT5A, TP63, VEGFA, WNT10B, WNT3, WNT7B, WWTR1, YAP1, ZHX3 | ACVR1, ACVR2A, ACVR2B, ADRB2, ANO6, ATP2B1, ATRAID, BMP2, BMPR1A, BMPR1B, BMPR2, CCN1, FAM20C, FBN2, GPM6B, OSR1, OSR2, RXRA, SLC20A2, SLC8A1, SMAD3, TENT5A, TFAP2A, VDR, WNT10B |
| hsa-miR-1246 | CEBPB, IL6ST, SMAD5, WWTR1 | ATP2B1 | |
| hsa-miR-1290 | ARHGAP12, CRK, FGD4, RABGAP1, RALBP1 | IL6ST, SUCO | |
| hsa-miR-1303 | NF1 | JAG1, PPP3CA | |
| hsa-miR-155-5p | ARHGAP12, CRK, EPHA4, EPHA5, EPHB3, NF1, NTRK2, PLXNA2, PLXNA3, PLXNB1, PLXND1, RAB3GAP1, RABGAP1, RABGAP1L, RAPGEF6, RDX, TBC1D15, VAV2 | ACVR2A, ACVR2B, BMP4, BMP6, BMPR1A, BMPR2, CCN1, CEBPB, CEBPD, CTNNB1, FBXO5, FERMT2, FGF2, FZD1, HGF, IFITM1, IGF1, IL6, IL6R, IL6ST, ILK, JAG1, MEF2C, RUNX2, SMAD1, SMAD5, TP63, VEGFA, WWTR1, YAP1 | ACVR2A, ACVR2B, ADRB2, ATP2B1, BMP4, BMP6, BMPR1A, BMPR2, CCN1, GPM6B, ISG15, MEF2C, RXRB, SMAD3, TFAP2A |
| hsa-miR-15a-5p | ADAP1, ADAP2, ALS2, ALS2CL, ARHGAP12, ARHGAP44, ARHGAP9, ARHGEF5, BVES, CRK, DAB2IP, EFNA5, EPHA1, EPHA3, EPHA4, EPHA5, EPHB3, FGD1, FGD3, FGD4, FGD5, FGD6, GPR137B, ITGB1BP1, NF1, PLXNA1, PLXNA2, PLXNA3, PLXNA4, PLXNB1, PLXNB2, PLXNB3, PLXNC1, PLXND1, PRKG1, PROM2, PTK2, RAB3GAP1, RAB3GAP2, RABGAP1, RABGAP1L, RALBP1, RAP1GAP, RAPGEF6, RASIP1, RDX, SIPA1L1, SOD1, STMN3, TBC1D10B, TBC1D15, VAV2, VAV3 | ACVR1, ACVR2A, ACVR2B, ATRAID, BMP2, BMP6, BMPR1A, BMPR1B, BMPR2, CCN1, CEBPB, CTHRC1, CTNNB1, CTNNBIP1, DDR2, FAM20C, FBN2, FBXO5, FERMT2, FGF2, FZD1, GLI3, HGF, IFITM1, IGF1, IL6, IL6R, IL6ST, JAG1, JUND, LRP3, LRP5, MEF2C, MSX2, NPNT, PPP3CA, RUNX2, SCUBE2, SCUBE3, SFRP2, SMAD1, SMAD5, SOX11, SUCO, TENT5A, VEGFA, WNT10B, WNT3, WNT7B, WWTR1, YAP1, ZHX3 | ACVR1, ACVR2A, ACVR2B, ADGRV1, ADRB2, ANO6, ATP2B1, ATRAID, BMP2, BMP6, BMPR1A, BMPR1B, BMPR2, CCN1, FAM20C, FBN2, GPM6B, KL, MEF2C, OSR2, P2RX7, PKDCC, RXRA, RXRB, SLC20A2, SLC8A1, SMAD3, TENT5A, TFAP2A, VDR, WNT10B |
| hsa-miR-181a-2-3p | ARHGAP9, CRK, PLXNA2, PLXNA4, PLXND1, RABGAP1, RAPGEF6, SIPA1L1, TBC1D15 | ACVR2A, ACVR2B, BMPR2, CTNNB1, CTNNBIP1, FERMT2, JAG1, LRP5, PPP3CA, SMAD5, SOX11, SUCO, VEGFA, YAP1 | ACVR2A, ACVR2B, BMPR2, RXRB |
| hsa-miR-195-5p | ALS2, ALS2CL, ARHGAP12, BVES, CRK, EFNA5, EPHA1, EPHA4, EPHA5, FGD1, NF1, PLXNA1, PLXNA3, PLXNB2, PLXNB3, PLXND1, PRKG1, PTK2, RAB3GAP1, RAB3GAP2, RABGAP1, RABGAP1L, RAP1GAP, SIPA1L1, SOD1, STMN3, TBC1D10B, TBC1D15, VAV2, VAV3 | ACVR2A, ACVR2B, BMPR2, CCN1, CEBPB, CTNNB1, CTNNBIP1, DDR2, FBN2, FBXO5, FERMT2, FGF2, FZD1, HGF, IGF1, IL6R, IL6ST, LRP3, LRP5, MEF2C, NPNT, PPP3CA, SMAD1, SMAD5, SOX11, SUCO, TENT5A, VEGFA, WNT3, WNT7B, WWTR1, YAP1, ZHX3 | ACVR2A, ACVR2B, ADGRV1, ADRB2, ANO6, ATP2B1, BMPR2, CCN1, FBN2, MEF2C, OSR1, OSR2, RXRA, RXRB, SLC20A2, SMAD3, TENT5A, TFAP2A, VDR |
| hsa-miR-224-5p | CRK, NTRK2, SOD1 | ATRAID, BMPR2, CTNNB1, FERMT2, IL6R, IL6ST, JUND, PPP3CA, SMAD5, SUCO, TENT5A, YAP1 | ATRAID, BMPR2, SLC20A2, TENT5A, VDR |
| hsa-miR-450a-5p | BVES, DAB2IP, EPHB3, FGD1, FGD6, NF1, PLXNA2, PLXNB2, PLXNC1, PLXND1, RABGAP1, RALBP1, RDX | ACVR1, ATRAID, BMP2, BMPR1A, BMPR2, CCN1, CEBPD, CTNNB1, DDR2, FAM20C, FBN2, FERMT2, FGF2, HGF, JAG1, JUND, MEF2C, SFRP2, SUCO, TENT5A, TP63, VEGFA, WWTR1, YAP1, ZHX3 | ACVR1, ATRAID, BMP2, BMPR1A, BMPR2, CCN1, FAM20C, FBN2, ISG15, MEF2C, PTN, RXRA, SLC20A2, SMAD3, TENT5A, TFAP2A |
| hsa-miR-8485 | ILK |
| miRNAs | Osteoblast Differentiation | Positive Regulation of Osteoblast Differentiation | Osteoclast Differentiation |
|---|---|---|---|
| hsa-miR-1303 | DDX21, GJA1, NF1, SEMA7A, SMAD4 | JAG1, PPP3CA | FOXP1, NF1, PIK3R1, TFRC |
| hsa-miR-17-3p | AKT1, BMPR2, CLTC, CTNNB1, DHX9, FASN, GABBR1, HIRA, HNRNPC, HNRNPU, LGR4, MAPK14, MEF2D, NF1, RPS15, RRBP1, RSL1D1, SMAD4, SMAD5, SNAI1, SND1, SNRNP200, SYNCRIP, TNC, UFL1, VCAN | ACVR1, ACVR2A, ACVR2B, BMPR2, CTNNB1, HGF, IGF1, IL6R, IL6ST, JAG1, LRP3, LRP5, PPP3CA, SMAD5, YAP1 | CSF1, CSF1R, CTNNB1, FARP2, FOXP1, GLO1, MAPK14, NF1, SLC4A2, TFRC, TOB2 |
| hsa-miR-30c-1-3p | ADAR, ATP5F1B, BMP2, BMPR1B, BMPR2, CAT, CBFB, CLTC, COL6A1, CREB3L1, CTNNB1, DDX21, DHX9, EPHA2, FASN, FBL, FGF2, FOSL2, GJA1, GLI3, GTPBP4, HIRA, HNRNPC, HNRNPU, IGFBP5, JUNB, LGR4, MAPK14, MEF2D, NF1, RDH14, RRBP1, RSL1D1, RUNX2, SEMA7A, SMAD4, SMAD5, SNAI2, SND1, SNRNP200, TNC, TP53INP2, UFL1, WWTR1 | ACVR2B, ATRAID, BMP2, BMPR1B, BMPR2, CLIC1, CTNNB1, DDR2, FERMT2, FGF2, GLI3, IL6R, IL6ST, ILK, JAG1, JUND, LRP5, NPNT, PPP3CA, RUNX2, SMAD1, SMAD5, SUCO, TENT5A, WWTR1, YAP1 | BMP2, CSF1, CTNNB1, EPHA2, FOSL2, FOXP1, GLO1, IREB2, JUNB, MAPK14, MITF, NF1, PIK3R1, SBNO2, SLC4A2, TFRC, TNF, TOB2 |
| hsa-miR-449a | ADAR, AKT1, ALPL, ATP5F1B, AXIN2, BMP4, BMPR1A, BMPR2, CAT, CCDC47, CLTC, DDX21, DNAJC13, EPHA2, FASN, FHL2, FIGNL1, GLI3, HIRA, HSPE1, IARS1, IGFBP3, IGFBP5, ITGA11, LGR4, MAPK14, MSX2, NF1, RSPO2, SEMA7A, SMAD4, SND1, SNRNP200, TNC, TP53INP2, VCAN | ACVR1, ATRAID, BMP4, BMPR1A, BMPR2, CEBPA, CEBPB, FBN2, FERMT2, GLI3, IL6R, ILK, JAG1, MSX2, NPNT, NPPC, SOX11, SUCO, VEGFA, YAP1 | EPHA2, FOS, MAPK14, NF1, OSTM1, PIK3R1, TOB2, TYROBP |
| hsa-miR-625-5p | ADAR, AKT1, BCAP29, BMP2, BMPR2, CBFB, CLTC, COL1A1, COL6A1, CTNNB1, DDX21, DNAJC13, EPHA2, FASN, FBL, FGF2, FOSL2, GABBR1, GTPBP4, HAND2, HIRA, HNRNPC, HNRNPU, MEF2D, NF1, RBMX, RPS15, RUNX2, SEMA7A, SMAD4, SMAD5, SNRNP200, SYNCRIP, VCAN | ACVR1, ACVR2B, BMP2, BMPR2, CEBPB, CLIC1, CTNNB1, DDR2, FERMT2, FGF2, IGF1, JAG1, PPP3CA, RUNX2, SMAD5, VEGFA, WNT3, YAP1 | BMP2, CREB1, CTNNB1, EPHA2, FOS, FOSL2, FOXP1, GLO1, IREB2, MITF, NF1, SBNO2, SLC4A2, TCIRG1, TFRC, TOB2, TRAF6 |
| hsa-miR-629-5p | ATP5F1B, BMP2, BMPR2, CLTC, COL1A1, CTNNB1, DDX21, FASN, FOSL2, HNRNPU, IFT80, IGFBP5, LGR4, MEF2C, MEF2D, PSMC2, RSL1D1, SEMA7A, SMAD5, SNRNP200, VCAN | ACVR2A, BMP2, BMPR2, CEBPB, CTNNB1, FERMT2, MEF2C, PPP3CA, SMAD5, YAP1 | BMP2, CTNNB1, FOSL2, FOXP1, PIK3R1, TMEM64 |
| miRNAs | Osteoblast Differentiation | Positive Regulation of Osteoblast Differentiation | Osteoclast Differentiation | Positive Regulation of Bone Mineralization |
|---|---|---|---|---|
| hsa-miR-1268a | CDK12, POLR2D, SCAF1 | |||
| hsa-miR-1268b | CCNT1, IRF9, SCAF1 | |||
| hsa-miR-155-5p | ABT1, ADRB2, ARID4A, ASH1L, ATRX, ATXN1L, BCL9, BMP4, CBFB, CCNT1, CCNT2, CDK12, CDK7, CDK9, CEBPB, CEBPD, COPS2, CTNNB1, DDX21, DEK, DRAP1, EDN1, ETS1, ETV1, FGF2, FOS, FOSB, GLIS3, GTF2A1, GTF2A2, GTF2H1, GTF2H3, IRF9, ISL1, JAK2, KLF4, MAF, MAPK14, MNT, NCAPG2, NFAT5, NFATC2, NFIX, NFKB1, NFX1, NFYA, NR4A1, PAXBP1, PIK3R1, POLR2B, POLR2C, POLR2J, PRDM4, PROX1, RPAP1, RREB1, SCAF1, SMAD1, SMARCC2, SNAPC3, SOX9, TAF1, TAF12, TAF5L, TAF7, TCEA1, TEAD2, TFAP2B, TFDP1, TP63, TRIM24, TRIP11, TRIP13, TSC22D1, XBP1 | ACVR2A, ACVR2B, BMP4, BMP6, BMPR1A, BMPR2, CCN1, CEBPB, CEBPD, CTNNB1, FBXO5, FERMT2, FGF2, FZD1, HGF, IFITM1, IGF1, IL6, IL6R, IL6ST, ILK, JAG1, MEF2C, RUNX2, SMAD1, SMAD5, TP63, VEGFA, WWTR1, YAP1 | CREB1, CSF1R, CTNNB1, FOS, FOSL2, FOXP1, GLO1, IREB2, JUNB, MAPK14, MITF, NF1, OSTM1, PIK3R1, SLC4A2, SNX10, TFRC, TMEM64, TNF | ACVR2A, ACVR2B, ADRB2, ATP2B1, BMP4, BMP6, BMPR1A, BMPR2, CCN1, GPM6B, ISG15, MEF2C, RXRB, SMAD3, TFAP2A |
| hsa-miR-224-5p | ATF4, BCL6, CCNT2, CTNNB1, ERCC2, ERCC3, GTF2H5, KLF13, NFAT5, NFIX, NR4A1, PARP1, POLR2F, PRDM4, SQSTM1, TAF1, TAF10, TAF5L, TFDP1, TSC22D1, XBP1 | ATRAID, BMPR2, CTNNB1, FERMT2, IL6R, IL6ST, JUND, PPP3CA, SMAD5, SUCO, TENT5A, YAP1 | CTNNB1, EPHA2, GLO1, IREB2 | ATRAID, BMPR2, SLC20A2, TENT5A, VDR |
| hsa-miR-30c-1-3p | ABLIM3, ADRB2, AR, ARID4A, ARRB2, ASH1L, ATF4, ATRX, ATXN1L, BCL9, BCL9L, BMP2, CBFB, CCNK, CCNT1, CCNT2, CDK12, CHD7, COPS2, CTNNB1, DDX21, FGF2, FOSB, FOXP3, GTF2A1, GTF2H1, HOXA13, INHBA, IRF9, JAK2, KLF4, KPNA6, MAPK14, MED16, MNT, MYEF2, NFAT5, NFIC, NFIX, NFX1, NFYA, PARP1, PAX5, PIK3R1, PKNOX1, POLR2A, POLR2B, POLR2E, PRDM4, RB1, RREB1, SCAF1, SMAD1, SMAD4, SMARCC2, SOX9, SPDEF, SQSTM1, SRF, TAF1, TAF10, TAF5L, TBP, TRIP13, TSC22D1, ZNF768 | ACVR2B, ATRAID, BMP2, BMPR1B, BMPR2, CLIC1, CTNNB1, DDR2, FERMT2, FGF2, GLI3, IL6R, IL6ST, ILK, JAG1, JUND, NPNT, PPP3CA, RUNX2, SMAD1, SMAD5, SUCO, TENT5A, WWTR1, YAP1 | BMP2, CSF1, CTNNB1, EPHA2, FOSL2, FOXP1, GLO1, IREB2, JUNB, MAPK14, MITF, NF1, PIK3R1, SBNO2, SLC4A2, TFRC, TNF, TOB2 | ACVR2B, ADRB2, ANO6, ATP2B1, ATRAID, BMP2, BMPR1B, BMPR2, RXRA, RXRB, SLC20A2, SMAD3, TENT5A, VDR |
| hsa-miR-30c-5p | ADRB2, AR, ARID4A, ARID4B, ARRB2, ASH1L, ATRX, ATXN1L, BCL11B, BCL9, CCNK, CCNT2, CDK12, CHD7, CTNNB1, DDX21, ETS1, GMEB2, GTF2A1, GTF2E2, GTF2H1, KLF13, KLF4, KPNA6, LHX1, MNT, MYEF2, NCAPG2, NFAT5, NFIC, NFKB1, NFX1, NOTCH1, PAXBP1, PIK3R1, PKNOX1, POLR2B, POLR2D, SLC40A1, SMAD4, SMARCC2, SRF, TBPL1, TFDP1, THRA, XBP1 | ACVR1, BMPR2, CTHRC1, CTNNB1, FZD1, JAG1, MEF2C, PPP3CA, RUNX2, SMAD5, SUCO, VEGFA, YAP1 | CTNNB1, EPHA2, FOSL2, FOXP1, IREB2, JUNB, NF1, PIK3R1, SLC4A2, SNX10, TFRC, TMEM64, TOB2 | ACVR1, ADRB2, ATP2B1, BMPR2, MEF2C, SLC20A2 |
| hsa-miR-374a-3p | AR, ATXN1L, CBFB, CDK12, CTNNB1, DDX21, ETS1, NFAT5, NFIC, NFIX, NFKB1, NFYA, PARP1, PIK3R1, SCAF1 | BMPR2, CTNNB1, IL6, SMAD5 | CTNNB1, GLO1, JUNB, PIK3R1, SBNO2, SLC4A2, TMEM64 | ATP2B1, BMPR2 |
| hsa-miR-450a-5p | ARID4A, ASH1L, ATXN1L, BCL9, BCL9L, BMP2, CBFB, CCNT2, CDK7, CDK9, CEBPD, CTNNB1, DRAP1, ERCC2, ETV1, FGF2, FOS, FOSB, GATA2, GTF2A1, GTF2H2, ISL1, JAK2, MBD1, MLIP, NFAT5, NFATC4, NFIC, NFIX, NFKB1, NOTCH1, NR4A1, PIK3R1, POLR2A, PROX1, RBMX, RREB1, SMAD4, SNAPC3, SRF, TAF10, TAF12, TEAD2, TP63, TSC22D1, ZNF141 | ACVR1, ATRAID, BMP2, BMPR1A, BMPR2, CCN1, CEBPD, CLIC1, CTNNB1, DDR2, FBN2, FERMT2, FGF2, HGF, JAG1, JUND, MEF2C, SFRP2, SUCO, TENT5A, TP63, VEGFA, WWTR1, YAP1, ZHX3 | BMP2, CTNNB1, EPHA2, FOS, FOSL2, GPR183, JUNB, MITF, NF1, PIK3R1, TFRC, TMEM64 | ACVR1, ATRAID, BMP2, BMPR1A, BMPR2, CCN1, FBN2, ISG15, MEF2C, PTN, RXRA, SLC20A2, SMAD3, TENT5A, TFAP2A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazanopoulos, N.; Sideris, C.D.; Xu, Y.; Konstantonis, D.; Vastardis, H.; Balmayor, E.R.; Wolf, M.; Apel, C. Identification of Salivary Exosome-Derived miRNAs as Potential Biomarkers of Bone Remodeling During Orthodontic Tooth Movement. Int. J. Mol. Sci. 2025, 26, 1228. https://doi.org/10.3390/ijms26031228
Kazanopoulos N, Sideris CD, Xu Y, Konstantonis D, Vastardis H, Balmayor ER, Wolf M, Apel C. Identification of Salivary Exosome-Derived miRNAs as Potential Biomarkers of Bone Remodeling During Orthodontic Tooth Movement. International Journal of Molecular Sciences. 2025; 26(3):1228. https://doi.org/10.3390/ijms26031228
Chicago/Turabian StyleKazanopoulos, Nikolaos, Constantinos D. Sideris, Yong Xu, Dimitrios Konstantonis, Heleni Vastardis, Elizabeth R. Balmayor, Michael Wolf, and Christian Apel. 2025. "Identification of Salivary Exosome-Derived miRNAs as Potential Biomarkers of Bone Remodeling During Orthodontic Tooth Movement" International Journal of Molecular Sciences 26, no. 3: 1228. https://doi.org/10.3390/ijms26031228
APA StyleKazanopoulos, N., Sideris, C. D., Xu, Y., Konstantonis, D., Vastardis, H., Balmayor, E. R., Wolf, M., & Apel, C. (2025). Identification of Salivary Exosome-Derived miRNAs as Potential Biomarkers of Bone Remodeling During Orthodontic Tooth Movement. International Journal of Molecular Sciences, 26(3), 1228. https://doi.org/10.3390/ijms26031228








