Is DEXI a Multiple Sclerosis Susceptibility Gene?
Abstract
1. Multiple Sclerosis Genetics
2. The 16p13 Genetic Region and MS
3. Autoimmune-Associated SNPs in CLEC16A Act as Expression Quantitative Trait Loci for DEXI
4. DEXI Is Ubiquitously Expressed
5. DEXI Function
6. Discussions and Future Studies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | Multiple sclerosis |
| SNP | Single nucleotide polymorphism |
| eQTL | Expression quantitative trait locus |
| GWAS | Genome-wide association study |
| LD | Linkage disequilibrium |
| CNS | Central nervous system |
| DEXI | Dexamethasone-induced protein |
| CLEC16A | C-type lectin like domain family 16, member A |
| EBV | Epstein–Barr virus |
| PMA | Phorbol 12-myristate 13-acetate |
| IO | Ionomycin |
| DMSO | Dimethyl sulfoxide |
| T1D | Type-1 diabetes |
| NOD | Non-obese diabetic |
References
- International Multiple Sclerosis Genetics Consortium; Beecham, A.H.; Patsopoulos, N.A.; Xifara, D.K.; Davis, M.F.; Kemppinen, A.; Cotsapas, C.; Shah, T.S.; Spencer, C.; Booth, D.; et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013, 45, 1353–1360. [Google Scholar] [PubMed]
- International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium; Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar] [PubMed]
- International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium; MultipleMS Consortium. Locus for severity implicates CNS resilience in progression of multiple sclerosis. Nature 2023, 619, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Isobe, N.; Madireddy, L.; Khankhanian, P.; Matsushita, T.; Caillier, S.J.; More, J.M.; Gourraud, P.A.; McCauley, J.L.; Beecham, A.H.; International Multiple Sclerosis Genetics Consortium; et al. An ImmunoChip study of multiple sclerosis risk in African Americans. Brain 2015, 138 Pt 6, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium; Hafler, D.A.; Compston, A.; Sawcer, S.; Lander, E.S.; Daly, M.J.; De Jager, P.L.; de Bakker, P.I.; Gabriel, S.B.; Mirel, D.B.; et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007, 357, 851–862. [Google Scholar]
- GTEx. The GTEx Portal. Available online: https://www.gtexportal.org/ (accessed on 1 December 2024).
- Rubio, J.P.; Stankovich, J.; Field, J.; Tubridy, N.; Marriott, M.; Chapman, C.; Bahlo, M.; Perera, D.; Johnson, L.J.; Tait, B.D.; et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008, 9, 624–630. [Google Scholar] [CrossRef]
- D’Netto, M.J.; Ward, H.; Morrison, K.M.; Ramagopalan, S.V.; Dyment, D.A.; DeLuca, G.C.; Handunnetthi, L.; Sadovnick, A.D.; Ebers, G.C. Risk alleles for multiple sclerosis in multiplex families. Neurology 2009, 72, 1984–1988. [Google Scholar] [CrossRef]
- Hoppenbrouwers, I.A.; Aulchenko, Y.S.; Janssens, A.C.; Ramagopalan, S.V.; Broer, L.; Kayser, M.; Ebers, G.C.; Oostra, B.A.; van Duijn, C.M.; Hintzen, R.Q. Replication of CD58 and CLEC16A as genome-wide significant risk genes for multiple sclerosis. J. Hum. Genet. 2009, 54, 676–680. [Google Scholar] [CrossRef]
- Perera, D.; Stankovich, J.; Butzkueven, H.; Taylor, B.V.; Foote, S.J.; Kilpatrick, T.J.; Rubio, J.P. Fine mapping of multiple sclerosis susceptibility genes provides evidence of allelic heterogeneity at the IL2RA locus. J. Neuroimmunol. 2009, 211, 105–109. [Google Scholar] [CrossRef]
- Johnson, B.A.; Wang, J.; Taylor, E.M.; Caillier, S.J.; Herbert, J.; Khan, O.A.; Cross, A.H.; De Jager, P.L.; Gourraud, P.A.; Cree, B.C.; et al. Multiple sclerosis susceptibility alleles in African Americans. Genes Immun. 2010, 11, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Perdigones, N.; Cenit, M.C.; Espino, L.; Varade, J.; Lamas, J.R.; Santiago, J.L.; Fernandez-Arquero, M.; de la Calle, H.; Arroyo, R.; et al. Chromosomal region 16p13: Further evidence of increased predisposition to immune diseases. Ann. Rheum. Dis. 2010, 69, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Mero, I.L.; Ban, M.; Lorentzen, A.R.; Smestad, C.; Celius, E.G.; Saether, H.; Saeedi, H.; Viken, M.K.; Skinningsrud, B.; Undlien, D.E.; et al. Exploring the CLEC16A gene reveals a MS-associated variant with correlation to the relative expression of CLEC16A isoforms in thymus. Genes Immun. 2011, 12, 191–198. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Browser, G. The Genome Browser. Available online: http://genome.ucsc.edu (accessed on 1 December 2024).
- Berge, T.; Leikfoss, I.S.; Harbo, H.F. From Identification to Characterization of the Multiple Sclerosis Susceptibility Gene CLEC16A. Int. J. Mol. Sci. 2013, 14, 4476–4497. [Google Scholar] [CrossRef]
- Lincoln, M.R.; Connally, N.; Axisa, P.P.; Gasperi, C.; Mitrovic, M.; van Heel, D.; Wijmenga, C.; Withoff, S.; Jonkers, I.H.; Padyukov, L.; et al. Genetic mapping across autoimmune diseases reveals shared associations and mechanisms. Nat. Genet. 2024, 56, 838–845. [Google Scholar] [CrossRef]
- Kim, S.; Naylor, S.A.; DiAntonio, A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc. Natl. Acad. Sci. USA 2012, 109, E1072–E1081. [Google Scholar] [CrossRef]
- Kim, S.; Wairkar, Y.P.; Daniels, R.W.; DiAntonio, A. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J. Cell Biol. 2010, 188, 717–734. [Google Scholar] [CrossRef]
- Schuster, C.; Gerold, K.D.; Schober, K.; Probst, L.; Boerner, K.; Kim, M.J.; Ruckdeschel, A.; Serwold, T.; Kissler, S. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity 2015, 42, 942–952. [Google Scholar] [CrossRef]
- Soleimanpour, S.A.; Gupta, A.; Bakay, M.; Ferrari, A.M.; Groff, D.N.; Fadista, J.; Spruce, L.A.; Kushner, J.A.; Groop, L.; Seeholzer, S.H.; et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 2014, 157, 1577–1590. [Google Scholar] [CrossRef]
- Tam, R.C.; Lee, A.L.; Yang, W.; Lau, C.S.; Chan, V.S. Systemic Lupus Erythematosus Patients Exhibit Reduced Expression of CLEC16A Isoforms in Peripheral Leukocytes. Int. J. Mol. Sci. 2015, 16, 14428–14440. [Google Scholar] [CrossRef] [PubMed]
- van Luijn, M.M.; Kreft, K.L.; Jongsma, M.L.; Mes, S.W.; Wierenga-Wolf, A.F.; van Meurs, M.; Melief, M.J.; der Kant, R.; Janssen, L.; Janssen, H.; et al. Multiple sclerosis-associated CLEC16A controls HLA class II expression via late endosome biogenesis. Brain 2015, 138, 1531–1547. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, A.M.; Leikfoss, I.S.; Abrahamsen, G.; Sundvold, V.; Isom, M.M.; Keshari, P.K.; Rognes, T.; Landsverk, O.J.B.; Bos, S.D.; Harbo, H.F.; et al. Exploring the role of the multiple sclerosis susceptibility gene CLEC16A in T cells. Scand. J. Immunol. 2021, 94, e13050. [Google Scholar] [CrossRef]
- Redmann, V.; Lamb, C.A.; Hwang, S.; Orchard, R.C.; Kim, S.; Razi, M.; Milam, A.; Park, S.; Yokoyama, C.C.; Kambal, A.; et al. Clec16a is Critical for Autolysosome Function and Purkinje Cell Survival. Sci. Rep. 2016, 6, 23326. [Google Scholar] [CrossRef]
- Pandey, R.; Bakay, M.; Hain, H.S.; Strenkowski, B.; Elsaqa, B.Z.B.; Roizen, J.D.; Kushner, J.A.; Orange, J.S.; Hakonarson, H. CLEC16A regulates splenocyte and NK cell function in part through MEK signaling. PLoS ONE 2018, 13, e0203952. [Google Scholar] [CrossRef]
- Pandey, R.; Bakay, M.; Hain, H.S.; Strenkowski, B.; Yermakova, A.; Kushner, J.A.; Orange, J.S.; Hakonarson, H. The Autoimmune Disorder Susceptibility Gene CLEC16A Restrains NK Cell Function in YTS NK Cell Line and Clec16a Knockout Mice. Front. Immunol. 2019, 10, 68. [Google Scholar] [CrossRef]
- Pandey, R.; Bakay, M.; Strenkowski, B.P.; Hain, H.S.; Hakonarson, H. JAK/STAT inhibitor therapy partially rescues the lipodystrophic autoimmune phenotype in Clec16a KO mice. Sci. Rep. 2021, 11, 7372. [Google Scholar] [CrossRef]
- Pearson, G.; Chai, B.; Vozheiko, T.; Liu, X.; Kandarpa, M.; Piper, R.C.; Soleimanpour, S.A. Clec16a, Nrdp1, and USP8 Form a Ubiquitin-Dependent Tripartite Complex That Regulates beta-Cell Mitophagy. Diabetes 2018, 67, 265–277. [Google Scholar] [CrossRef]
- Hain, H.S.; Pandey, R.; Bakay, M.; Strenkowski, B.P.; Harrington, D.; Romer, M.; Motley, W.W.; Li, J.; Lancaster, E.; Roth, L.; et al. Inducible knockout of Clec16a in mice results in sensory neurodegeneration. Sci. Rep. 2021, 11, 9319. [Google Scholar] [CrossRef]
- Smits, D.J.; Dekker, J.; Schot, R.; Tabarki, B.; Alhashem, A.; Demmers, J.A.A.; Dekkers, D.H.W.; Romito, A.; van der Spek, P.J.; van Ham, T.J.; et al. CLEC16A interacts with retromer and TRIM27, and its loss impairs endosomal trafficking and neurodevelopment. Hum. Genet. 2023, 142, 379–397. [Google Scholar] [CrossRef]
- Pandey, R.; Bakay, M.; Hakonarson, H. CLEC16A-An Emerging Master Regulator of Autoimmunity and Neurodegeneration. Int. J. Mol. Sci. 2023, 24, 8224. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Flavell, R.A. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J. Exp. Med. 1995, 181, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroeck, K.; Alvarez, J.; Swaminathan, B.; Alloza, I.; Matesanz, F.; Urcelay, E.; Comabella, M.; Alcina, A.; Fedetz, M.; Ortiz, M.A.; et al. A cytokine gene screen uncovers SOCS1 as genetic risk factor for multiple sclerosis. Genes Immun. 2011, 13, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Davison, L.J.; Wallace, C.; Cooper, J.D.; Cope, N.F.; Wilson, N.K.; Smyth, D.J.; Howson, J.M.; Saleh, N.; Al-Jeffery, A.; Angus, K.L.; et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum. Mol. Genet. 2012, 21, 322–333. [Google Scholar] [CrossRef]
- Tomlinson, M.J.t.; Pitsillides, A.; Pickin, R.; Mika, M.; Keene, K.; Hou, X.; Mychaleckyj, J.; Chen, W.M.; Concannon, P.; Onengut-Gumuscu, S. Fine Mapping and Functional Studies of Risk Variants for Type 1 Diabetes at Chromosome 16p13.13. Diabetes 2014, 63, 4360–4368. [Google Scholar] [CrossRef]
- Wallace, C.; Rotival, M.; Cooper, J.D.; Rice, C.M.; Yang, J.H.; McNeill, M.; Smyth, D.J.; Niblett, D.; Cambien, F.; Cardiogenics, C.; et al. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum. Mol. Genet. 2012, 21, 2815–2824. [Google Scholar] [CrossRef]
- Leikfoss, I.S.; Mero, I.L.; Dahle, M.K.; Lie, B.A.; Harbo, H.F.; Spurkland, A.; Berge, T. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013, 14, 62–66. [Google Scholar] [CrossRef]
- Martin, P.; McGovern, A.; Orozco, G.; Duffus, K.; Yarwood, A.; Schoenfelder, S.; Cooper, N.J.; Barton, A.; Wallace, C.; Fraser, P.; et al. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 2015, 6, 10069. [Google Scholar] [CrossRef]
- Edgar, A.J.; Birks, E.J.; Yacoub, M.H.; Polak, J.M. Cloning of dexamethasone-induced transcript: A novel glucocorticoid-induced gene that is upregulated in emphysema. Am. J. Respir. Cell Mol. Biol. 2001, 25, 119–124. [Google Scholar] [CrossRef]
- Grundberg, E.; Adoue, V.; Kwan, T.; Ge, B.; Duan, Q.L.; Lam, K.C.; Koka, V.; Kindmark, A.; Weiss, S.T.; Tantisira, K.; et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 2011, 7, e1001279. [Google Scholar] [CrossRef]
- Wang, J.C.; Derynck, M.K.; Nonaka, D.F.; Khodabakhsh, D.B.; Haqq, C.; Yamamoto, K.R. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15603–15608. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lu, F.; Soldan, S.S.; Lamontagne, R.J.; Tang, H.Y.; Napoletani, G.; Farrell, P.J.; Tempera, I.; Kossenkov, A.V.; Lieberman, P.M. EBNA2 driven enhancer switching at the CIITA-DEXI locus suppresses HLA class II gene expression during EBV infection of B-lymphocytes. PLoS Pathog. 2021, 17, e1009834. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.S.; Marroqui, L.; Velayos, T.; Olazagoitia-Garmendia, A.; Jauregi-Miguel, A.; Castellanos-Rubio, A.; Eizirik, D.L.; Castano, L.; Santin, I. DEXI, a candidate gene for type 1 diabetes, modulates rat and human pancreatic beta cell inflammation via regulation of the type I IFN/STAT signalling pathway. Diabetologia 2019, 62, 459–472. [Google Scholar] [CrossRef]
- Leikfoss, I.S.; Keshari, P.K.; Gustavsen, M.W.; Bjolgerud, A.; Brorson, I.S.; Celius, E.G.; Spurkland, A.; Bos, S.D.; Harbo, H.F.; Berge, T. Multiple Sclerosis Risk Allele in CLEC16A Acts as an Expression Quantitative Trait Locus for CLEC16A and SOCS1 in CD4+ T Cells. PLoS ONE 2015, 10, e0132957. [Google Scholar] [CrossRef][Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Dinkel, H.; Van Roey, K.; Michael, S.; Kumar, M.; Uyar, B.; Altenberg, B.; Milchevskaya, V.; Schneider, M.; Kuhn, H.; Behrendt, A.; et al. ELM 2016—Data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016, 44, D294–D300. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Miyaki, Y.; Suzuki, K.; Koizumi, K.; Kato, T.; Saito, M.; Kamiyama, H.; Maeda, T.; Shibata, K.; Shiya, N.; Konishi, F. Identification of a potent epigenetic biomarker for resistance to camptothecin and poor outcome to irinotecan-based chemotherapy in colon cancer. Int. J. Oncol. 2012, 40, 217–226. [Google Scholar] [PubMed]
- Nieves-Bonilla, J.M.; Kiaf, B.; Schuster, C.; Kissler, S. The type 1 diabetes candidate gene Dexi does not affect disease risk in the nonobese diabetic mouse model. Genes Immun. 2020, 21, 71–77. [Google Scholar] [CrossRef]
- Davison, L.J.; Wallace, M.D.; Preece, C.; Hughes, K.; Todd, J.A.; Davies, B.; O’Callaghan, C.A. Dexi disruption depletes gut microbial metabolites and accelerates autoimmune diabetes. bioRxiv, 2018; preprint. [Google Scholar]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Gregory, A.P.; Dendrou, C.A.; Attfield, K.E.; Haghikia, A.; Xifara, D.K.; Butter, F.; Poschmann, G.; Kaur, G.; Lambert, L.; Leach, O.A.; et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 2012, 488, 508–511. [Google Scholar] [CrossRef]
- Laaksonen, H.; Guerreiro-Cacais, A.O.; Adzemovic, M.Z.; Parsa, R.; Zeitelhofer, M.; Jagodic, M.; Olsson, T. The multiple sclerosis risk gene IL22RA2 contributes to a more severe murine autoimmune neuroinflammation. Genes Immun. 2014, 15, 457–465. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Cortes, A.; Shipman, L.; Evans, H.G.; Attfield, K.E.; Jostins, L.; Barber, T.; Kaur, G.; Kuttikkatte, S.B.; Leach, O.A.; et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci. Transl. Med. 2016, 8, 363ra149. [Google Scholar] [CrossRef]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef]
- Zuvich, R.L.; Bush, W.S.; McCauley, J.L.; Beecham, A.H.; De Jager, P.L.; Ivinson, A.J.; Compston, A.; Hafler, D.A.; Hauser, S.L.; Sawcer, S.J.; et al. Interrogating the complex role of chromosome 16p13.13 in multiple sclerosis susceptibility: Independent genetic signals in the CIITA-CLEC16A-SOCS1 gene complex. Hum. Mol. Genet. 2011, 20, 3517–3524. [Google Scholar] [CrossRef]
- Chan, S.L.; Lindquist, L.D.; Hansen, M.J.; Girtman, M.A.; Pease, L.R.; Bram, R.J. Calcium-Modulating Cyclophilin Ligand Is Essential for the Survival of Activated T Cells and for Adaptive Immunity. J. Immunol. 2015, 195, 5648–5656. [Google Scholar] [CrossRef]
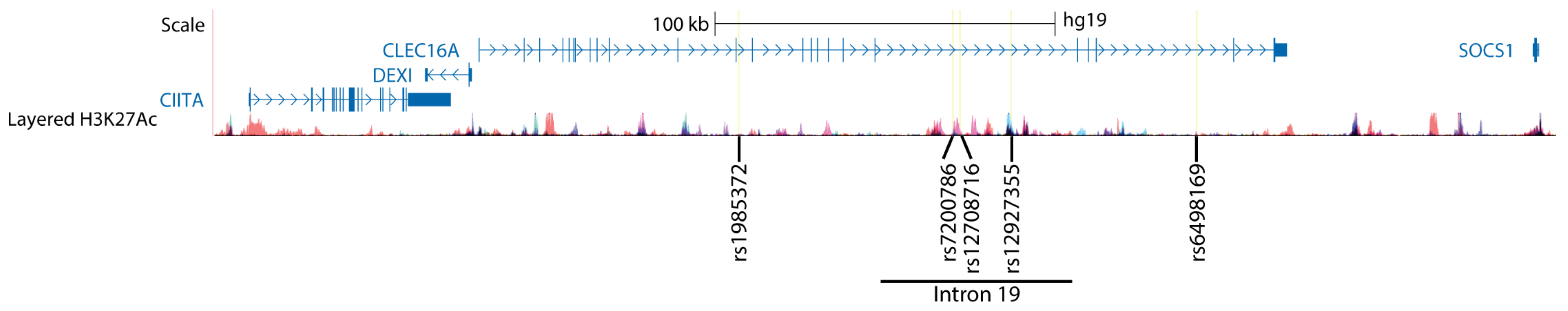
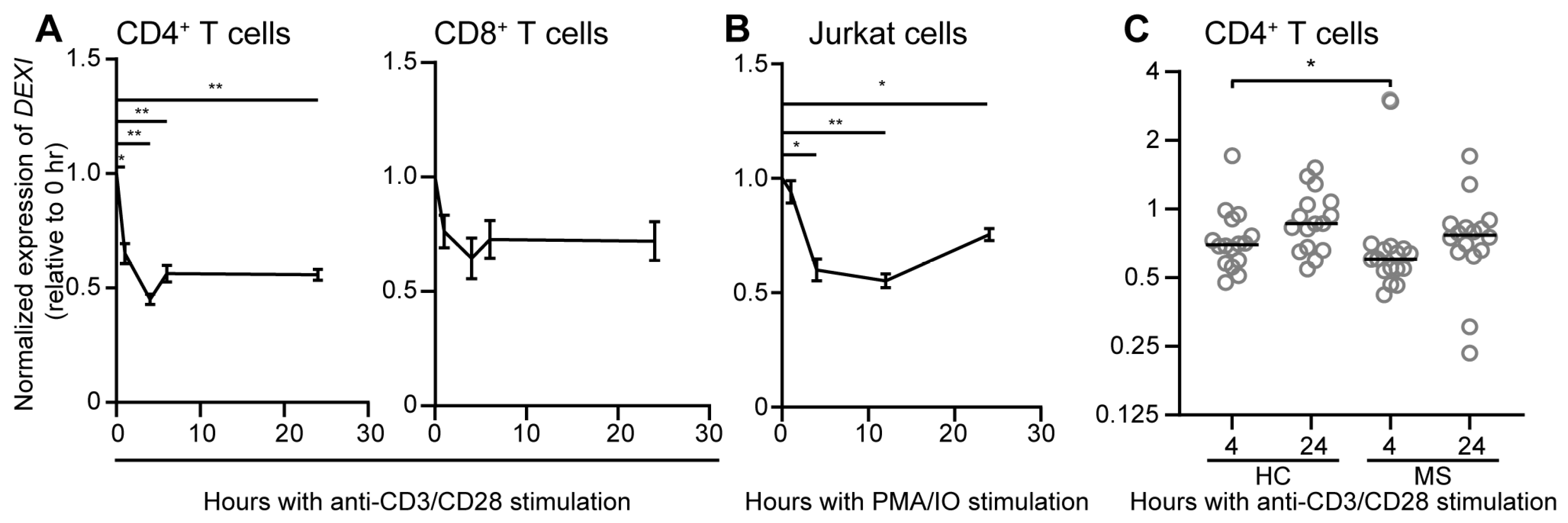
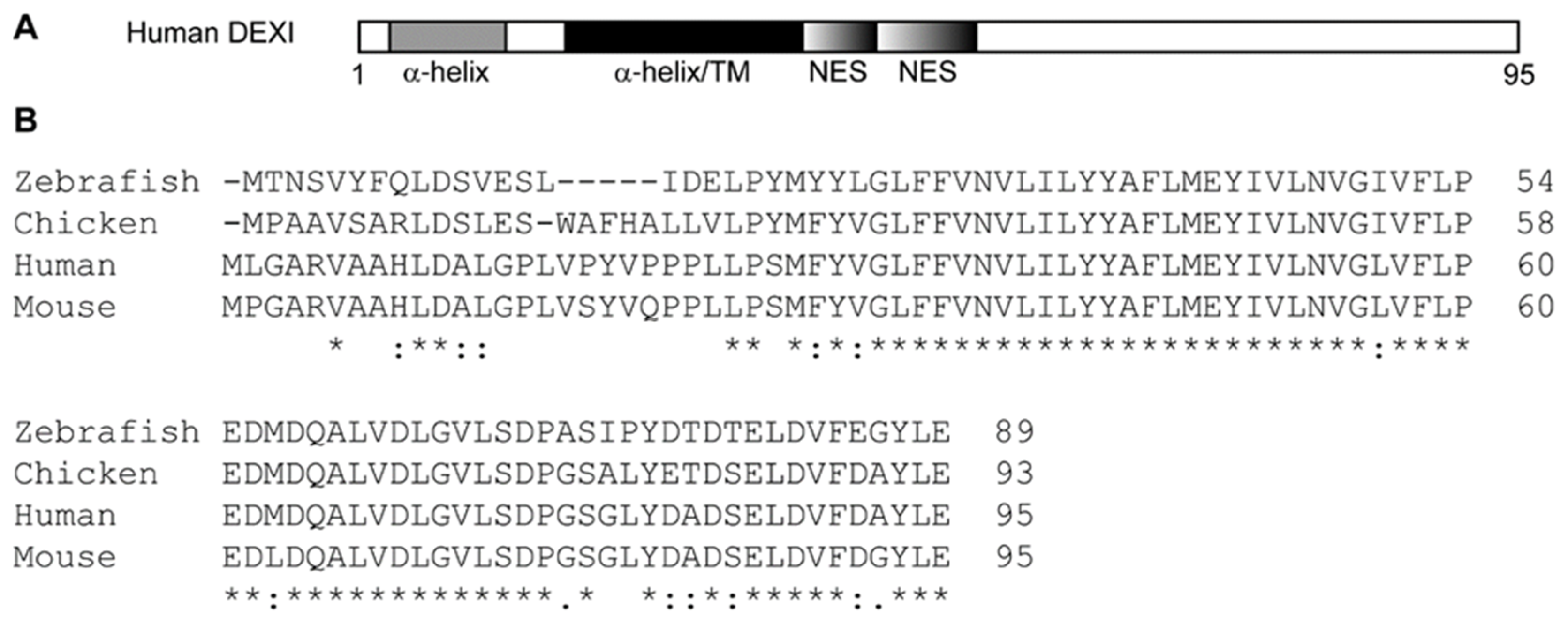
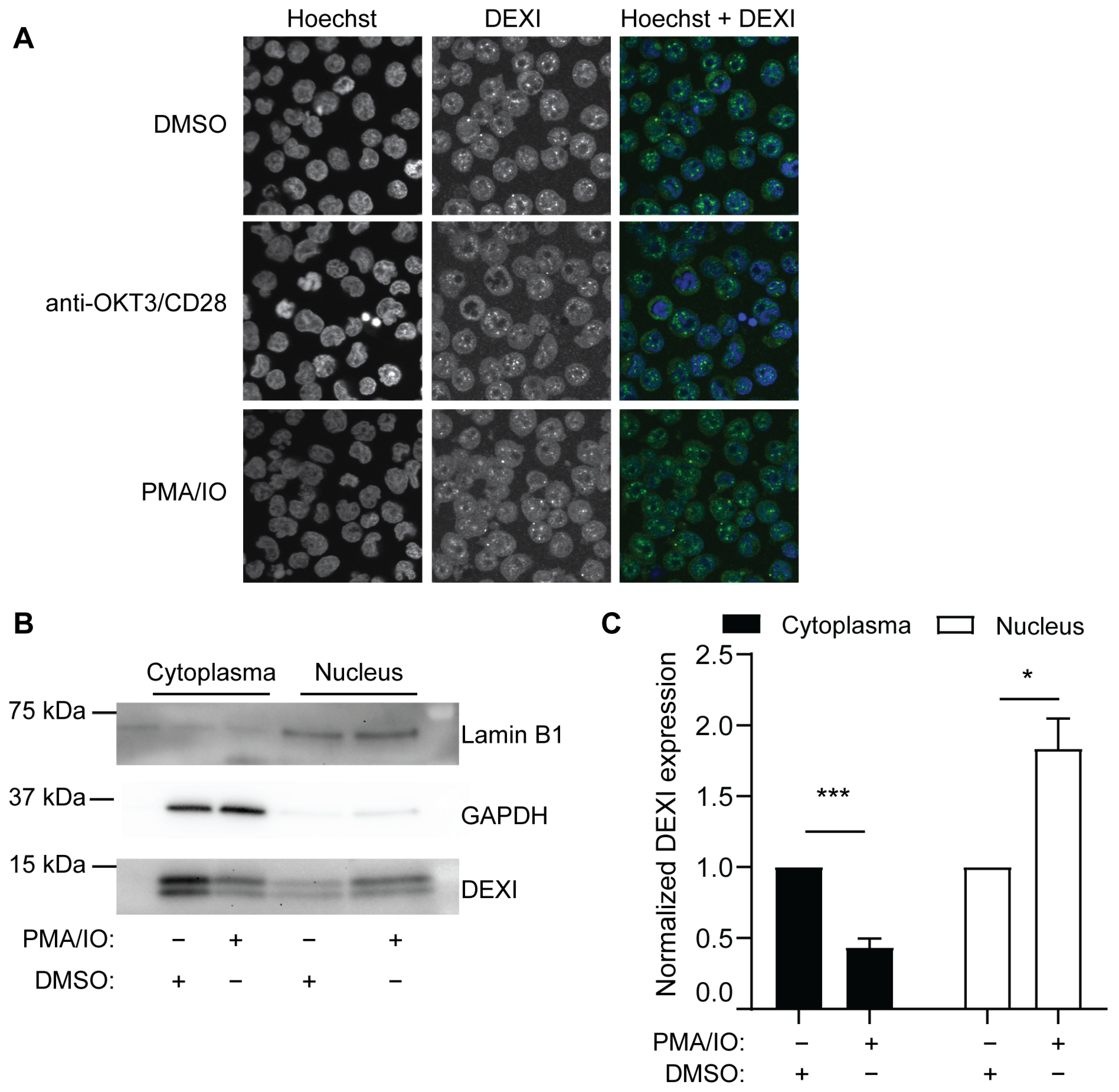
| SNP ID | Localization in CLEC16A | Disease | DEXI expr. | Cells | Study |
|---|---|---|---|---|---|
| rs12708716 | Intron 19 | T1D, MS | Lower | Brain, monocytes, B lymphoblastoid cell line | [3,36,37,38] |
| rs1985372 | Intron 12 | MS | Lower | Brain, CD4+ T cells | [3] |
| rs725613 | Intron 19 | T1D | Lower | Monocytes | [36] |
| rs3901386 | Intron 17 | T1D | Lower | Monocytes | [38] |
| rs7403919 | Intron 10 | T1D | Lower | B lymphoblastoid cell line | [37] |
| rs34306440 | Intron 20 | T1D | Lower | B lymphoblastoid cell line | [37] |
| rs6498169 | Intron 22 | MS | Lower | Thymic tissue | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eriksson, A.M.; Emini, N.; Harbo, H.F.; Berge, T. Is DEXI a Multiple Sclerosis Susceptibility Gene? Int. J. Mol. Sci. 2025, 26, 1175. https://doi.org/10.3390/ijms26031175
Eriksson AM, Emini N, Harbo HF, Berge T. Is DEXI a Multiple Sclerosis Susceptibility Gene? International Journal of Molecular Sciences. 2025; 26(3):1175. https://doi.org/10.3390/ijms26031175
Chicago/Turabian StyleEriksson, Anna M., Nora Emini, Hanne F. Harbo, and Tone Berge. 2025. "Is DEXI a Multiple Sclerosis Susceptibility Gene?" International Journal of Molecular Sciences 26, no. 3: 1175. https://doi.org/10.3390/ijms26031175
APA StyleEriksson, A. M., Emini, N., Harbo, H. F., & Berge, T. (2025). Is DEXI a Multiple Sclerosis Susceptibility Gene? International Journal of Molecular Sciences, 26(3), 1175. https://doi.org/10.3390/ijms26031175






