Blood-Based miRNA Panels for Timely Detection of Non-Small-Cell Lung Cancer: From Biomarker Discovery to Clinical Translation
Abstract
1. Introduction
2. Cell-Free miRNAs as Promising Biomarkers for Early-Stage NSCLC
| miRNA | Study Model | Sample Type | Analytical Technique | Normalization | Ref. | Regulation | Biological Processes Regulated | Target | Expression Changes | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-21 | 63 NSCLC patients. Stages: I–IV Mouse model | Plasma | RT-qPCR | miR-16 | [31] | ↑ | Cell cycle DNA repair Apoptosis Angiogenesis Proteolysis Cell adhesion MAPK/ERK pathway TGFβ pathway G-protein pathway cell growth and resistance to apoptosis | STAG2, KIF6 MSH2, FANCC, CHD7 PDCD4, APAF1, STAT3, MALT1, SGK3 SOS2, JAG1, MAP3K1, STAT3 WWP1 CCL1, MATN2, TGFBI, VCL MAP3K1, STAT3, SOS2, NKIRAS1, SPRY1, SPRY2 BMPR2, SMAD7 SOS2, TIAM2, GPR64, KRIT1 PTEN | ↑ in stage I compared to control group. ↑ as NSCLC progressed. | [31,32,43,50,51] |
| Retrospective analysis of three cohorts included 317 NSCLC patients. Stages: I–III | Tissue samples | TaqMan miRNA assay | RNU66 | [30] | ||||||
| miR-145 | 80 NSCLC patients. Stages: I–IV | Plasma, Tissue samples | TaqMan miRNA assay | RNU48 | [34] | ↓ | Proliferation, EMT, Migration, Invasion Metastasis, Immune evasion Cell cycle Apoptosis Tumor growth | GOLM1, RTKN, SOX9 CXCL3 c-Myc Caspase-3/-9, MTDH EGFR, NUDT1 | Study A: ↓ in all patients, regardless of clinical stage [36]. Study B: ↓ as NSCLC progressed [38]. | [34,52,53,54,55,56] |
| 70 paired normal and NSCLC tissues. Stages: I–III | Tissue samples | TaqMan miRNA assay | miR-191 | [35] | ||||||
| miR-126 | 45 NSCLC patients. Stages: I–IV | Serum, Exosomal, Exosomal-free | TaqMan miRNA assay and RT-qPCR | U6 cel miR-39 | [39] | ↓ | Angiogenesis VEGF-A/VEGFR-2/ERK PI3K/AKT signaling mTOR signaling EMT Invasion | EGFL7, IRS-1, Crk, VEGFA PIK3R2 LAT1 SOX11, PLOD2 | ↓ in advanced stage patients compared to early-stage patients [39]. | [57,58,59] |
| miR-126-3p | 182 NSCLC patients. Stages: IA–IIIA | Serum | RT-qPCR | cel-miR-39 | [40] | |||||
| miR-155 | 50 NSCLC patients and 50 healthy volunteers. Early stages | Serum | RT-qPCR | Mean of CT of all healthy controls | [42] | ↑ | Tumor growth Metastasis Migration Invasion | PTEN, SOCS1, SOCS6 Smad2 | ↑ as NSCLC progressed. | [43,60] |
| 80 paired normal and NSCLC tissues. Stages: I–III | Tissue samples | RT-qPCR | U6 | [43] | ||||||
| miR-205 | 265 NSCLC patients. Stage: I | Tissue samples | TaqMan miRNA assay | RNU6B | [61] | ↑ | Growth metastasis EMT Apoptosis NF-κB signaling Proliferation | PTEN, TP53INP1 Cripto-1 APBB2 EGFR | ↑ as NSCLC progressed. | [49,61,62,63,64] |
| miR-150 | 171 NSCLC patients. Stages: I–II | Serum | Stem-loop array reverse transcription PCR (SLA-RT-PCR) | U6 snRNA | [65] | ↑ | Proliferation Metastasis | SRCIN1, P-STAT3, ROS, EPG5 FOXO4 | ↑ as NSCLC progressed. | [65,66,67,68,69] |
| miR-141 | 155 NSCLC patients. Stages: I–III | Tissue samples | TaqMan miRNA assay | RNU44 RNU48 | [47] | ↑ | Angiogenesis | KLF6, VEGFA | Study A: no correlations between the expression level and TNM stage [70]. Study B: ↑ as NSCLC progressed [71]. | [47] |
| miR-146a | 101 NSCLC patients. Stages: I–IV | Tissue samples | RT-qPCR | RNU6B | [49] | ↓ | Proliferation EMT Migration Survivability | EGFR, TNF-α, NF-κB and MEK-1/2, and JNK-1/2. Notch2 TRAF6 | ↓ in advanced stage patients compared to early-stage patients. | [49,72,73] |
| ↑ | ||||||||||
| miR-223 | 75 NSCLC patients and 111 tumor-free controls. Stages: I–II | Serum | droplet digital PCR (ddPCR) | UniSp6/cel-miR-39-3p | [74] | ↓ | PI3K/AKT pathway Proliferation and invasion Migration | EGFR IGF-1R NLRP3, E2F8 | ↓ in advanced stage patients compared to early-stage patients. | [74,75,76,77,78] |
| 31 NSCLC patients. Stages: I–IV 3 NSCLC cell lines. Mouse model. | Serum | Real-Time PCR | U6 snRNA | [78] | ||||||
| miR-34b/c | 140 AC patients. Stages: I–II 15 human lung AC cell lines. | Tissue samples | RT-qPCR | RNU48 | [79] | ↓ | Tumor growth Cell cycle Apoptosis EMT | TP53 CDK4, CDK6, CCND1 BCL2, SIRT1 Zeb1 | N/A | [80] |
| miR-183 | 33 AD patients. Stage: I 2 NSCLC cell lines. | Tissue samples | RT-qPCR | U6 snRNA | [81] | ↑ | mTOR regulation Proliferation, migration and cell cycle Metastasis | SESN1 PTEN, FOXO1 LOXL4 | N/A | [81,82,83,84] |
| miR-1246 | 105 NSCLC patients, 50 patients with NMRD and 50 healthy volunteers. Stages: I–IV | Serum | RT-qPCR | cel-miR-39 | [85] | ↑ | Stemness Metastasis, Wnt/β-Catenin Pathway Radioresistance | TRIM17 CPEB4, GSK-3β DR5 | ↑ in advanced stage patients compared to early-stage patients. | [85,86,87,88,89] |
| miR-328-5P | 86 NSCLC patients and 24 healthy donors. Stages: I–IV. | Peripheral blood cells | TaqMan miRNA assay | RNU38B, RNU58A | [90] | ↑ | Migration | PRKCA, IL-1beta, c-Raf1, LOXL4 | N/A | [90,91] |
| miR-328-3P | ↓ | Genomic stability | H2AX | ↓ as NSCLC progressed. | [92] | |||||
| miR-200 | 168 NSCLC patients and 128 patients with benign lung nodules. Stages: I–II | Plasma (EV-derived) | RT-qPCR | cel-miR-39-3p | [93] | ↓ | EMT through Notch signaling | Notch ligand Jagged1 and Jagged2, Flt1 PTEN, ABCA1 | Stage II patients exhibited the highest expression level compared to the other stages. | [94,95,96,97,98,99,100,101,102] |
| Let-7b | 220 NSCLC patients and 220 healthy controls. Stages: IA–IIB | Plasma | TaqMan miRNA assay | Cel-miR-54 Cel-miR-238 | [46] | ↓ | MAPK/ERK pathway Immune response | BRF2 PD-L1 | N/A | [103,104,105] |
| Serum | ||||||||||
| miR-4732-5p | 18 AD patients and 18 BPN patients. Stage: I. | Serum (EV-derived) | RT-qPCR | miR-20a | [106] | ↓ | Migration EMT | TSPAN13 XPR1, PI3K/Akt/GSK3β/Snail pathway | ↑ as NSCLC progressed. | [107,108] |
| miR-374a | 38 NSCLC patients and 27 Heathy controls Stages: IA–IIB | Tissue samples | TaqMan miRNA assay | RNU48, miR-16 and miR-26b | [109] | ↓ | Metastasis Proliferation | γ-adducin NCK1 | N/A | [110,111] |
3. Blood-Based microRNA Biomarkers: Functional Implications in NSCLC
4. Multi-miRNA Panels: Improving Early NSCLC Detection
| Panels | miRNAs | Study Cohort | Sample Type | AUC | Sensitivity | Specificity | Panel Score | Ref. |
|---|---|---|---|---|---|---|---|---|
| Zhong, Y. et al. (2021) | miR-520c-3p, miR-1274b | 207 NSCLC patients, 168 healthy controls, 31 benign nodule patients | Serum and plasma | 0.823 | 82.3 | 73.9 | 2.385 | [132] |
| Wang, P. et al. (2015) | miR-125a-5p, miR-25, miR-126 | 94 NSCLC patients and 48 stage III–IV NSCLC patients and 111 healthy controls | Serum | 0.936 | 87.5 | 87.5 | 2.686 | [137] |
| Zheng, D. et al. (2011) | miR-155, miR-197, miR-182 | 74 NSCLC patients and 68 healthy controls | Plasma | 0.9012 | 81.33 | 86.76 | 2.5821 | [138] |
| Lv, S. et al. (2017) | miR-146a, miR-222, miR-223 | 180 AD patients and 180 healthy controls | Serum | 0.951 | 84.35 | 90.83 | 2.7028 | [139] |
| Peng, H. et al. (2016) | miR-1254, miR-485-5p, miR-574-5p | Training set: 36 NSCLCs vs. 36 controls validation set: 120 NSCLCs and 71 controls | Serum | 0.844 | 93.3 | 73.2 | 2.509 | [140] |
| Aiso, T. et al. (2018) | miR-145-5p, miR-20a-5p, miR-21-5p | 56 NSCLC patients and 26 healthy controls | Serum | 0.893 | 85.7 | 80 | 2.55 | [36] |
| Ying, L. et al. (2020) | let-7a-5p, miR-1-3p, miR-1291, miR-214-3p, miR-375 | 744 NSCLC patients and 944 healthy controls | Serum | 0.935 | 82.9 | 90.7 | 2.671 | [133] |
| Yang, X. et al. (2019) | miR-146b, miR-205, miR-29c and miR-30b | 128 NSCLC patients and 30 healthy controls | Serum | 0.96 | 95.31 | 82.98 | 2.7429 | [141] |
| Poh, K.C. et al. (2025) | miR-196a-5p, miR-1268, miR-130b-5p, miR-1290, miR-106b-5p, miR-1246 | 82 NSCLC patients and 123 healthy controls | Serum | 0.989 | 92.1 | 97.5 | 2.885 | [13] |
| Wang, Y. et al. (2016) | miR-532, miR-628-3p, and miR-425-3p | 201 early-stage and 25 late-stage AD patients and 43 patients with lung benign disease and 178 healthy controls | Plasma | 0.976 | 90.2 | 98.9 | 2.867 | [136] |
| Leng, Q. et al. (2017) | miR-21, 210, and 486-5p | 92 LC patients and 88 cancer-free smokers | Plasma | 0.85 | 75.5 | 85.3 | 2.458 | [142] |
| miR-126, miR-145, miR-210, and miR-205-5p | 0.96 | 91.5 | 96.2 | 2.837 | ||||
| Abdipourbozorgbaghi, M. et al. (2024) | miR-9-3p, miR-96-5p, miR-147b-3p, miR-196a-5p, miR-708-3p, miR-708-5p, miR-4652-5p | 78 NSCLC patients and 44 healthy controls | Plasma | 0.85 | 83 | 78 | 2.46 | [143] |
| miR-130b-3p, miR-269-3p, miR-301a-5p, miR-301b-5p, miR-744-3p, miR-760, miR-767-5p, miR-4652-5p, miR-6499-3p | 0.88 | 92 | 73 | 2.53 |
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Adenocarcinoma |
| BAL | Bronchoalveolar lavage |
| Cf-miRNA | Cell-free microRNA |
| LC | Lung Cancer |
| miRNA | MicroRNA |
| NSCLC | Non-Small-Cell Lung Cancer |
| RT-qPCR | Quantitative real-time Polymerase Chain Reaction |
| SSC | Squamous Cell Carcinoma |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Verbraecken, J.; Van de Heyning, P.; De Backer, W.; Van Gaal, L. Body surface area in normal-weight, overweight, and obese adults. A comparison study. Metabolism 2006, 55, 515–524. [Google Scholar] [CrossRef]
- Ozlu, T.; Bulbul, Y. Smoking and lung cancer. Tuberk. Toraks 2005, 53, 200–209. [Google Scholar] [PubMed]
- Possenti, I.; Romelli, M.; Carreras, G.; Biffi, A.; Bagnardi, V.; Specchia, C.; Gallus, S.; Lugo, A. Association between second-hand smoke exposure and lung cancer risk in never-smokers: A systematic review and meta-analysis. Eur. Respir. Rev. 2024, 33, 240077. [Google Scholar] [CrossRef]
- Saller, J.J.; Boyle, T.A. Molecular Pathology of Lung Cancer. Cold Spring Harb. Perspect. Med. 2022, 12, a037812. [Google Scholar] [CrossRef]
- Guo, H.; Li, H.; Zhu, L.; Feng, J.; Huang, X.; Baak, J.P.A. “How Long Have I Got?” in Stage IV NSCLC Patients with at Least 3 Months Up to 10 Years Survival, Accuracy of Long-, Intermediate-, and Short-Term Survival Prediction Is Not Good Enough to Answer This Question. Front. Oncol. 2021, 11, 761042. [Google Scholar] [CrossRef] [PubMed]
- Jovanoski, N.; Bowes, K.; Brown, A.; Belleli, R.; Di Maio, D.; Chadda, S.; Abogunrin, S. Survival and quality-of-life outcomes in early-stage NSCLC patients: A literature review of real-world evidence. Lung Cancer Manag. 2023, 12, LMT60. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, J.B.; Keum, D.Y.; Hwang, I.; Park, C.K. Long term survival of patients with unsuspected n2 disease in non-small cell lung cancer. Korean J. Thorac. Cardiovasc. Surg. 2013, 46, 49–55. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.H.; Chang, Y.S. Recent advances in diagnostic technologies in lung cancer. Korean J. Intern. Med. 2020, 35, 257–268. [Google Scholar] [CrossRef]
- Jeon, H.; Wang, S.; Song, J.; Gill, H.; Cheng, H. Update 2025: Management of Non-Small-Cell Lung Cancer. Lung 2025, 203, 53. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- Force, U.S.P.S.T.; Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962–970. [Google Scholar] [CrossRef]
- Poh, K.C.; Ren, T.M.; Ling, G.L.; Goh, J.S.Y.; Rose, S.; Wong, A.; Mehta, S.S.; Goh, A.; Chong, P.Y.; Cheng, S.W.; et al. Development of a miRNA-Based Model for Lung Cancer Detection. Cancers 2025, 17, 942. [Google Scholar] [CrossRef]
- Wadowska, K.; Bil-Lula, I.; Trembecki, L.; Sliwinska-Mosson, M. Genetic Markers in Lung Cancer Diagnosis: A Review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef]
- Zou, Y.; Wang, Y.; Jiang, Z.; Zhou, Y.; Chen, Y.; Hu, Y.; Jiang, G.; Xie, D. Breath profile as composite biomarkers for lung cancer diagnosis. Lung Cancer 2021, 154, 206–213. [Google Scholar] [CrossRef]
- Lv, W.; Shi, W.; Zhang, Z.; Ru, L.; Feng, W.; Tang, H.; Wang, X. Identification of volatile biomarkers for lung cancer from different histological sources: A comprehensive study. Anal. Biochem. 2024, 690, 115527. [Google Scholar] [CrossRef]
- Janssens, E.; van Meerbeeck, J.P.; Lamote, K. Volatile organic compounds in human matrices as lung cancer biomarkers: A systematic review. Crit. Rev. Oncol. Hematol. 2020, 153, 103037. [Google Scholar] [CrossRef]
- Bazan Russo, T.D.; Pepe, F.; Gristina, V.; Gottardo, A.; Russo, G.; Scimone, C.; Palumbo, L.; Busuito, G.; Incorvaia, L.; Guerry, J.A.; et al. Recent advances in liquid biopsy for precision oncology: Emerging biomarkers and clinical applications in lung cancer. Future Oncol. 2025, 21, 2803–2821. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Herman, J.G. Noninvasive Diagnostics for Early Detection of Lung Cancer: Challenges and Potential with a Focus on Changes in DNA Methylation. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2416–2422. [Google Scholar] [CrossRef] [PubMed]
- Chow, Y.P.; Zainul Abidin, N.; Kow, K.S.; Tho, L.M.; Wong, C.L. Analytical and clinical validation of a custom 15-gene next-generation sequencing panel for the evaluation of circulating tumor DNA mutations in patients with advanced non-small-cell lung cancer. PLoS ONE 2022, 17, e0276161. [Google Scholar] [CrossRef] [PubMed]
- Kumbrink, J.; Demes, M.C.; Jeroch, J.; Brauninger, A.; Hartung, K.; Gerstenmaier, U.; Marienfeld, R.; Hillmer, A.; Bohn, N.; Lehning, C.; et al. Development, testing and validation of a targeted NGS-panel for the detection of actionable mutations in lung cancer (NSCLC) using anchored multiplex PCR technology in a multicentric setting. Pathol. Oncol. Res. 2024, 30, 1611590. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, D.; Guo, H.; Ma, W. Beyond blood: Advancing the frontiers of liquid biopsy in oncology and personalized medicine. Cancer Sci. 2024, 115, 1060–1072. [Google Scholar] [CrossRef]
- Bryzgunova, O.; Konoshenko, M.; Zaporozhchenko, I.; Yakovlev, A.; Laktionov, P. Isolation of Cell-Free miRNA from Biological Fluids: Influencing Factors and Methods. Diagnostics 2021, 11, 865. [Google Scholar] [CrossRef]
- Liao, J.; Shen, J.; Leng, Q.; Qin, M.; Zhan, M.; Jiang, F. MicroRNA-based biomarkers for diagnosis of non-small cell lung cancer (NSCLC). Thorac. Cancer 2020, 11, 762–768. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Zhang, Q.; Tang, L.; Liu, X.; Dai, Y.; Xiao, L.; Huang, S.; Chen, L.; Guo, Z.; et al. MicroRNA-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS ONE 2015, 10, e0134220, Correction in PLoS ONE 2016, 11, e0148589. https://doi.org/10.1371/journal.pone.0148589. [Google Scholar] [CrossRef]
- Yi, M.; Liao, Z.; Deng, L.; Xu, L.; Tan, Y.; Liu, K.; Chen, Z.; Zhang, Y. High diagnostic value of miRNAs for NSCLC: Quantitative analysis for both single and combined miRNAs in lung cancer. Ann. Med. 2021, 53, 2178–2193. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Guan, Y.; Zhang, Z.; Wang, H. Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer. BMC Cancer 2020, 20, 329. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Saito, M.; Schetter, A.J.; Mollerup, S.; Kohno, T.; Skaug, V.; Bowman, E.D.; Mathe, E.A.; Takenoshita, S.; Yokota, J.; Haugen, A.; et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: A retrospective analysis of three cohorts. Clin. Cancer Res. 2011, 17, 1875–1882. [Google Scholar] [CrossRef]
- Wei, J.; Gao, W.; Zhu, C.J.; Liu, Y.Q.; Mei, Z.; Cheng, T.; Shu, Y.Q. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin. J. Cancer 2011, 30, 407–414. [Google Scholar] [CrossRef]
- Qi, Z.; Yang, D.Y.; Cao, J. Increased micro-RNA 17, 21, and 192 gene expressions improve early diagnosis in non-small cell lung cancer. Med. Oncol. 2014, 31, 195. [Google Scholar] [CrossRef]
- Geng, Q.; Fan, T.; Zhang, B.; Wang, W.; Xu, Y.; Hu, H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir. Res. 2014, 15, 149. [Google Scholar] [CrossRef]
- Cho, W.C.; Wong, C.F.; Li, K.P.; Fong, A.H.; Fung, K.Y.; Au, J.S. miR-145 as a Potential Biomarker and Therapeutic Target in Patients with Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 10022. [Google Scholar] [CrossRef] [PubMed]
- Campayo, M.; Navarro, A.; Vinolas, N.; Diaz, T.; Tejero, R.; Gimferrer, J.M.; Molins, L.; Cabanas, M.L.; Ramirez, J.; Monzo, M.; et al. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur. Respir. J. 2013, 41, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Aiso, T.; Ohtsuka, K.; Ueda, M.; Karita, S.; Yokoyama, T.; Takata, S.; Matsuki, N.; Kondo, H.; Takizawa, H.; Okada, A.A.; et al. Serum levels of candidate microRNA diagnostic markers differ among the stages of non-small-cell lung cancer. Oncol. Lett. 2018, 16, 6643–6651. [Google Scholar] [CrossRef] [PubMed]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016, 7, 72084–72098. [Google Scholar] [CrossRef]
- Shen, H.; Shen, J.; Wang, L.; Shi, Z.; Wang, M.; Jiang, B.H.; Shu, Y. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed. Pharmacother. 2015, 69, 301–305. [Google Scholar] [CrossRef]
- Grimolizzi, F.; Monaco, F.; Leoni, F.; Bracci, M.; Staffolani, S.; Bersaglieri, C.; Gaetani, S.; Valentino, M.; Amati, M.; Rubini, C.; et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci. Rep. 2017, 7, 15277. [Google Scholar] [CrossRef]
- Ulivi, P.; Petracci, E.; Marisi, G.; Baglivo, S.; Chiari, R.; Billi, M.; Canale, M.; Pasini, L.; Racanicchi, S.; Vagheggini, A.; et al. Prognostic Role of Circulating miRNAs in Early-Stage Non-Small Cell Lung Cancer. J. Clin. Med. 2019, 8, 131. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, F.; Shen, T.; Luo, Q.; Ding, Z.; Qian, L.; Huang, J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol. Lett. 2017, 13, 669–676. [Google Scholar] [CrossRef]
- Azimi, S.A.; Sadegh Nia, H.R.; Bahrami, N.; Dargahi, H.; Jamaati, H.; Pasdar, A.; Kazempour Dizaji, M.; Shirian, S.; Zerehsaz, B.; Mohamadnia, A. Expression of miR-155 and CEA and VEGF Proteins as Diagnostic Markers in Early Stages of Non-Small Cell Lung Cancer in Peripheral Blood. Tanaffos 2024, 23, 58–64. [Google Scholar]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef]
- Zhang, W.T.; Zhang, G.X.; Gao, S.S. The Potential Diagnostic Accuracy of Let-7 Family for Cancer: A Meta-Analysis. Technol. Cancer Res. Treat. 2021, 20, 15330338211033061. [Google Scholar] [CrossRef]
- Tulinsky, L.; Dzian, A.; Matakova, T.; Ihnat, P. Overexpression of the miR-143/145 and reduced expression of the let-7 and miR-126 for early lung cancer diagnosis. J. Appl. Biomed. 2022, 20, 1–6. [Google Scholar] [CrossRef]
- Heegaard, N.H.; Schetter, A.J.; Welsh, J.A.; Yoneda, M.; Bowman, E.D.; Harris, C.C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer 2012, 130, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Tejero, R.; Navarro, A.; Campayo, M.; Vinolas, N.; Marrades, R.M.; Cordeiro, A.; Ruiz-Martinez, M.; Santasusagna, S.; Molins, L.; Ramirez, J.; et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS ONE 2014, 9, e101899. [Google Scholar] [CrossRef]
- Zeybek, A.; Oz, N.; Kalemci, S.; Edgunlu, T.; Kiziltug, M.T.; Tosun, K.; Tunc, M.; Tekin, L.; Erdal, M.E. Diagnostic Value of MiR-125b as a Potential Biomarker for Stage I Lung Adenocarcinoma. Curr. Mol. Med. 2019, 19, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Umelo, I.A.; Lv, S.; Teugels, E.; Fostier, K.; Kronenberger, P.; Dewaele, A.; Sadones, J.; Geers, C.; De Greve, J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS ONE 2013, 8, e60317. [Google Scholar] [CrossRef]
- Capodanno, A.; Boldrini, L.; Proietti, A.; Ali, G.; Pelliccioni, S.; Niccoli, C.; D’Incecco, A.; Cappuzzo, F.; Chella, A.; Lucchi, M.; et al. Let-7g and miR-21 expression in non-small cell lung cancer: Correlation with clinicopathological and molecular features. Int. J. Oncol. 2013, 43, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Cho, W.C.; Chow, A.S.; Au, J.S. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011, 8, 125–131. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, H.; Guo, Y.; Liu, P.; Pan, H.; Deng, A.; Hu, J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. 2010, 29, 151. [Google Scholar] [CrossRef]
- Zhang, G.; Lei, S.; Zhang, K. The effect of microRNA-145 on proliferation and apoptosis of cutaneous squamous cell carcinoma cells. Discov. Oncol. 2025, 16, 1201. [Google Scholar] [CrossRef]
- Pei, X.; Chen, S.W.; Long, X.; Zhu, S.Q.; Qiu, B.Q.; Lin, K.; Lu, F.; Xu, J.J.; Zhang, P.F.; Wu, Y.B. circMET promotes NSCLC cell proliferation, metastasis, and immune evasion by regulating the miR-145-5p/CXCL3 axis. Aging 2020, 12, 13038–13058. [Google Scholar] [CrossRef]
- Alinejad, T.; Hao, Z.; Zhou, W.; Zareh, D.; Farajtabrizi, E.; Mossahebi-Mohammadi, M.; Chen, C.S. Knockdown of long noncoding RNA MALAT1 enhances the anti-cancer effects of polysaccharides Glehnia littoralis in lung cancer cells possibly via the regulation of miR-145/SOX9 axis. Int. Immunopharmacol. 2025, 164, 115323. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, S.; Zhao, J.; Zhou, Y.; Xu, L. MicroRNA-126: A new and promising player in lung cancer. Oncol. Lett. 2021, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Mikosz, A.M.; Ringsby, A.J.; Anderson, K.C.; Beatman, E.L.; Koike, K.; Petrache, I. MicroRNA-126-3p Inhibits Angiogenic Function of Human Lung Microvascular Endothelial Cells via LAT1 (L-Type Amino Acid Transporter 1)-Mediated mTOR (Mammalian Target of Rapamycin) Signaling. Arter. Thromb. Vasc. Biol. 2020, 40, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Gu, Y.Y.; Ma, F.C.; He, R.Q.; Li, Z.Y.; Zhai, G.Q.; Lin, X.; Hu, X.H.; Pan, L.J.; Chen, G. Expression levels and co-targets of miRNA-126-3p and miRNA-126-5p in lung adenocarcinoma tissues: Alphan exploration with RT-qPCR, microarray and bioinformatic analyses. Oncol. Rep. 2019, 41, 939–953. [Google Scholar] [CrossRef]
- Lin, J.; Chen, Y.; Liu, L.; Shen, A.; Zheng, W. MicroRNA-155-5p suppresses the migration and invasion of lung adenocarcinoma A549 cells by targeting Smad2. Oncol. Lett. 2018, 16, 2444–2452. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Moon, Y.W.; Raffeld, M.; Lee, D.H.; Wang, Y.; Giaccone, G. High cripto-1 and low miR-205 expression levels as prognostic markers in early stage non-small cell lung cancer. Lung Cancer 2018, 116, 38–45. [Google Scholar] [CrossRef]
- Lei, L.; Huang, Y.; Gong, W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol. Rep. 2013, 30, 2897–2902. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.B.; Xiong, J.; Zhang, Y.H.; Dai, Y.; Ren, X.P.; Ren, Y.J.; Han, D.; Wei, S.H.; Qi, M. miR-205-3p promotes lung cancer progression by targeting APBB2. Mol. Med. Rep. 2021, 24, 588. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Zhang, J.X.; Yang, J.J.; Wei, Y.B.; Peng, J.F.; Fu, C.J.; Huang, M.H.; Wang, R.; Wang, P.Y.; Sun, G.B.; et al. MiR-205-5p promotes lung cancer progression and is valuable for the diagnosis of lung cancer. Thorac. Cancer 2022, 13, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, J.; Ye, Y.; Oba, T.; Gentile, E.; Lian, J.; Wang, J.; Zhao, Y.; Gu, J.; Wistuba, I.I.; et al. Serum MicroRNA-150 Predicts Prognosis for Early-Stage Non-Small Cell Lung Cancer and Promotes Tumor Cell Proliferation by Targeting Tumor Suppressor Gene SRCIN1. Clin. Pharmacol. Ther. 2018, 103, 1061–1073. [Google Scholar] [CrossRef]
- Qin, A.; Chen, H.; Xu, F.; Li, W.; Guo, S.; Zhang, G.; Zhang, A.; Zheng, A.; Tian, F.; Zheng, Q. MiR-150 deletion promotes lung tumor growth by upregulating P-STAT3 and ROS in MDSCs. Sci. Rep. 2025, 15, 12988. [Google Scholar] [CrossRef]
- Li, H.; Liu, J.; Cao, W.; Xiao, X.; Liang, L.; Liu-Smith, F.; Wang, W.; Liu, H.; Zhou, P.; Ouyang, R.; et al. C-myc/miR-150/EPG5 axis mediated dysfunction of autophagy promotes development of non-small cell lung cancer. Theranostics 2019, 9, 5134–5148. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ouyang, R.; Wang, Z.; Zhou, W.; Chen, H.; Jiang, Y.; Zhang, Y.; Li, H.; Liao, M.; Wang, W.; et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 2016, 6, 39001. [Google Scholar] [CrossRef]
- Yin, Q.W.; Sun, X.F.; Yang, G.T.; Li, X.B.; Wu, M.S.; Zhao, J. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 842–846. [Google Scholar]
- Zhao, Y. The diagnostic and prognostic role of circulating miR-141 expression in non-small-cell lung cancer patients. Int. J. Clin. Exp. Pathol. 2018, 11, 2597–2604. [Google Scholar]
- Zhang, X.; Li, P.; Rong, M.; He, R.; Hou, X.; Xie, Y.; Chen, G. MicroRNA-141 is a biomarker for progression of squamous cell carcinoma and adenocarcinoma of the lung: Clinical analysis of 125 patients. Tohoku J. Exp. Med. 2015, 235, 161–169. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Zhang, L.; Chen, X.; Liu, F.; Zhang, J.; Guan, S.; Sun, Y.; Chen, P.; Wang, D.; et al. miR-146a-5p mediates epithelial-mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br. J. Cancer 2016, 115, 1548–1554, Correction in Br. J. Cancer 2018, 118, e12. https://doi.org/10.1038/bjc.2017.471. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, B.; Li, R.; Wang, F.; Wang, N.; Zhang, M.; Bai, Y.; Wu, J.; Liu, L.; Han, D.; et al. miR-146a-5p Plays an Oncogenic Role in NSCLC via Suppression of TRAF6. Front. Cell Dev. Biol. 2020, 8, 847. [Google Scholar] [CrossRef] [PubMed]
- D’Antona, P.; Cattoni, M.; Dominioni, L.; Poli, A.; Moretti, F.; Cinquetti, R.; Gini, E.; Daffre, E.; Noonan, D.M.; Imperatori, A.; et al. Serum miR-223: A Validated Biomarker for Detection of Early-Stage Non-Small Cell Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.P.; Kong, W.X.; Li, X.Y.; Li, W.; Zhang, Y.; Wu, Y. miRNA-223 is an anticancer gene in human non-small cell lung cancer through the PI3K/AKT pathway by targeting EGFR. Oncol. Rep. 2019, 41, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Nian, W.; Ao, X.; Wu, Y.; Huang, Y.; Shao, J.; Wang, Y.; Chen, Z.; Chen, F.; Wang, D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol. Lett. 2013, 6, 359–366. [Google Scholar] [CrossRef]
- Zhu, S.; Kong, X.; Song, M.; Chi, M.; Liu, Y.; Zhang, P.; Zhang, Q.; Shang, P.; Feng, F. MiR-223-3p attenuates the migration and invasion of NSCLC cells by regulating NLRP3. Front. Oncol. 2022, 12, 985962. [Google Scholar] [CrossRef]
- Dou, L.; Han, K.; Xiao, M.; Lv, F. miR-223-5p Suppresses Tumor Growth and Metastasis in Non-Small Cell Lung Cancer by Targeting E2F8. Oncol. Res. 2019, 27, 261–268. [Google Scholar] [CrossRef]
- Nadal, E.; Chen, G.; Gallegos, M.; Lin, L.; Ferrer-Torres, D.; Truini, A.; Wang, Z.; Lin, J.; Reddy, R.M.; Llatjos, R.; et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin. Cancer Res. 2013, 19, 6842–6852. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, E.J.; Lee, S.; Tan, X.; Liu, X.; Park, S.; Kang, K.; Yoon, J.S.; Ko, Y.H.; Kurie, J.M.; et al. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Y.; Li, L.; Li, B.; Jiang, T.; Cao, Y.; Yang, X.; Liu, X.; Qu, H.; Li, S.; et al. Integration analysis of microRNAs as potential biomarkers in early-stage lung adenocarcinoma: The diagnostic and therapeutic significance of miR-183-3p. Front. Oncol. 2024, 14, 1508715. [Google Scholar] [CrossRef]
- Zhang, T.; Li, W.; Gu, M.; Wang, Z.; Zhou, S.; Hao, X.; Li, W.; Xu, S. Clinical Significance of miR-183-3p and miR-182-5p in NSCLC and Their Correlation. Cancer Manag. Res. 2021, 13, 3539–3550. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, D.; Liao, H.; Li, Y. miR-183-5p regulates ECM and EMT to promote non-small cell lung cancer progression by targeting LOXL4. J. Thorac. Dis. 2023, 15, 1734–1748. [Google Scholar] [CrossRef]
- Zhang, L.; Quan, H.; Wang, S.; Li, X.; Che, X. MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015, 36, 8121–8126. [Google Scholar] [CrossRef]
- Huang, D.; Qu, D. Early diagnostic and prognostic value of serum exosomal miR-1246 in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2020, 13, 1601–1607. [Google Scholar] [PubMed]
- Liu, Z.; Zhao, W.; Yang, R. MiR-1246 is responsible for lung cancer cells-derived exosomes-mediated promoting effects on lung cancer stemness via targeting TRIM17. Environ. Toxicol. 2022, 37, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Li, F.; Wang, L.; Chen, C.; Huang, X.; Wu, X.; She, W.; Zhou, L.; Tao, Z. Downregulated cytoplasmic polyadenylation element-binding protein-4 is associated with the carcinogenesis of head and neck squamous cell carcinoma. Oncol. Lett. 2018, 15, 3226–3232. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiong, H.; Duan, L.; Li, Q.; Li, X.; Zhou, Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3beta-Mediated Wnt/beta-Catenin Pathway. Cancer Res. Treat. 2019, 51, 1420–1429. [Google Scholar] [CrossRef]
- Yuan, D.; Xu, J.; Wang, J.; Pan, Y.; Fu, J.; Bai, Y.; Zhang, J.; Shao, C. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016, 7, 32707–32722. [Google Scholar] [CrossRef]
- Ulivi, P.; Foschi, G.; Mengozzi, M.; Scarpi, E.; Silvestrini, R.; Amadori, D.; Zoli, W. Peripheral blood miR-328 expression as a potential biomarker for the early diagnosis of NSCLC. Int. J. Mol. Sci. 2013, 14, 10332–10342. [Google Scholar] [CrossRef]
- Ji, Y.; You, Y.; Wu, Y.; Wang, M.; He, Q.; Zhou, X.; Chen, L.; Sun, X.; Liu, Y.; Fu, X.; et al. Overexpression of miR-328-5p influences cell growth and migration to promote NSCLC progression by targeting LOXL4. Ann. Transl. Med. 2022, 10, 301. [Google Scholar] [CrossRef]
- Ma, W.; Ma, C.N.; Zhou, N.N.; Li, X.D.; Zhang, Y.J. Up-regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Sci. Rep. 2016, 6, 31651. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, F.; Niu, D.; Guo, X.; Lei, T.; Liu, H. Diagnostic value of microRNA-200 expression in peripheral blood-derived extracellular vesicles in early-stage non-small cell lung cancer. Clin. Exp. Med. 2024, 24, 214. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Bao, T.; Li, Z.; Ji, G.; Zhang, L. Function of miR-200a in proliferation and apoptosis of non-small cell lung cancer cells. Oncol. Lett. 2020, 20, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; He, X.; Fang, C.; Wu, H.; Hu, L.; Xue, Y. MicroRNA-200a-3p and GATA6 are abnormally expressed in patients with non-small cell lung cancer and exhibit high clinical diagnostic efficacy. Exp. Ther. Med. 2022, 23, 281. [Google Scholar] [CrossRef]
- Li, W.; Jia, M.X.; Deng, J.; Wang, J.H.; Lin, Q.L.; Tang, J.X.; Zeng, X.X.; Cai, F.; Ma, L.; Su, W.; et al. Down-regulation of microRNA-200b is a potential prognostic marker of lung cancer in southern-central Chinese population. Saudi J. Biol. Sci. 2019, 26, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, W.; Tan, J.; Ma, J.; Zhao, J. MiR-200b-3p Functions as an Oncogene by Targeting ABCA1 in Lung Adenocarcinoma. Technol. Cancer Res. Treat. 2019, 18, 1533033819892590. [Google Scholar] [CrossRef]
- Chen, S.; Tu, Y.; Yuan, H.; Shi, Z.; Guo, Y.; Gong, W.; Tu, S. Regulatory functions of miR-200b-3p in tumor development (Review). Oncol. Rep. 2022, 47, 96. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, N.; Huang, T.; Shen, N. MicroRNA-200c in Cancer Generation, Invasion, and Metastasis. Int. J. Mol. Sci. 2025, 26, 710. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, J.; Xiong, D.; Zheng, J.; McPherson, K.N.; Lee, S.; Huang, M.; Xu, Y.; Chen, S.H.; Wang, Y.; et al. Aerosolized miR-138-5p and miR-200c targets PD-L1 for lung cancer prevention. Front. Immunol. 2023, 14, 1166951. [Google Scholar] [CrossRef]
- Shen, Y.; Zhu, Y.; Rong, F. miR-200c-3p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting PTEN. Mol. Med. Rep. 2020, 22, 1605–1612. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Li, Q.; Xu, R.; Huang, N. MiR-200c-3p and miR-485-5p overexpression elevates cisplatin sensitivity and suppresses the malignant phenotypes of non-small cell lung cancer cells through targeting RRM2. Thorac. Cancer 2022, 13, 1974–1985. [Google Scholar] [CrossRef] [PubMed]
- Pop-Bica, C.; Pintea, S.; Magdo, L.; Cojocneanu, R.; Gulei, D.; Ferracin, M.; Berindan-Neagoe, I. The Clinical Utility of miR-21 and let-7 in Non-small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 516850. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, R.; Lu, M.; Cheng, C.; Feng, Z.; Zhao, R.; Liang, J.; Han, J.; Jiang, J.; Xu-Welliver, M.; et al. Let-7b-3p inhibits tumor growth and metastasis by targeting the BRF2-mediated MAPK/ERK pathway in human lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 1841–1856. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, J.; Xiong, D.; Wang, Y.; Miller, M.S.; Sei, S.; Shoemaker, R.H.; Izzotti, A.; You, M. Pulmonary Aerosol Delivery of Let-7b microRNA Confers a Striking Inhibitory Effect on Lung Carcinogenesis through Targeting the Tumor Immune Microenvironment. Adv. Sci. 2021, 8, e2100629. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Chen, J.; Yu, L.; Li, J.; You, S.; Zheng, Y.; Zhuang, W.; Qiu, B.; Huang, Y. Detection of fucosylated extracellular vesicles miR-4732-5p related to diagnosis of early lung adenocarcinoma by the electrochemical biosensor. Sci. Rep. 2024, 14, 11217. [Google Scholar] [CrossRef]
- Tang, X.; Liu, S.; Cui, Y.; Zhao, Y. MicroRNA-4732 is downregulated in non-small cell lung cancer and inhibits tumor cell proliferation, migration, and invasion. Respir. Med. Res. 2021, 80, 100865. [Google Scholar] [CrossRef]
- Hu, Y.; Bai, J.; Zhou, D.; Zhang, L.; Chen, X.; Chen, L.; Liu, Y.; Zhang, B.; Li, H.; Yin, C. The miR-4732-5p/XPR1 axis suppresses the invasion, metastasis, and epithelial-mesenchymal transition of lung adenocarcinoma via the PI3K/Akt/GSK3beta/Snail pathway. Mol. Omics 2022, 18, 417–429. [Google Scholar] [CrossRef]
- Vosa, U.; Vooder, T.; Kolde, R.; Fischer, K.; Valk, K.; Tonisson, N.; Roosipuu, R.; Vilo, J.; Metspalu, A.; Annilo, T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer 2011, 50, 812–822. [Google Scholar] [CrossRef]
- Jin, J.; Cui, Y.; Niu, H.; Lin, Y.; Wu, X.; Qi, X.; Bai, K.; Zhang, Y.; Wang, Y.; Bu, H. NSCLC Extracellular Vesicles Containing miR-374a-5p Promote Leptomeningeal Metastasis by Influencing Blood–Brain Barrier Permeability. Mol. Cancer Res. 2024, 22, 699–710. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, H.; Xu, Y.; Wang, M.; Tian, Z. miR-374a-5p inhibits non-small cell lung cancer cell proliferation and migration via targeting NCK1. Exp. Ther. Med. 2021, 22, 943. [Google Scholar] [CrossRef]
- Ling, D.J.; Chen, Z.S.; Zhang, Y.D.; Liao, Q.D.; Feng, J.X.; Zhang, X.Y.; Shi, T.S. MicroRNA-145 inhibits lung cancer cell metastasis. Mol. Med. Rep. 2015, 11, 3108–3114. [Google Scholar] [CrossRef]
- Li, B.; Ding, C.M.; Li, Y.X.; Peng, J.C.; Geng, N.; Qin, W.W. MicroRNA-145 inhibits migration and induces apoptosis in human non-small cell lung cancer cells through regulation of the EGFR/PI3K/AKT signaling pathway. Oncol. Rep. 2018, 40, 2944–2954. [Google Scholar] [CrossRef]
- Jiang, L.P.; He, C.Y.; Zhu, Z.T. Role of microRNA-21 in radiosensitivity in non-small cell lung cancer cells by targeting PDCD4 gene. Oncotarget 2017, 8, 23675–23689. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, X.; Tian, W.; Yin, X.; Wang, J.; Yang, H. The role of TGF-beta1-miR-21-ROS pathway in bystander responses induced by irradiated non-small-cell lung cancer cells. Br. J. Cancer 2014, 111, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, Y.; Liu, M.; Bi, X.; Bao, J.; Zeng, N.; Zhu, Z.; Mo, Z.; Wu, C.; Chen, X. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1alpha expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 2012, 103, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Cao, L.; Zhang, J. miR-21 modulates paclitaxel sensitivity and hypoxia-inducible factor-1alpha expression in human ovarian cancer cells. Oncol. Lett. 2013, 6, 795–800. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, R.; Yan, H.; Jin, L.; Dou, X.; Chen, D. MicroRNA-21 modulates radiation resistance through upregulation of hypoxia-inducible factor-1alpha-promoted glycolysis in non-small cell lung cancer cells. Mol. Med. Rep. 2016, 13, 4101–4107. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Bhawal, U.K.; Ueda, N.; Shimomoto, T.; Yamamoto, K.; Kirita, T.; Kuniyasu, H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br. J. Cancer 2012, 107, 700–706. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L.; Chen, J.; Ruan, L.; Tan, A.; Wang, P. miR-34a inhibits tumorigenesis of NSCLC via targeting SIRT6. Int. J. Clin. Exp. Pathol. 2018, 11, 1135–1145. [Google Scholar]
- Zhang, T.; Hu, Y.; Yang, N.; Yu, S.; Pu, X. The microRNA-34 Family and Its Functional Role in Lung Cancer. Am. J. Clin. Oncol. 2024, 47, 448–457. [Google Scholar] [CrossRef]
- Ballou, L.M.; Lin, R.Z. Rapamycin and mTOR kinase inhibitors. J. Chem. Biol. 2008, 1, 27–36. [Google Scholar] [CrossRef]
- Costa, D.B.; Nguyen, K.S.; Cho, B.C.; Sequist, L.V.; Jackman, D.M.; Riely, G.J.; Yeap, B.Y.; Halmos, B.; Kim, J.H.; Janne, P.A.; et al. Effects of erlotinib in EGFR mutated non-small cell lung cancers with resistance to gefitinib. Clin. Cancer Res. 2008, 14, 7060–7067. [Google Scholar] [CrossRef]
- Hu, Y.; Hong, Y.; Xu, Y.; Liu, P.; Guo, D.H.; Chen, Y. Inhibition of the JAK/STAT pathway with ruxolitinib overcomes cisplatin resistance in non-small-cell lung cancer NSCLC. Apoptosis 2014, 19, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Hillan, K.J.; Novotny, W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 2005, 333, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Giaccone, G.; de Marinis, F.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.; Geater, S.; et al. Atezolizumab for First-Line Treatment of PD-L1-Selected Patients with NSCLC. N. Engl. J. Med. 2020, 383, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Loke, S.Y.; Tan, V.K.; Quek, S.T.; Jagmohan, P.; Tang, Y.C.; Madhukumar, P.; Tan, B.K.; Yong, W.S.; Sim, Y.; et al. Development of a microRNA Panel for Classification of Abnormal Mammograms for Breast Cancer. Cancers 2021, 13, 2130. [Google Scholar] [CrossRef]
- Dama, E.; Colangelo, T.; Cuttano, R.; Dziadziuszko, R.; Dandekar, T.; Widlak, P.; Rzyman, W.; Veronesi, G.; Bianchi, F. A plasma 9-microRNA signature for lung cancer early detection: A multicenter analysis. Biomark. Res. 2025, 13, 74. [Google Scholar] [CrossRef]
- Sur, D.; Advani, S.; Braithwaite, D. MicroRNA panels as diagnostic biomarkers for colorectal cancer: A systematic review and meta-analysis. Front. Med. 2022, 9, 915226. [Google Scholar] [CrossRef]
- Zhong, Y.; Ding, X.; Bian, Y.; Wang, J.; Zhou, W.; Wang, X.; Li, P.; Shen, Y.; Wang, J.J.; Li, J.; et al. Discovery and validation of extracellular vesicle-associated miRNAs as noninvasive detection biomarkers for early-stage non-small-cell lung cancer. Mol. Oncol. 2021, 15, 2439–2452. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Du, L.; Zou, R.; Shi, L.; Zhang, N.; Jin, J.; Xu, C.; Zhang, F.; Zhu, C.; Wu, J.; et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 25036–25042. [Google Scholar] [CrossRef]
- Kamkar, L.; Saberi, S.; Totonchi, M.; Kavousi, K. Circulating microRNA panels for multi-cancer detection and gastric cancer screening: Leveraging a network biology approach. BMC Med. Genom. 2025, 18, 27. [Google Scholar] [CrossRef]
- Wu, P.; Li, D.; Zhang, C.; Dai, B.; Tang, X.; Liu, J.; Wu, Y.; Wang, X.; Shen, A.; Zhao, J.; et al. A unique circulating microRNA pairs signature serves as a superior tool for early diagnosis of pan-cancer. Cancer Lett. 2024, 588, 216655. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Gao, X.; Wei, F.; Zhang, X.; Su, Y.; Wang, C.; Li, H.; Ren, X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget 2016, 7, 26070–26086. [Google Scholar] [CrossRef]
- Wang, P.; Yang, D.; Zhang, H.; Wei, X.; Ma, T.; Cheng, Z.; Hong, Q.; Hu, J.; Zhuo, H.; Song, Y.; et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer 2015, 16, 313–319.e1. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Haddadin, S.; Wang, Y.; Gu, L.Q.; Perry, M.C.; Freter, C.E.; Wang, M.X. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 2011, 4, 575–586. [Google Scholar]
- Lv, S.; Xue, J.; Wu, C.; Wang, L.; Wu, J.; Xu, S.; Liang, X.; Lou, J. Identification of A Panel of Serum microRNAs as Biomarkers for Early Detection of Lung Adenocarcinoma. J. Cancer 2017, 8, 48–56. [Google Scholar] [CrossRef]
- Peng, H.; Wang, J.; Li, J.; Zhao, M.; Huang, S.K.; Gu, Y.Y.; Li, Y.; Sun, X.J.; Yang, L.; Luo, Q.; et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016, 151, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Q.; Zhang, M.; Su, W.; Wang, Z.; Li, Y.; Zhang, J.; Beer, D.G.; Yang, S.; Chen, G. Serum microRNA Signature Is Capable of Early Diagnosis for Non-Small Cell Lung Cancer. Int. J. Biol. Sci. 2019, 15, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Lin, Y.; Jiang, F.; Lee, C.J.; Zhan, M.; Fang, H.; Wang, Y.; Jiang, F. A plasma miRNA signature for lung cancer early detection. Oncotarget 2017, 8, 111902–111911. [Google Scholar] [CrossRef]
- Abdipourbozorgbaghi, M.; Vancura, A.; Radpour, R.; Haefliger, S. Circulating miRNA panels as a novel non-invasive diagnostic, prognostic, and potential predictive biomarkers in non-small cell lung cancer (NSCLC). Br. J. Cancer 2024, 131, 1350–1362. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.O.; Gazala, S.; Razzak, R.; Guo, L.; Ghosh, S.; Roa, W.H.; Bedard, E.L. Non-small cell lung cancer detection using microRNA expression profiling of bronchoalveolar lavage fluid and sputum. Anticancer Res. 2015, 35, 1873–1880. [Google Scholar]
- Gahlawat, A.W.; Fahed, L.; Witte, T.; Schott, S. Total circulating microRNA level as an independent prognostic marker for risk stratification in breast cancer. Br. J. Cancer 2022, 127, 156–162. [Google Scholar] [CrossRef]
- Fehlmann, T.; Lehallier, B.; Schaum, N.; Hahn, O.; Kahraman, M.; Li, Y.; Grammes, N.; Geffers, L.; Backes, C.; Balling, R.; et al. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun. 2020, 11, 5958. [Google Scholar] [CrossRef]
- Rao, Y.S.; Mott, N.N.; Wang, Y.; Chung, W.C.; Pak, T.R. MicroRNAs in the aging female brain: A putative mechanism for age-specific estrogen effects. Endocrinology 2013, 154, 2795–2806. [Google Scholar] [CrossRef]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef]
- Maiocchi, S.; Collins, E.N.; Peterson, A.R.; Alexander, K.C.; McGlamery, D.J.; Cassidy, N.A.; Ikonomidis, J.S.; Akerman, A.W. Plasma microrna quantification protocol. Vessel Plus 2023, 7, 27. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Kauko, O.; Laajala, T.D.; Jumppanen, M.; Hintsanen, P.; Suni, V.; Haapaniemi, P.; Corthals, G.; Aittokallio, T.; Westermarck, J.; Imanishi, S.Y. Label-free quantitative phosphoproteomics with novel pairwise abundance normalization reveals synergistic RAS and CIP2A signaling. Sci. Rep. 2015, 5, 13099. [Google Scholar] [CrossRef] [PubMed]
- Marabita, F.; de Candia, P.; Torri, A.; Tegner, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 2016, 17, 204–212. [Google Scholar] [CrossRef]
- Dhilipkannah, P.; Sachdeva, A.; Holden, V.K.; Jiang, F. Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA. Curr. Oncol. 2024, 31, 5949–5959. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, L.; Gu, J.; Qu, K.; Wang, Y. Identification of microRNA differentially expressed in three subtypes of non-small cell lung cancer and in silico functional analysis. Oncotarget 2017, 8, 74554–74566. [Google Scholar] [CrossRef] [PubMed]
- Sens, A.; Rischke, S.; Hahnefeld, L.; Dorochow, E.; Schafer, S.M.G.; Thomas, D.; Kohm, M.; Geisslinger, G.; Behrens, F.; Gurke, R. Pre-analytical sample handling standardization for reliable measurement of metabolites and lipids in LC-MS-based clinical research. J. Mass. Spectrom. Adv. Clin. Lab. 2023, 28, 35–46. [Google Scholar] [CrossRef]
- Zafar, S.; Hafeez, A.; Shah, H.; Mutiullah, I.; Ali, A.; Khan, K.; Figueroa-Gonzalez, G.; Reyes-Hernandez, O.D.; Quintas-Granados, L.I.; Pena-Corona, S.I.; et al. Emerging biomarkers for early cancer detection and diagnosis: Challenges, innovations, and clinical perspectives. Eur. J. Med. Res. 2025, 30, 760. [Google Scholar] [CrossRef]
- Metcalf, G.A.D. MicroRNAs: Circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene 2024, 43, 2135–2142. [Google Scholar] [CrossRef]
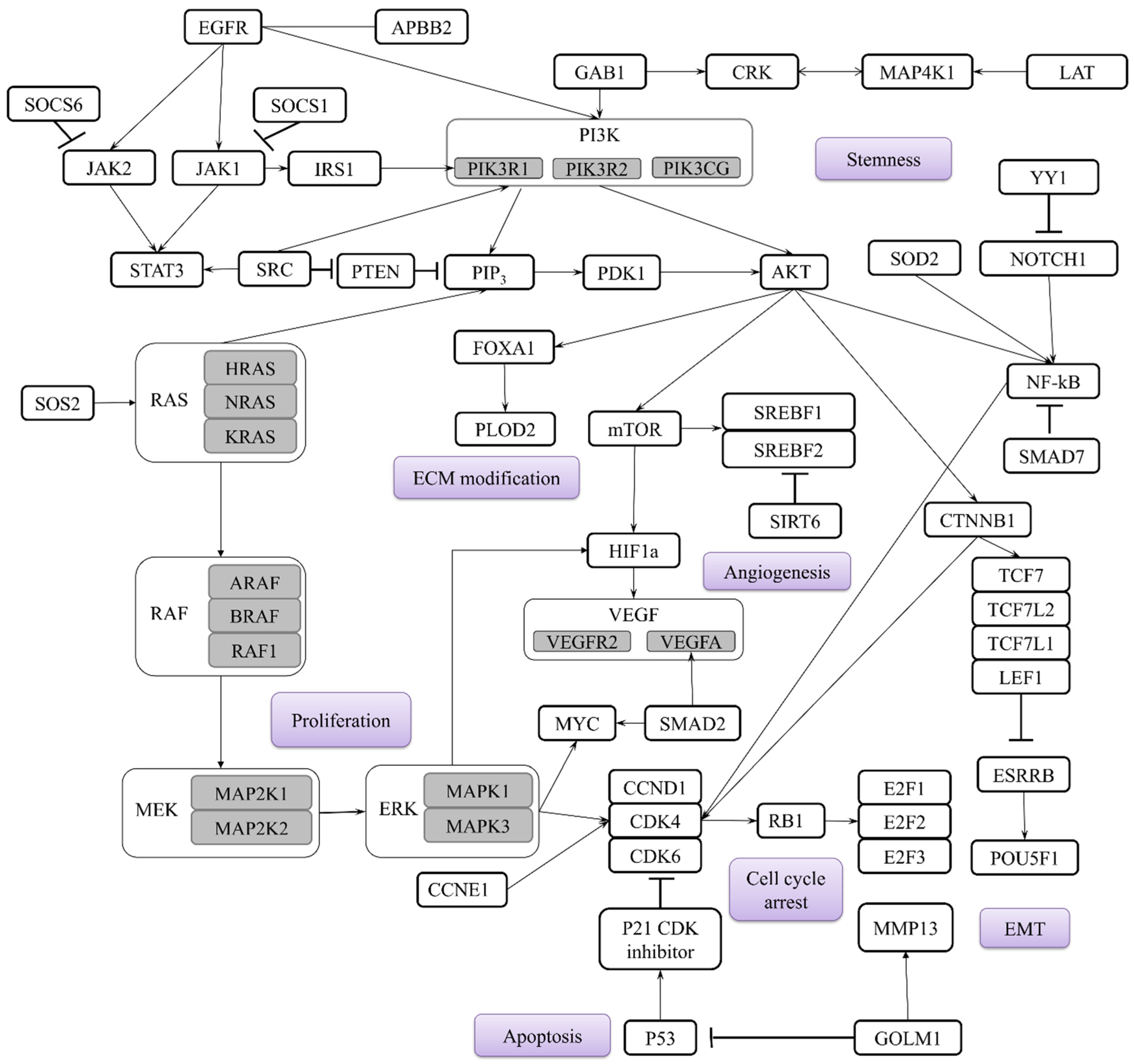

| Number of Validating Studies | miRNAs |
|---|---|
| ≥11 | miR-145 |
| ≥10 | miR-21 |
| ≥5 | miR-126 |
| ≥4 | miR-20a, miR-223, miR-155, miR-205, miR-210, miR-1246 |
| ≥3 | miR-150, miR-205, miR-574-5p, miR-146a, miR-486 |
| ≥2 | let-7a, Let-7b, miR-29c, miR-34b, miR-125a-5p, miR-125b, miR-141, miR-182, miR-183, miR-222, miR-429, miR-886, miR-1254, miR-1290, miR-106b |
| ≥1 | More than 66 different miRNAs have been reported across the studies |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zedan, Y.; Konoshenko, M.Y.; Bryzgunova, O.E.; Ilyushchenko, A.A.; Danilova, Y.M.; Gorbunkov, S.D.; Zykov, K.A.; Laktionov, P.P. Blood-Based miRNA Panels for Timely Detection of Non-Small-Cell Lung Cancer: From Biomarker Discovery to Clinical Translation. Int. J. Mol. Sci. 2025, 26, 12035. https://doi.org/10.3390/ijms262412035
Zedan Y, Konoshenko MY, Bryzgunova OE, Ilyushchenko AA, Danilova YM, Gorbunkov SD, Zykov KA, Laktionov PP. Blood-Based miRNA Panels for Timely Detection of Non-Small-Cell Lung Cancer: From Biomarker Discovery to Clinical Translation. International Journal of Molecular Sciences. 2025; 26(24):12035. https://doi.org/10.3390/ijms262412035
Chicago/Turabian StyleZedan, Yazan, Maria Yurievna Konoshenko, Olga Evgenievna Bryzgunova, Antonina Aleksandrovna Ilyushchenko, Yaroslava Mikhailovna Danilova, Stanislav Dmitrievich Gorbunkov, Kirill Alekseevich Zykov, and Pavel Petrovich Laktionov. 2025. "Blood-Based miRNA Panels for Timely Detection of Non-Small-Cell Lung Cancer: From Biomarker Discovery to Clinical Translation" International Journal of Molecular Sciences 26, no. 24: 12035. https://doi.org/10.3390/ijms262412035
APA StyleZedan, Y., Konoshenko, M. Y., Bryzgunova, O. E., Ilyushchenko, A. A., Danilova, Y. M., Gorbunkov, S. D., Zykov, K. A., & Laktionov, P. P. (2025). Blood-Based miRNA Panels for Timely Detection of Non-Small-Cell Lung Cancer: From Biomarker Discovery to Clinical Translation. International Journal of Molecular Sciences, 26(24), 12035. https://doi.org/10.3390/ijms262412035






