Fibrinogen Alpha Chain as a Potential Serum Biomarker for Predicting Response to Cisplatin and Gemcitabine Doublet Chemotherapy in Lung Adenocarcinoma: Integrative Transcriptome and Proteome Analyses
Abstract
1. Introduction
2. Results
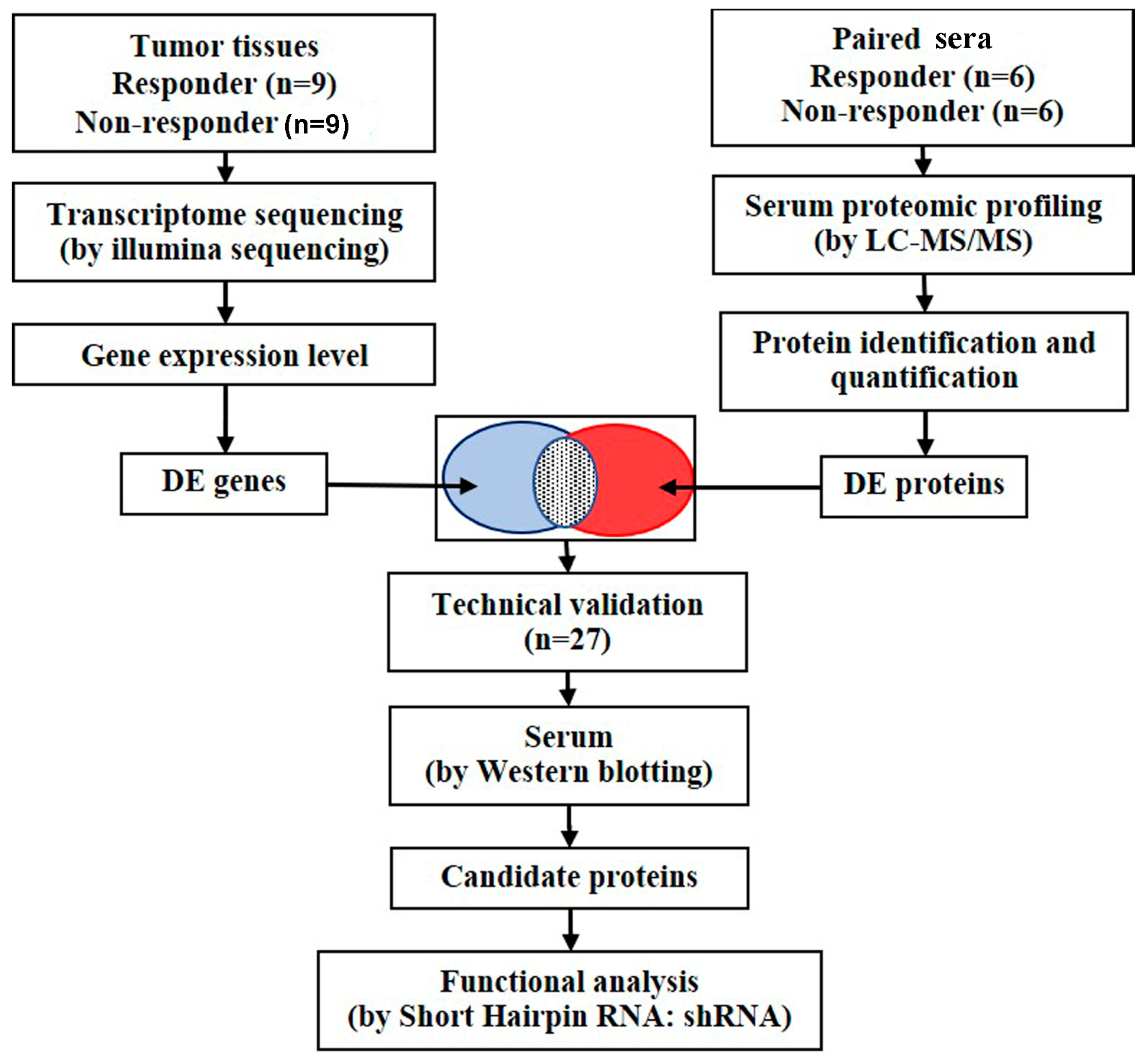
2.1. Study Design and Patient Characteristics
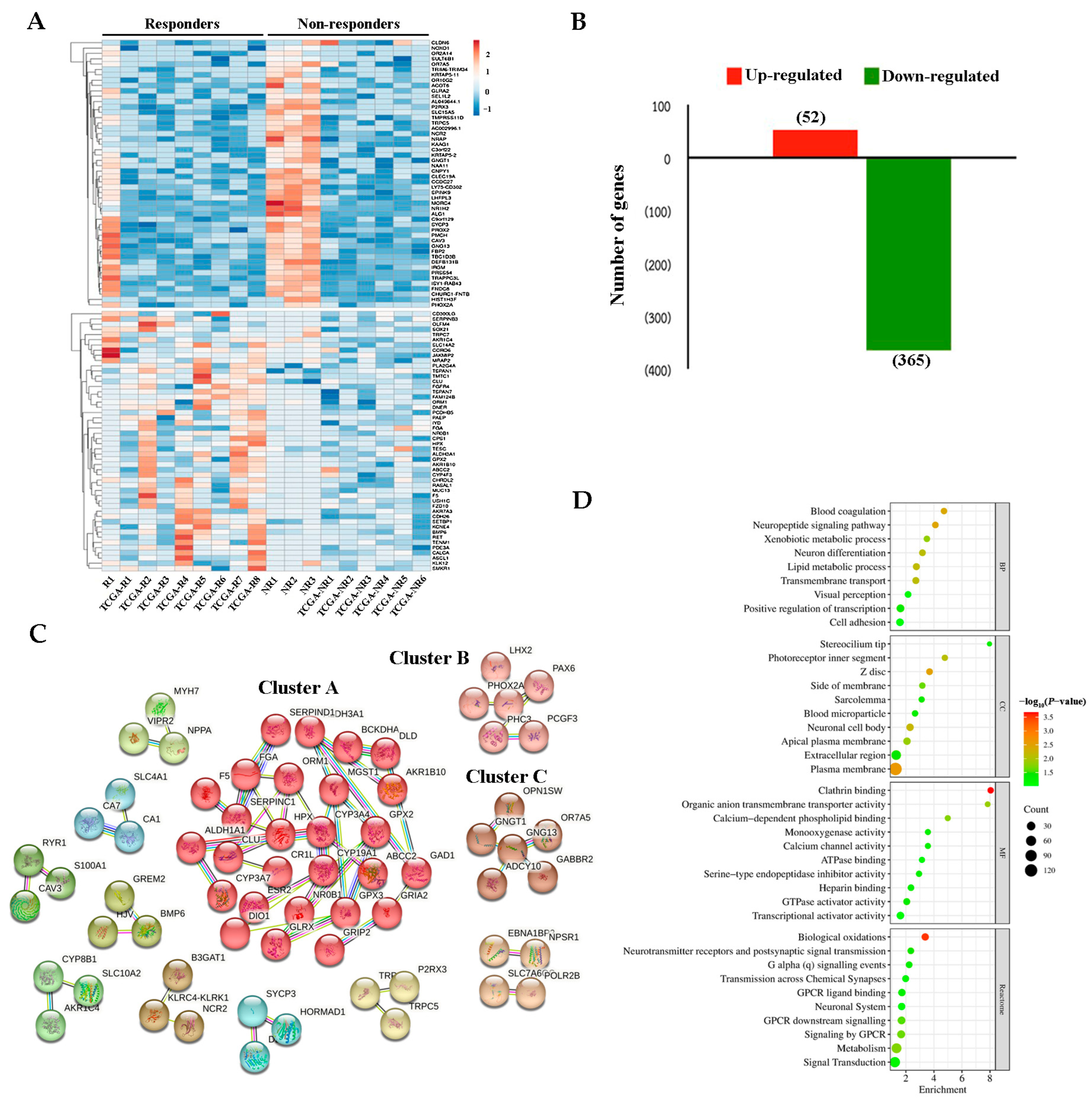
2.2. DEGs Between Responders and Non-Responders
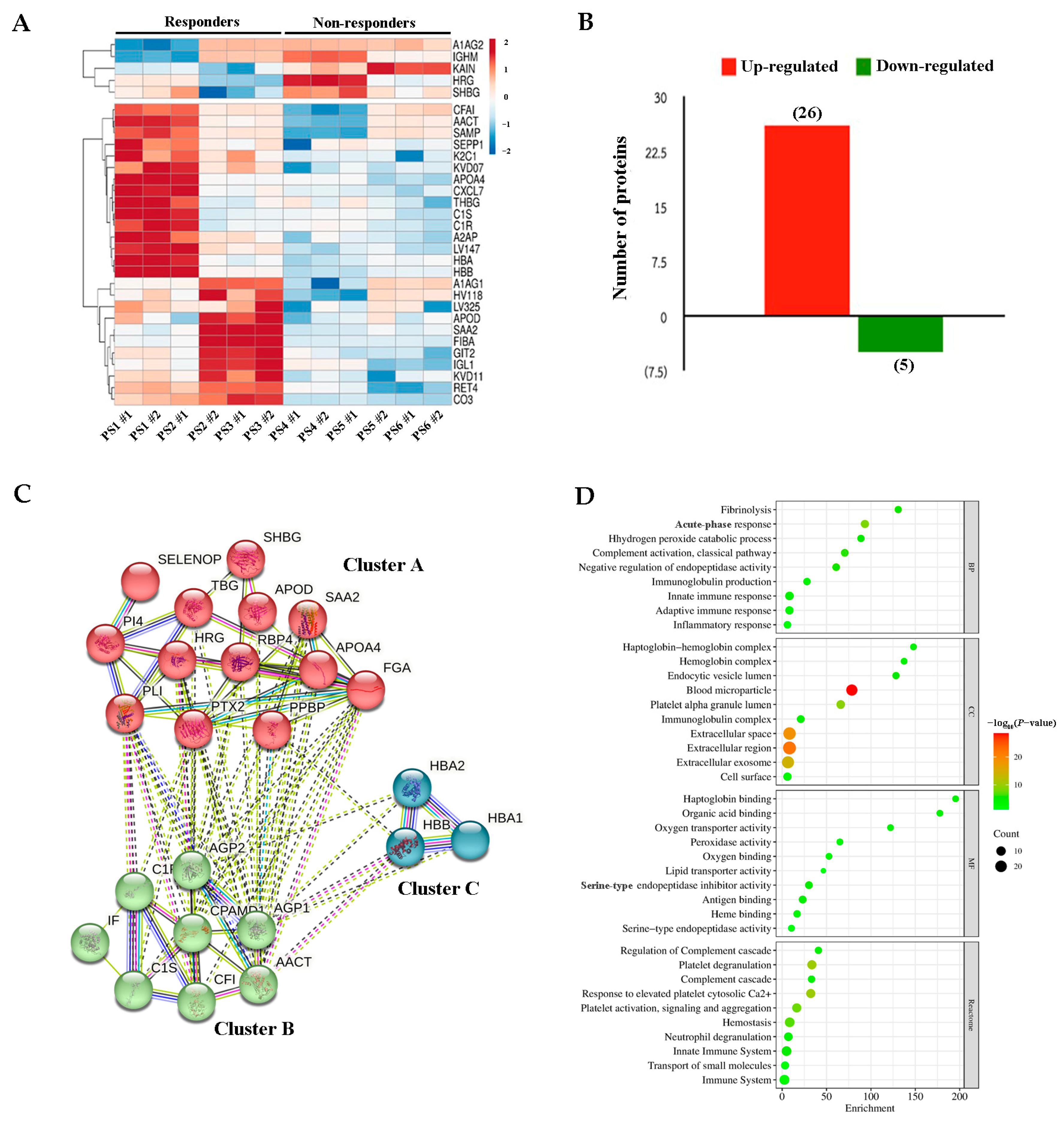
2.3. DEPs Between Responders and Non-Responders
2.4. Identification of Blood-Secretory Proteins via Integrative Analysis and Validation
2.5. Association of A1AG1 and FGA with Clinicopathological Variables
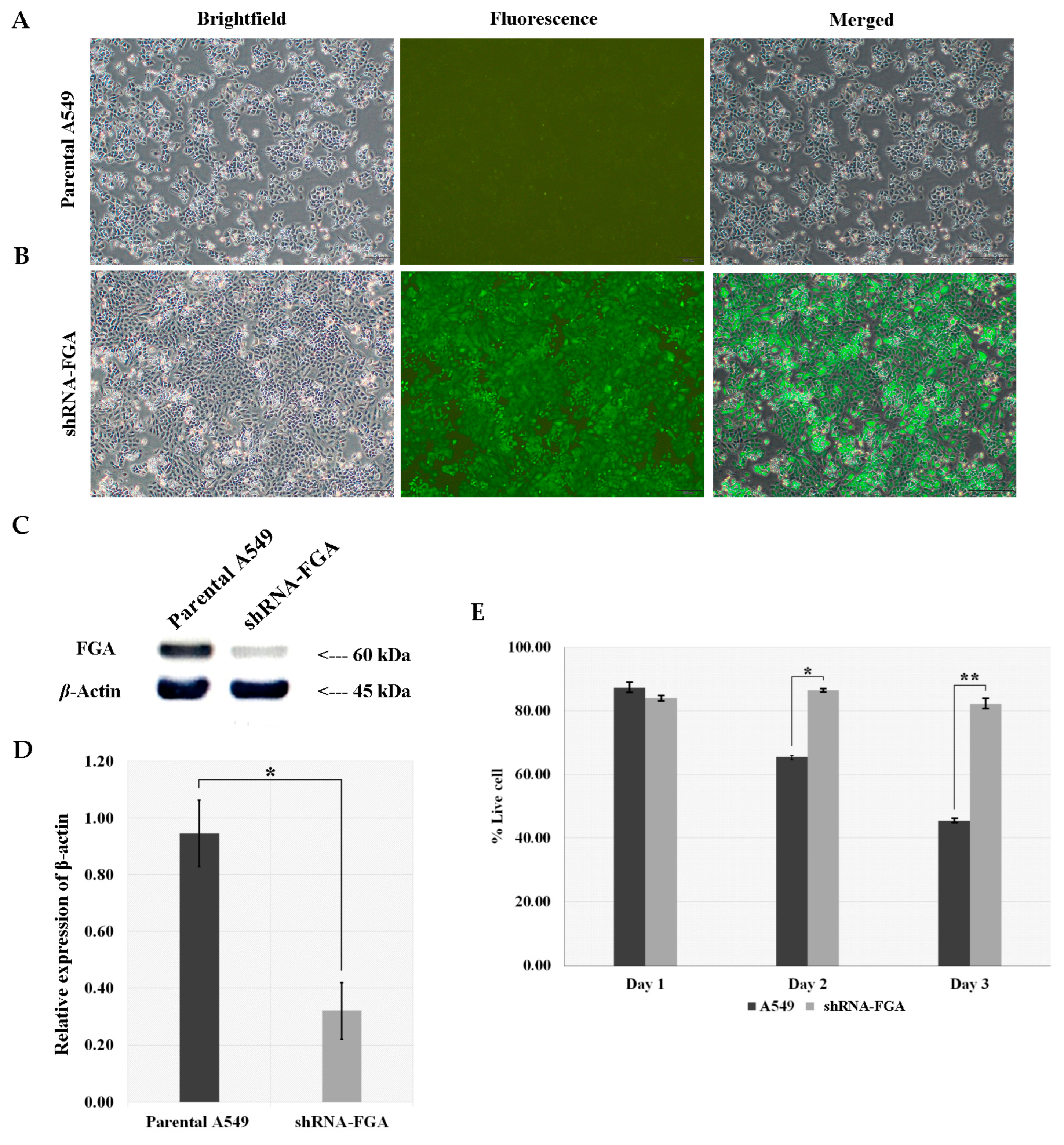
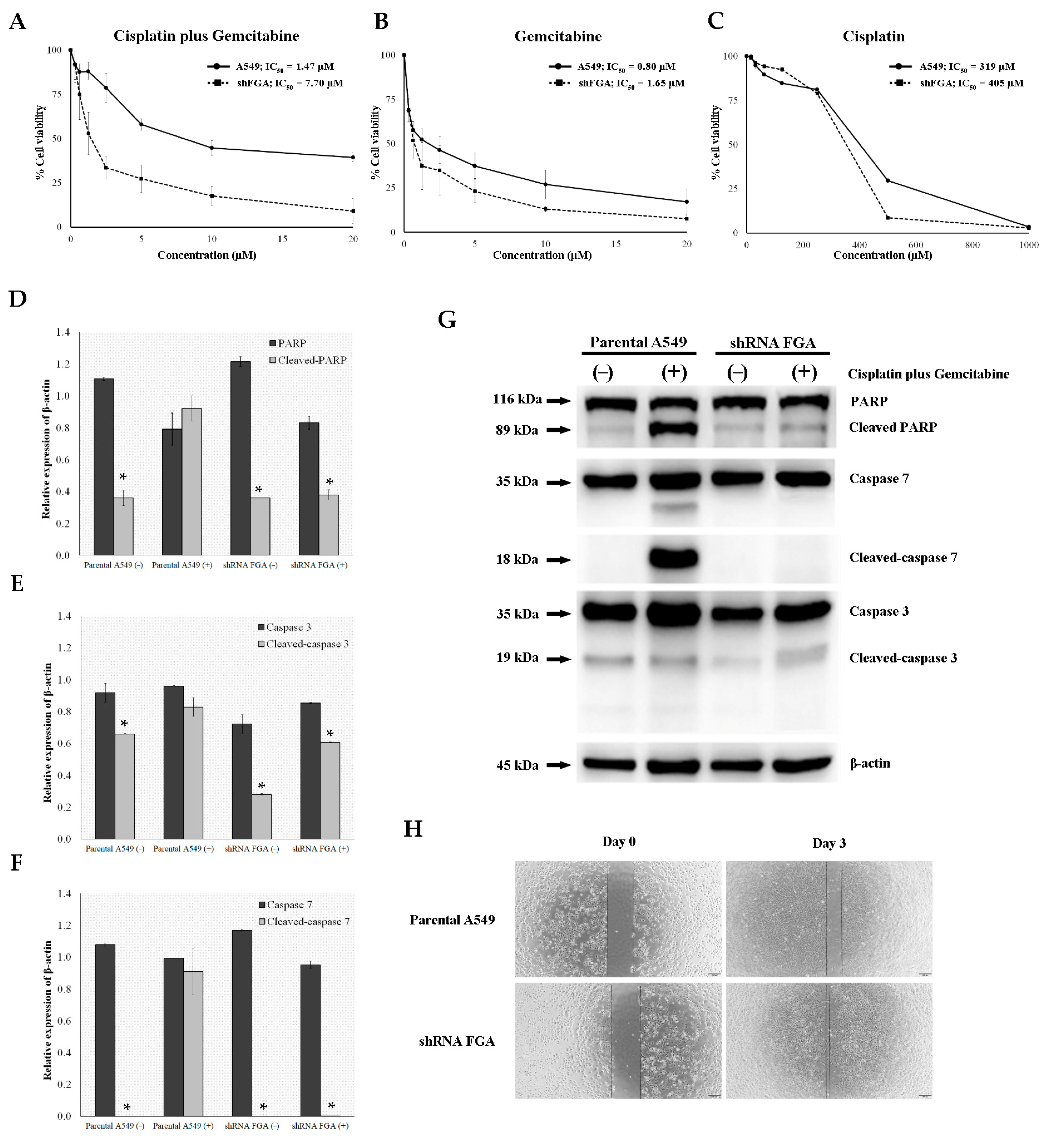
2.6. FGA Knockdown Constructed by Lentivirus-Mediated shRNA Infection of A549 Cells
3. Discussion
4. Materials and Methods
4.1. Subject Selection
4.2. Collection of Tissue-Based Transcriptome Data from the Cancer Genome Atlas (TCGA)
4.3. Sample Preparation, Treatment, and Clinical Follow-Up
4.4. Transcriptome Sequencing
4.5. Gene Identification and Data Analysis
4.6. Serum Proteomics
4.7. Protein Identification and Data Analysis
4.8. Bioinformatics Analysis
4.9. Western Blotting
4.10. Plasmid Preparation and Lentiviral shRNA Packaging
4.11. Generation of Lentivirus-Mediated shRNA Interference Targeting FGA
4.12. Trypan Blue Dye Exclusion Assay
4.13. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) Assay
4.14. Wound Healing Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Vaccarella, S.; Morgan, E.; Li, M.; Etxeberria, J.; Chokunonga, E.; Manraj, S.S.; Kamate, B.; Omonisi, A.; Bray, F. Global variations in lung cancer incidence by histological subtype in 2020: A population-based study. Lancet Oncol. 2023, 24, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- SEER Cancer Statistics Review, 1975–2018, National Cancer Institute. Bethesda, MD, [Internet]. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 10 April 2021).
- Spira, A.; Ettinger, D.S. Multidisciplinary management of lung cancer. N. Engl. J. Med. 2004, 350, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Rudd, R. Gemcitabine and carboplatin: Is this the best combination for non-small cell lung cancer? Expert. Rev. Anticancer. Ther. 2006, 6, 165–173. [Google Scholar] [CrossRef]
- Schiller, J.H.; Harrington, D.; Belani, C.P.; Langer, C.; Sandler, A.; Krook, J.; Zhu, J.; Johnson, D.H. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 2002, 346, 92–98. [Google Scholar] [CrossRef]
- Ohe, Y.; Ohashi, Y.; Kubota, K.; Tamura, T.; Nakagawa, K.; Negoro, S.; Nishiwaki, Y.; Saijo, N.; Ariyoshi, Y.; Fukuoka, M. Randomized phase III study of cisplatin plus irinotecan versus carboplatin plus paclitaxel, cisplatin plus gemcitabine, and cisplatin plus vinorelbine for advanced non-small-cell lung cancer: Four-Arm Cooperative Study in Japan. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2007, 18, 317–323. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Hellweg, R.; Mooneyham, A.; Chang, Z.; Shetty, M.; Emmings, E.; Iizuka, Y.; Clark, C.; Starr, T.; Abrahante, J.H.; Schütz, F.; et al. RNA Sequencing of Carboplatin- and Paclitaxel-Resistant Endometrial Cancer Cells Reveals New Stratification Markers and Molecular Targets for Cancer Treatment. Horm. Cancer 2018, 9, 326–337. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, C.; Wu, T.; Wang, Q.; Liu, J.; Dai, P. Transcriptome Sequencing Reveals Key Pathways and Genes Associated with Cisplatin Resistance in Lung Adenocarcinoma A549 Cells. PLoS ONE 2017, 12, e0170609. [Google Scholar] [CrossRef]
- Shen, H.; Fang, X.-F.; Yuan, Y.; Yang, J.; Zheng, S. Serum Protein Pattern Could Predict the Therapeutic Effect of First-Line Pemetrexed/Cisplatin Chemotherapy in Patients with Lung Adenocarcinoma. World J. Oncol. 2015, 6, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Liu, Q.; Yu, J.; Zheng, S. Identification of candidate molecular markers predicting chemotherapy resistance in non-small cell lung cancer. Clin. Chem. Lab. Med. 2010, 48, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, G.; Aplin, A.C.; Dunn, B.E.; Morishita, A.; Nicosia, R.F. The acute phase reactant orosomucoid-1 is a bimodal regulator of angiogenesis with time- and context-dependent inhibitory and stimulatory properties. PLoS ONE 2012, 7, e41387. [Google Scholar] [CrossRef]
- Bachtiar, I.; Santoso, J.M.; Atmanegara, B.; Gani, R.A.; Hasan, I.; Lesmana, L.A.; Sulaiman, A.; Gu, J.; Tai, S. Combination of alpha-1-acid glycoprotein and alpha-fetoprotein as an improved diagnostic tool for hepatocellular carcinoma. Clin. Chim. Acta Int. J. Clin. Chem. 2009, 399, 97–101. [Google Scholar] [CrossRef]
- Uslu, C.; Taysi, S.; Akcay, F.; Sutbeyaz, M.Y.; Bakan, N. Serum free and bound sialic acid and alpha-1-acid glycoprotein in patients with laryngeal cancer. Ann. Clin. Lab. Sci. 2003, 33, 156–159. [Google Scholar]
- Zhou, Q.; Andersson, R.; Hu, D.; Bauden, M.; Sasor, A.; Bygott, T.; PawŁowski, K.; Pla, I.; Marko-Varga, G.; Ansari, D. Alpha-1-acid glycoprotein 1 is upregulated in pancreatic ductal adenocarcinoma and confers a poor prognosis. Transl. Res. J. Lab. Clin. Med. 2019, 212, 67–79. [Google Scholar] [CrossRef]
- Ayyub, A.; Saleem, M.; Fatima, I.; Tariq, A.; Hashmi, N.; Musharraf, S.G. Glycosylated Alpha-1-acid glycoprotein 1 as a potential lung cancer serum biomarker. Int. J. Biochem. Cell Biol. 2016, 70, 68–75. [Google Scholar] [CrossRef]
- Hyung, S.-W.; Lee, M.Y.; Yu, J.-H.; Shin, B.; Jung, H.-J.; Park, J.-M.; Han, W.; Lee, K.-M.; Moon, H.-G.; Zhang, H.; et al. A serum protein profile predictive of the resistance to neoadjuvant chemotherapy in advanced breast cancers. Mol. Cell. Proteom. MCP 2011, 10, M111.011023. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, Z.; Sun, Z.; Zhang, X.; Lu, L.; Wang, Y.; Zhang, M. S100A9 and ORM1 serve as predictors of therapeutic response and prognostic factors in advanced extranodal NK/T cell lymphoma patients treated with pegaspargase/gemcitabine. Sci. Rep. 2016, 6, 23695. [Google Scholar] [CrossRef]
- Mon, M.M.; Srisomsap, C.; Chokchaichamnankit, D.; Watcharatanyatip, K.; Weeraphan, C.; Svasti, J.; Maneechai, K.; Thomgsuksai, P.; Raungrut, P. Serum Proteomic Profiling Reveals Differentially Expressed IGHG3 and A1AG1 as Potential Predictors of Chemotherapeutic Response in Advanced Non-small Cell Lung Cancer. Anticancer Res. 2021, 41, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Cao, Z.; Chung, D.W.; Davie, E.W. The role of betagamma and alphagamma complexes in the assembly of human fibrinogen. J. Biol. Chem. 1996, 271, 27942–27947. [Google Scholar] [CrossRef] [PubMed]
- Pacchiarotta, T.; Hensbergen, P.J.; Wuhrer, M.; van Nieuwkoop, C.; Nevedomskaya, E.; Derks, R.J.; Schoenmaker, B.; Koeleman, C.A.; van Dissel, J.; Deelder, A.M.; et al. Fibrinogen alpha chain O-glycopeptides as possible markers of urinary tract infection. J. Proteom. 2012, 75, 1067–1073. [Google Scholar] [CrossRef]
- Lei, T.; Zhao, X.; Jin, S.; Meng, Q.; Zhou, H.; Zhang, M. Discovery of potential bladder cancer biomarkers by comparative urine proteomics and analysis. Clin. Genitourin. Cancer 2013, 11, 56–62. [Google Scholar] [CrossRef]
- Duan, S.; Gong, B.; Wang, P.; Huang, H.; Luo, L.; Liu, F. Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol. Med. Rep. 2018, 18, 3727–3736. [Google Scholar] [CrossRef]
- Liu, G.; Xu, X.; Geng, H.; Li, J.; Zou, S.; Li, X. FGA inhibits metastases and induces autophagic cell death in gastric cancer via inhibiting ITGA5 to regulate the FAK/ERK pathway. Tissue Cell 2022, 76, 101767. [Google Scholar] [CrossRef]
- Liu, C.; Pan, C.; Liang, Y. Screening and identification of serum proteomic biomarkers for gastric adenocarcinoma. Exp. Ther. Med. 2012, 3, 1005–1009. [Google Scholar] [CrossRef]
- Li, X.-J.; Wu, Q.-F.; He, D.-L.; Fu, J.-K.; Jin, X. Proteomic profiling of serum from stage I lung squamous cell carcinoma patients. Asian Pac. J. Cancer Prev. 2013, 14, 2273–2276. [Google Scholar] [CrossRef]
- Wang, H.; Luo, C.; Zhu, S.; Fang, H.; Gao, Q.; Ge, S.; Qu, H.; Ma, Q.; Ren, H.; Wang, W.; et al. Serum peptidome profiling for the diagnosis of colorectal cancer: Discovery and validation in two independent cohorts. Oncotarget 2017, 8, 59376–59386. [Google Scholar] [CrossRef]
- Li, H.; Cai, E.; Cheng, H.; Ye, X.; Ma, R.; Zhu, H.; Chang, X. FGA Controls VEGFA Secretion to Promote Angiogenesis by Activating the VEGFR2-FAK Signalling Pathway. Front. Endocrinol. 2022, 13, 791860. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, G.; Zhang, Y.; Cui, X.; Wang, S.; Gao, S.; Wang, Y.; Liu, Y.; Bae, J.H.; Yang, W.-H.; et al. Fibrinogen Alpha Chain Knockout Promotes Tumor Growth and Metastasis through Integrin-AKT Signaling Pathway in Lung Cancer. Mol. Cancer Res. 2020, 18, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

| Variables | Category | Number (%) |
|---|---|---|
| Sex | ||
| Female | 13 (48.1) | |
| Male | 14 (51.9) | |
| Age (years) | ||
| <60 | 13 (48.1) | |
| ≥60 | 14 (51.9) | |
| Smoke | ||
| No | 17 (63) | |
| Yes | 10 (37) | |
| Drink | ||
| No | 19 (70.4) | |
| Yes | 8 (29.6) | |
| Stage | ||
| 3 | 3 (11.1) | |
| 4 | 24 (88.9) | |
| Response | ||
| CR | 1 (3.7) | |
| PR | 11 (40.7) | |
| SD | 9 (33.3) | |
| PD | 6 (22.2) |
| Gene Symbol | Ensembl ID | Annotated Protein Symbol | log2FC | p-Value * | p-Adj ** |
|---|---|---|---|---|---|
| CALCA | ENSG00000110680.11 | CALC, CALCA | 7.9954 | 2.04 × 10−7 | 8.61 × 10−5 |
| TRPC7 | ENSG00000069018.16 | TRPM2, TRPC7 | 7.9069 | 3.09 × 10−4 | 1.65 × 10−2 |
| AKR1B10 | ENSG00000198074.8 | AK1BA | 7.8999 | 2.15 × 10−7 | 8.83 × 10−5 |
| CPS1 | ENSG00000021826.13 | CPSM | 6.5426 | 2.06 × 10−8 | 1.62 × 10−5 |
| ALDH3A1 | ENSG00000108602.16 | AL3A1 | 6.4751 | 6.02 × 10−8 | 3.63 × 10−5 |
| OLFM4 | ENSG00000102837.6 | OLFM4 | 6.3730 | 4.74 × 10−6 | 6.94 × 10−4 |
| NR0B1 | ENSG00000169297.7 | NR0B1 | 5.9363 | 2.32 × 10−5 | 2.34 × 10−3 |
| ASCL1 | ENSG00000139352.3 | ASCL1 | 5.8997 | 1.25 × 10−5 | 1.48 × 10−3 |
| SLC14A2 | ENSG00000132874.12 | UT2 | 5.8886 | 2.63 × 10−6 | 4.78 × 10−4 |
| MUC13 | ENSG00000173702.6 | MUC13 | 5.3260 | 2.56 × 10−6 | 4.71 × 10−4 |
| PAEP | ENSG00000122133.15 | PAEP | 5.1279 | 4.68 × 10−5 | 3.93 × 10−3 |
| AKR1C4 | ENSG00000198610.9 | AK1C4 | 5.0082 | 1.20 × 10−4 | 7.82 × 10−3 |
| USH1C | ENSG00000006611.14 | USH1C | 4.7127 | 1.09 × 10−4 | 7.30 × 10−3 |
| ABCC2 | ENSG00000023839.9 | MRP2 | 4.6668 | 9.99 × 10−6 | 1.24 × 10−3 |
| FZD10 | ENSG00000111432.4 | FZD10 | 4.6245 | 5.58 × 10−5 | 4.42 × 10−3 |
| FGA | ENSG00000171560.13 | FIBA | 4.3066 | 7.00 × 10−4 | 3.17 × 10−2 |
| KLK12 | ENSG00000186474.14 | KLK12 | 4.2535 | 6.15 × 10−4 | 2.90 × 10−2 |
| CD300LG | ENSG00000161649.11 | CLM9 | 4.1614 | 5.25 × 10−4 | 2.55 × 10−2 |
| SERPINB3 | ENSG00000057149.13 | SPB3 | 4.0722 | 6.23 × 10−4 | 2.91 × 10−2 |
| BMP6 | ENSG00000153162.8 | BMP6 | 4.0414 | 4.29 × 10−6 | 6.65 × 10−4 |
| NOXO1 | ENSG00000196408.10 | NOXO1 | −10.9752 | 2.06 × 10−4 | 1.22 × 10−2 |
| NCR2 | ENSG00000267261.4 | NCTR2 | −10.5805 | 1.38 × 10−11 | 7.97 × 10−8 |
| PRSS54 | ENSG00000096264.12 | PRS54 | −9.7211 | 1.96 × 10−9 | 2.60 × 10−6 |
| LHFPL3 | ENSG00000279018.1 | LHPL3 | −9.4202 | 5.39 × 10−9 | 5.54 × 10−6 |
| NR1H2 | ENSG00000103023.10 | NR1H2 | −9.0295 | 2.00 × 10−14 | 3.46 × 10−10 |
| FNDC8 | ENSG00000268643.1 | FNDC8 | −8.7288 | 5.10 × 10−11 | 2.20 × 10−7 |
| CLEC19A | ENSG00000187416.10 | CL19A | −8.3502 | 5.45 × 10−9 | 5.54 × 10−6 |
| HIST1H3F | ENSG00000131408.12 | H31 | −8.1709 | 3.31 × 10−7 | 1.19 × 10−4 |
| NRAP | ENSG00000280778.1 | NRAP | −8.1458 | 1.30 × 10−9 | 2.17 × 10−6 |
| TRIM6-TRIM34 | ENSG00000251357.4 | B2RNG4 | −8.1102 | 2.62 × 10−5 | 2.53 × 10−3 |
| Protein Symbol | Accession | Protein Name | Unique Peptides | log2FC | p-Value * |
|---|---|---|---|---|---|
| SAA2 | SAA2_HUMAN | Serum amyloid A-2 protein | 2 | 3.0683 | 3.04 × 10−2 |
| RET4 | RET4_HUMAN | Retinol-binding protein 4 | 3 | 2.2992 | 3.81 × 10−5 |
| LV147 | LV147_HUMAN | Immunoglobulin lambda variable 1–47 | 1 | 2.1425 | 5.03 × 10−3 |
| K2C1 | K2C1_HUMAN | Keratin, type II cytoskeletal 1 | 1 | 2.1402 | 6.41 × 10−3 |
| HBA | HBA_HUMAN | Hemoglobin subunit alpha | 1 | 2.0593 | 2.96 × 10−2 |
| A1AG1 | A1AG1_HUMAN | Alpha-1-acid glycoprotein 1 | 3 | 1.9118 | 4.96 × 10−2 |
| FIBA | FIBA_HUMAN | Fibrinogen alpha chain | 2 | 1.8921 | 2.34 × 10−2 |
| HBB | HBB_HUMAN | Hemoglobin subunit beta | 7 | 1.8515 | 1.44 × 10−2 |
| CO3 | CO3_HUMAN | Complement C3 | 6 | 1.8333 | 3.04 × 10−4 |
| GIT2 | GIT2_HUMAN | ARF GTPase-activating protein GIT2 | 1 | 1.7255 | 5.27 × 10−3 |
| SEPP1 | SEPP1_HUMAN | Selenoprotein P | 1 | 1.6813 | 2.24 × 10−2 |
| APOA4 | APOA4_HUMAN | Apolipoprotein A-IV | 11 | 1.6533 | 1.85 × 10−2 |
| CXCL7 | CXCL7_HUMAN | Platelet basic protein | 3 | 1.5721 | 1.82 × 10−2 |
| KVD07 | KVD07_HUMAN | Immunoglobulin kappa variable 3D-7 | 1 | 1.5575 | 1.08 × 10−3 |
| IGL1 | IGL1_HUMAN | Immunoglobulin lambda-1 light chain | 2 | 1.3985 | 1.22 × 10−2 |
| THBG | THBG_HUMAN | Thyroxine-binding globulin | 4 | 1.3594 | 4.55 × 10−2 |
| KVD11 | KVD11_HUMAN | Immunoglobulin kappa variable 3D-11 | 1 | 1.3542 | 2.21 × 10−3 |
| AACT | AACT_HUMAN | Alpha-1-antichymotrypsin | 11 | 1.3511 | 1.04 × 10−2 |
| APOD | APOD_HUMAN | Apolipoprotein D | 1 | 1.3155 | 2.38 × 10−2 |
| A2AP | A2AP_HUMAN | Alpha-2-antiplasmin | 4 | 1.2936 | 3.82 × 10−3 |
| SAMP | SAMP_HUMAN | Serum amyloid P-component | 5 | 1.2844 | 7.54 × 10−3 |
| LV325 | LV325_HUMAN | Immunoglobulin lambda variable 3–25 | 2 | 1.2722 | 6.86 × 10−3 |
| C1S | C1S_HUMAN | Complement C1s subcomponent | 5 | 1.2685 | 4.35 × 10−2 |
| C1R | C1R_HUMAN | Complement C1r subcomponent | 8 | 1.2652 | 3.95 × 10−2 |
| HV118 | HV118_HUMAN | Immunoglobulin heavy variable 1–18 | 1 | 1.2640 | 1.90 × 10−2 |
| CFAI | CFAI_HUMAN | Complement factor I | 7 | 1.2558 | 1.13 × 10−2 |
| KAIN | KAIN_HUMAN | Kallistatin | 2 | −2.0818 | 9.60 × 10−4 |
| A1AG2 | A1AG2_HUMAN | Alpha-1-acid glycoprotein 2 | 1 | −1.7496 | 4.82 × 10−2 |
| HRG | HRG_HUMAN | Histidine-rich glycoprotein | 7 | −1.6026 | 3.59 × 10−2 |
| SHBG | SHBG_HUMAN | Sex hormone-binding globulin | 1 | −1.5059 | 2.96 × 10−2 |
| IGHM | IGHM_HUMAN | Immunoglobulin heavy constant mu | 7 | −1.3817 | 4.62 × 10−2 |
| Variables | A1AG1 Expression | p-Value | FIBA Expression | p-Value | ||
|---|---|---|---|---|---|---|
| High (%) | Low (%) | High (%) | Low (%) | |||
| Sex | 1.000 | 1.000 | ||||
| Female | 10 (76.9) | 3 (23.1) | 3 (23.1) | 10 (76.9) | ||
| Male | 10 (71.4) | 4 (28.6) | 4 (28.6) | 10 (71.4) | ||
| Age, years | 0.385 | 0.678 | ||||
| <60 | 11 (84.6) | 2 (15.4) | 4 (30.8) | 9 (69.2) | ||
| ≥60 | 9 (64.3) | 5 (35.7) | 3 (21.4) | 11 (78.6) | ||
| Smoking | 1.000 | 0.365 | ||||
| No | 13 (76.5) | 4 (23.5) | 3 (17.6) | 14 (82.4) | ||
| Yes | 7 (70.0) | 3 (30) | 4 (40.0) | 6 (60.0) | ||
| Drinking | 1.000 | 0.011 * | ||||
| No | 14 (73.7) | 5 (26.3) | 2 (10.5) | 17 (89.5) | ||
| Yes | 6 (75.0) | 2 (25.0) | 5 (62.5) | 3 (37.5) | ||
| Chemotherapy response | 0.018 * | 0.029 * | ||||
| Response | 10 (100.0) | 0 (0.0) | 5 (50.0) | 5 (50.0) | ||
| Non-response | 10 (58.8) | 7 (41.2) | 2 (11.8) | 15 (88.2) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raungrut, P.; Jirapongsak, J.; Tanyapattrapong, S.; Bunsong, T.; Ruklert, T.; Kueakool, K.; Thongsuksai, P.; Nakwan, N. Fibrinogen Alpha Chain as a Potential Serum Biomarker for Predicting Response to Cisplatin and Gemcitabine Doublet Chemotherapy in Lung Adenocarcinoma: Integrative Transcriptome and Proteome Analyses. Int. J. Mol. Sci. 2025, 26, 1010. https://doi.org/10.3390/ijms26031010
Raungrut P, Jirapongsak J, Tanyapattrapong S, Bunsong T, Ruklert T, Kueakool K, Thongsuksai P, Nakwan N. Fibrinogen Alpha Chain as a Potential Serum Biomarker for Predicting Response to Cisplatin and Gemcitabine Doublet Chemotherapy in Lung Adenocarcinoma: Integrative Transcriptome and Proteome Analyses. International Journal of Molecular Sciences. 2025; 26(3):1010. https://doi.org/10.3390/ijms26031010
Chicago/Turabian StyleRaungrut, Pritsana, Jirapon Jirapongsak, Suchanan Tanyapattrapong, Thitaya Bunsong, Thidarat Ruklert, Kannika Kueakool, Paramee Thongsuksai, and Narongwit Nakwan. 2025. "Fibrinogen Alpha Chain as a Potential Serum Biomarker for Predicting Response to Cisplatin and Gemcitabine Doublet Chemotherapy in Lung Adenocarcinoma: Integrative Transcriptome and Proteome Analyses" International Journal of Molecular Sciences 26, no. 3: 1010. https://doi.org/10.3390/ijms26031010
APA StyleRaungrut, P., Jirapongsak, J., Tanyapattrapong, S., Bunsong, T., Ruklert, T., Kueakool, K., Thongsuksai, P., & Nakwan, N. (2025). Fibrinogen Alpha Chain as a Potential Serum Biomarker for Predicting Response to Cisplatin and Gemcitabine Doublet Chemotherapy in Lung Adenocarcinoma: Integrative Transcriptome and Proteome Analyses. International Journal of Molecular Sciences, 26(3), 1010. https://doi.org/10.3390/ijms26031010





