BGN Secreted by Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression via Activation of TLR4-Mediated Erk and NF-κB Signaling Pathways
Abstract
1. Introduction
2. Results
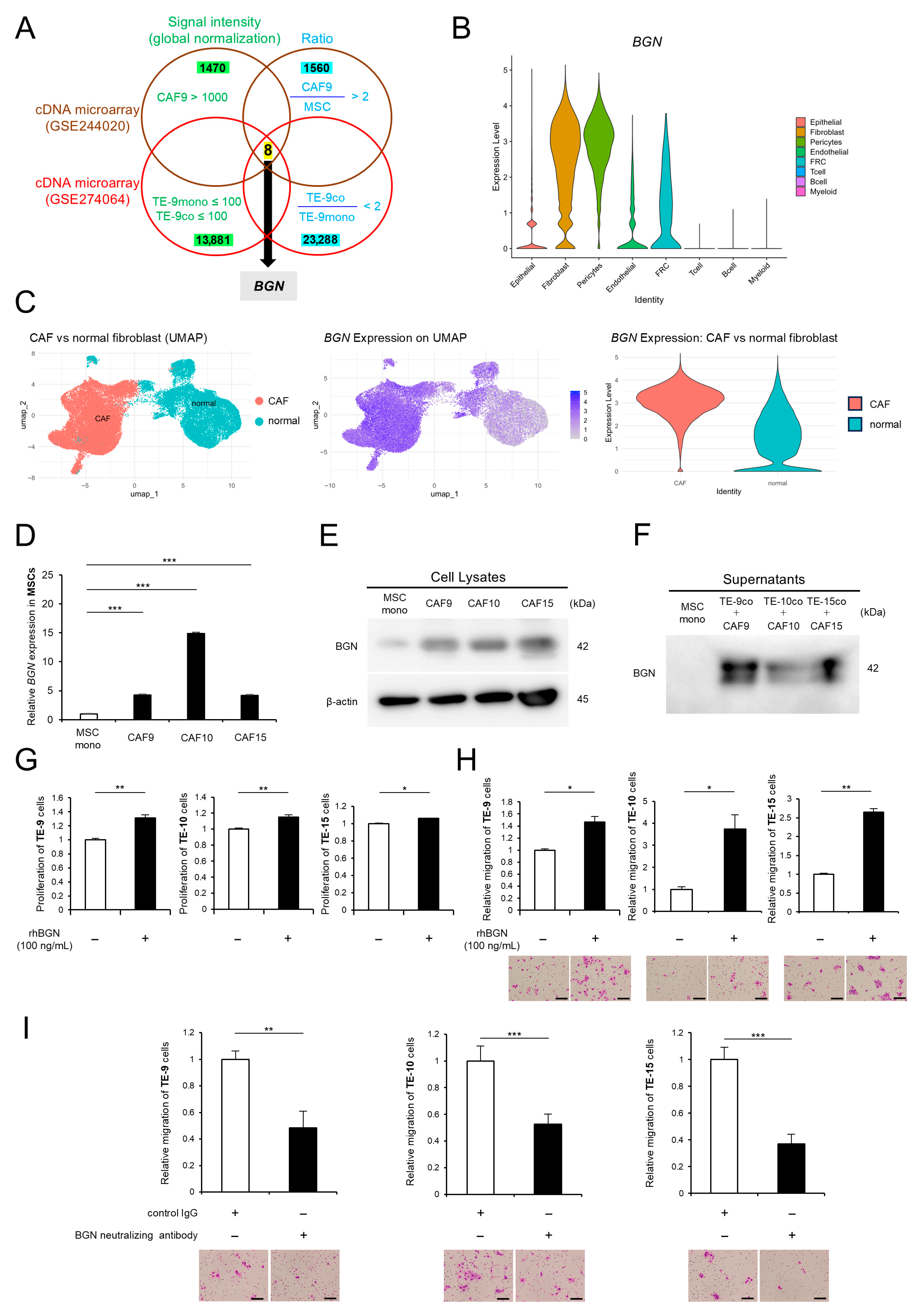
2.1. Direct Co-Culture Induces the Upregulation of BGN Gene Expression and Protein Expression/Secretion Levels in CAF-like Cells, and BGN Promotes ESCC’s Malignant Phenotypes
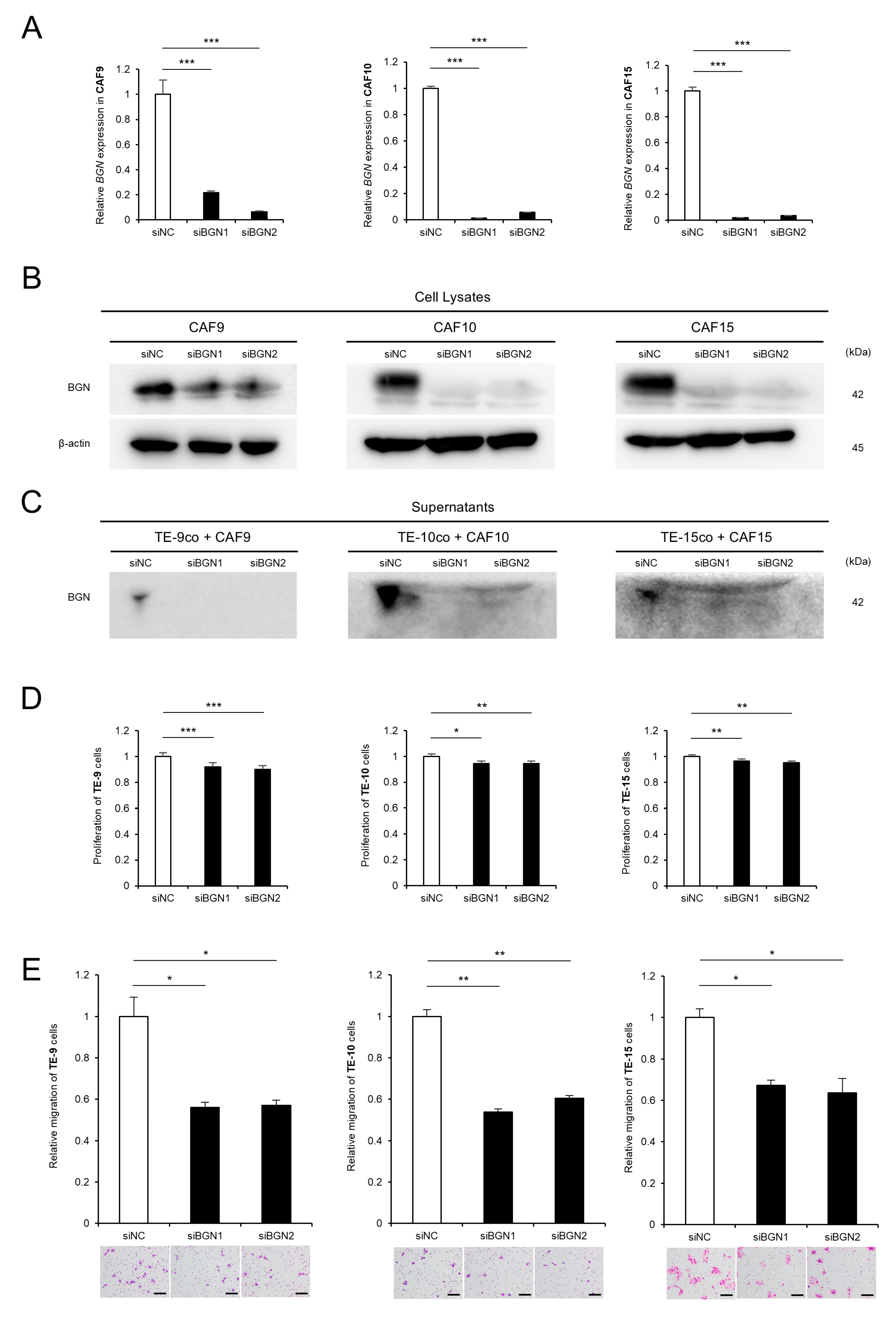
2.2. Knockdown of BGN in CAF-like Cells Attenuates CAF-Induced Proliferation and Migration of ESCC Cells
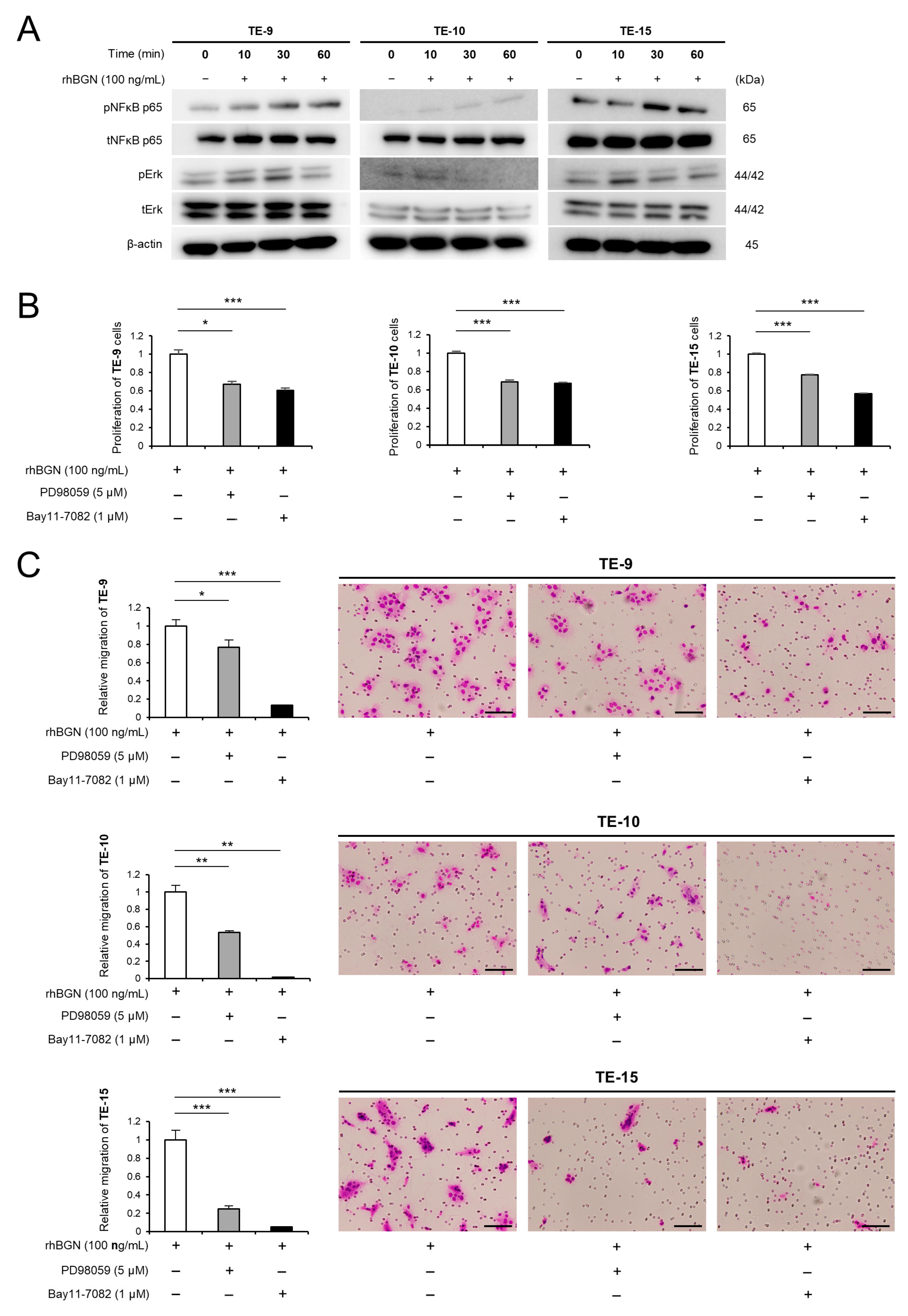
2.3. BGN Promotes the Proliferation and Migration of ESCC Cells Through the Erk and NF-κB Signaling Pathways
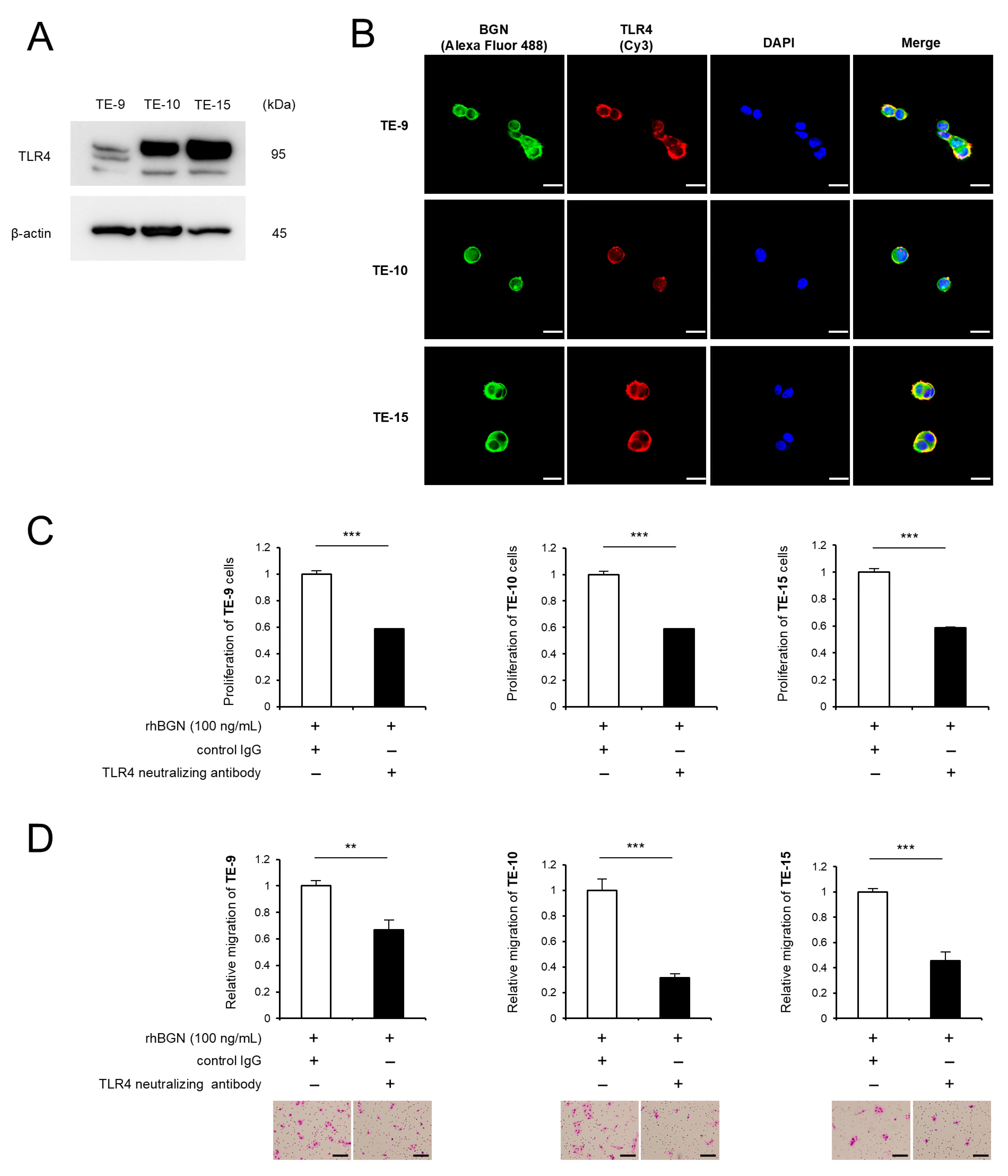
2.4. BGN Promotes Proliferation and Migration Through Its Receptor TLR4
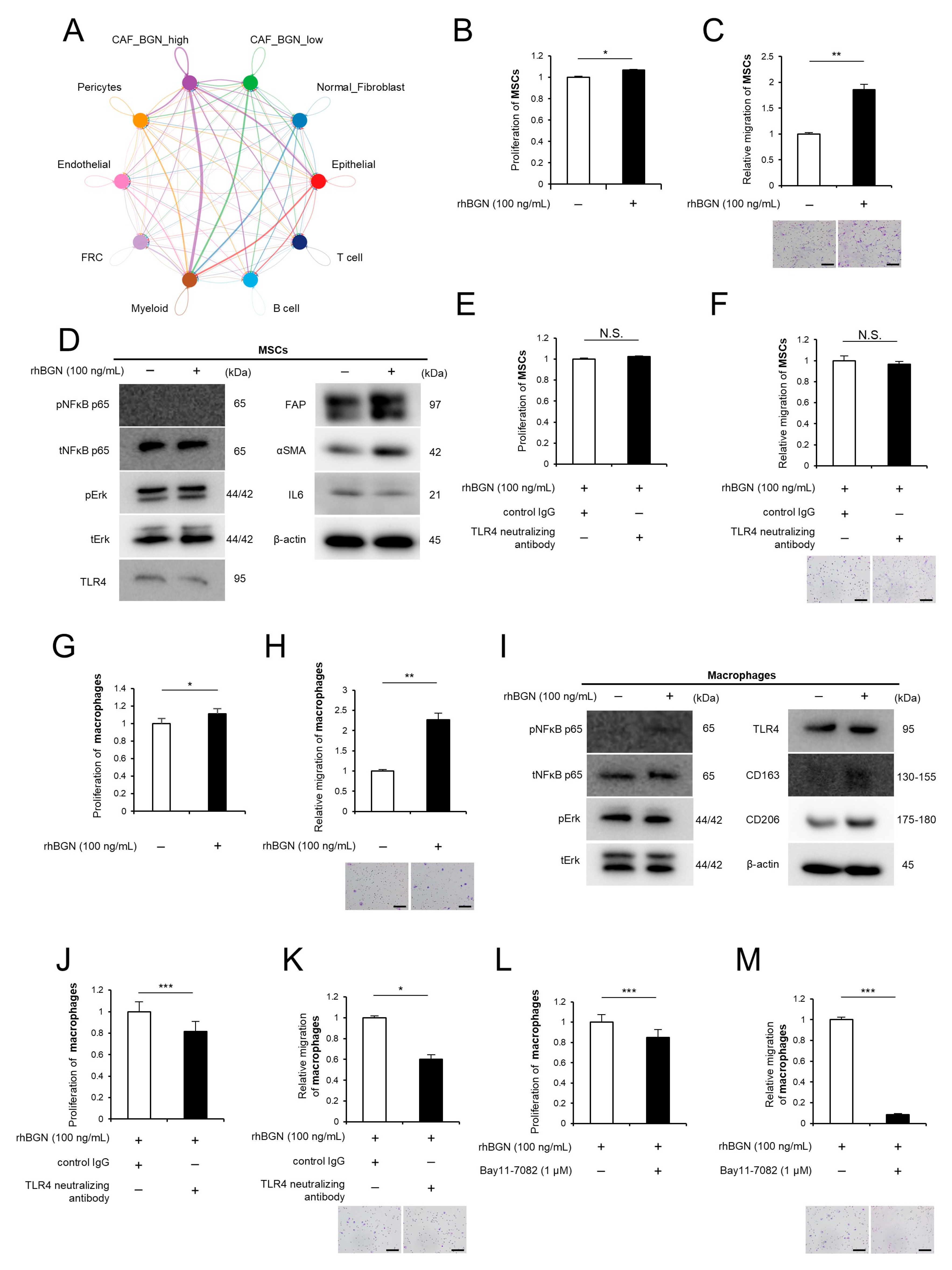
2.5. BGN Contributes to the Activation Phenotypes of MSCs and Macrophages
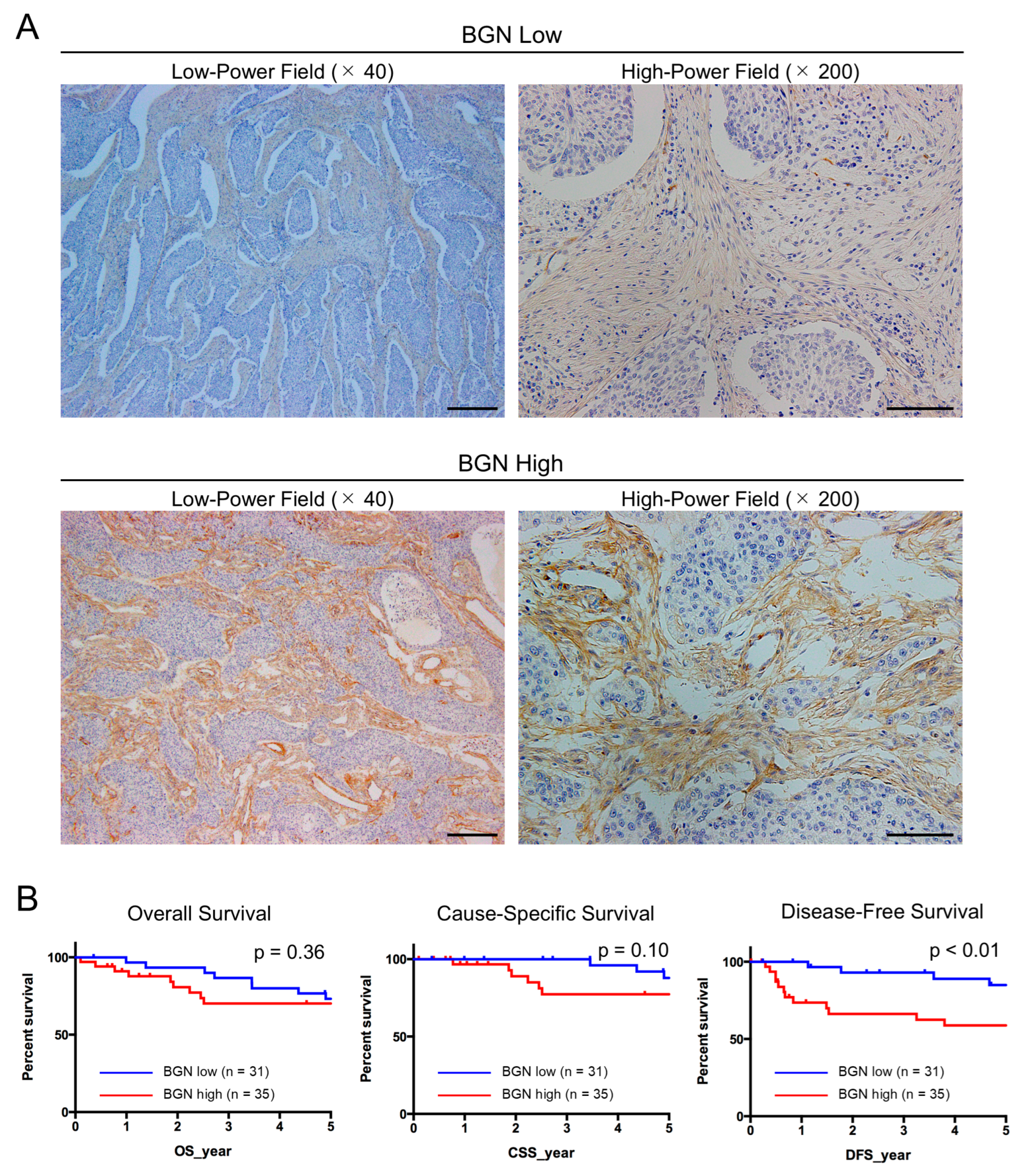
2.6. BGN Expression Is Elevated in CAFs Within ESCC Tissues and Is Correlated with Tumor Progression in Disease-Free Survival
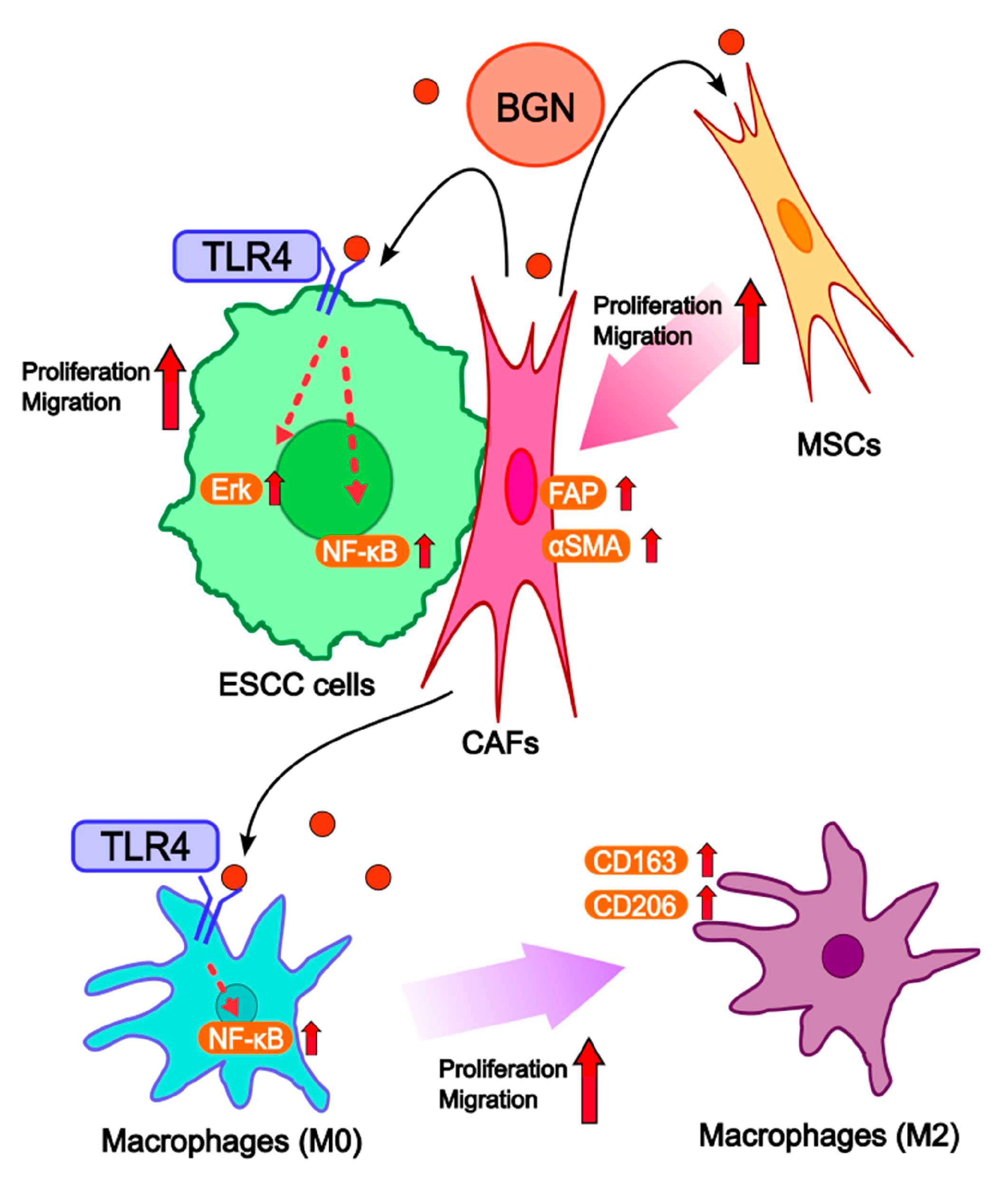
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Direct Co-Culture Model
4.3. cDNA Microarray Analysis
4.4. Single-Cell RNA Sequencing (scRNA-Seq)
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Western Blotting
4.7. Cell Proliferation Assay
4.8. Transwell Migration Assay
4.9. Knockdown of BGN in the Direct Co-Culture Model
4.10. Immunofluorescence
4.11. Tissue Samples
4.12. Immunohistochemistry
4.13. Bioinformatics Analysis
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lander, S.; Lander, E.; Gibson, M.K. Esophageal Cancer: Overview, Risk Factors, and Reasons for the Rise. Curr. Gastroenterol. Rep. 2023, 25, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.; Roshandel, G.; McCormack, V.; Malekzadeh, R. Current Status and Future Prospects for Esophageal Cancer. Cancers 2023, 15, 765. [Google Scholar] [CrossRef]
- Tirumani, H.; Rosenthal, M.H.; Tirumani, S.H.; Shinagare, A.B.; Krajewski, K.M.; Ramaiya, N.H. Esophageal Carcinoma: Current Concepts in the Role of Imaging in Staging and Management. Can. Assoc. Radiol. J. 2015, 66, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.; Yu, H.; Ding, Z.; Chen, L.; Zeng, X.; He, S.; Liao, Q.; Zhao, Y.; Yuan, Y. Comprehensive landscape of resistance mechanisms for neoadjuvant therapy in esophageal squamous cell carcinoma by single-cell transcriptomics. Signal Transduct. Target. Ther. 2023, 8, 298. [Google Scholar] [CrossRef]
- An, L.; Li, M.; Jia, Q. Mechanisms of radiotherapy resistance and radiosensitization strategies for esophageal squamous cell carcinoma. Mol. Cancer 2023, 22, 140. [Google Scholar] [CrossRef]
- Maman, S.; Witz, I.P. A history of exploring cancer in context. Nat. Rev. Cancer 2018, 18, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Olumi, A.F.; Grossfeld, G.D.; Hayward, S.W.; Carroll, P.R.; Tlsty, T.D.; Cunha, G.R. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999, 59, 5002–5011. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Raz, Y.; Cohen, N.; Shani, O.; Bell, R.E.; Novitskiy, S.V.; Abramovitz, L.; Levy, C.; Milyavsky, M.; Leider-Trejo, L.; Moses, H.L.; et al. Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J. Exp. Med. 2018, 215, 3075–3093. [Google Scholar] [CrossRef]
- Quante, M.; Tu, S.P.; Tomita, H.; Gonda, T.; Wang, S.S.; Takashi, S.; Baik, G.H.; Shibata, W.; Diprete, B.; Betz, K.S.; et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 2011, 19, 257–272. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Higashino, N.; Koma, Y.I.; Hosono, M.; Takase, N.; Okamoto, M.; Kodaira, H.; Nishio, M.; Shigeoka, M.; Kakeji, Y.; Yokozaki, H. Fibroblast activation protein-positive fibroblasts promote tumor progression through secretion of CCL2 and interleukin-6 in esophageal squamous cell carcinoma. Lab. Investig. 2019, 99, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Koma, Y.I.; Sakamoto, H.; Tsukamoto, S.; Kitamura, Y.; Urakami, S.; Tanigawa, K.; Kodama, T.; Higashino, N.; Nishio, M.; et al. Metallothionein 2A Expression in Cancer-Associated Fibroblasts and Cancer Cells Promotes Esophageal Squamous Cell Carcinoma Progression. Cancers 2021, 13, 4552. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Koma, Y.I.; Higashino, N.; Kodama, T.; Tanigawa, K.; Shimizu, M.; Fujikawa, M.; Nishio, M.; Shigeoka, M.; Kakeji, Y.; et al. PAI-1 derived from cancer-associated fibroblasts in esophageal squamous cell carcinoma promotes the invasion of cancer cells and the migration of macrophages. Lab. Investig. 2021, 101, 353–368. [Google Scholar] [CrossRef]
- Miyako, S.; Koma, Y.I.; Nakanishi, T.; Tsukamoto, S.; Yamanaka, K.; Ishihara, N.; Azumi, Y.; Urakami, S.; Shimizu, M.; Kodama, T.; et al. Periostin in Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression by Enhancing Cancer and Stromal Cell Migration. Am. J. Pathol. 2024, 194, 828–848. [Google Scholar] [CrossRef]
- Nakanishi, T.; Koma, Y.I.; Miyako, S.; Torigoe, R.; Yokoo, H.; Omori, M.; Yamanaka, K.; Ishihara, N.; Tsukamoto, S.; Kodama, T.; et al. AREG Upregulation in Cancer Cells via Direct Interaction with Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression Through EGFR-Erk/p38 MAPK Signaling. Cells 2024, 13, 1733. [Google Scholar] [CrossRef]
- Maishi, N.; Ohba, Y.; Akiyama, K.; Ohga, N.; Hamada, J.; Nagao-Kitamoto, H.; Alam, M.T.; Yamamoto, K.; Kawamoto, T.; Inoue, N.; et al. Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Sci. Rep. 2016, 6, 28039. [Google Scholar] [CrossRef]
- Wu, H.; Xiang, Z.; Huang, G.; He, Q.; Song, J.; Dou, R.; Yang, C.; Wang, S.; Xiong, B. BGN/FAP/STAT3 positive feedback loop mediated mutual interaction between tumor cells and mesothelial cells contributes to peritoneal metastasis of gastric cancer. Int. J. Biol. Sci. 2023, 19, 465–483. [Google Scholar] [CrossRef]
- Hu, L.; Zang, M.D.; Wang, H.X.; Li, J.F.; Su, L.P.; Yan, M.; Li, C.; Yang, Q.M.; Liu, B.Y.; Zhu, Z.G. Biglycan stimulates VEGF expression in endothelial cells by activating the TLR signaling pathway. Mol. Oncol. 2016, 10, 1473–1484. [Google Scholar] [CrossRef]
- Babelova, A.; Moreth, K.; Tsalastra-Greul, W.; Zeng-Brouwers, J.; Eickelberg, O.; Young, M.F.; Bruckner, P.; Pfeilschifter, J.; Schaefer, R.M.; Gröne, H.J.; et al. Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors. J. Biol. Chem. 2009, 284, 24035–24048. [Google Scholar] [CrossRef] [PubMed]
- Moreth, K.; Brodbeck, R.; Babelova, A.; Gretz, N.; Spieker, T.; Zeng-Brouwers, J.; Pfeilschifter, J.; Young, M.F.; Schaefer, R.M.; Schaefer, L. The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. J. Clin. Investig. 2010, 120, 4251–4272. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Babelova, A.; Kiss, E.; Hausser, H.J.; Baliova, M.; Krzyzankova, M.; Marsche, G.; Young, M.F.; Mihalik, D.; Götte, M.; et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J. Clin. Investig. 2005, 115, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Sheyhidin, I.; Nabi, G.; Hasim, A.; Zhang, R.P.; Ainiwaer, J.; Ma, H.; Wang, H. Overexpression of TLR3, TLR4, TLR7 and TLR9 in esophageal squamous cell carcinoma. World J. Gastroenterol. 2011, 17, 3745–3751. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Ping, W.; Deng, T.; Zhang, N.; Fu, X.; Sun, W. Lipopolysaccharide-induced toll-like receptor 4 signaling in esophageal squamous cell carcinoma promotes tumor proliferation and regulates inflammatory cytokines expression. Dis. Esophagus 2017, 30, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Motoyama, S.; Wakita, A.; Kawakita, Y.; Liu, J.; Nagaki, Y.; Nanjo, H.; Ito, S.; Terata, K.; Imai, K.; et al. High TLR4 expression predicts a poor prognosis after esophagectomy for advanced thoracic esophageal squamous cell carcinoma. Esophagus 2020, 17, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Mine, S.; Tanaka, K.; Kawachi, H.; Shirakawa, Y.; Kitagawa, Y.; Toh, Y.; Yasuda, T.; Watanabe, M.; Kamei, T.; Oyama, T.; et al. Japanese Classification of Esophageal Cancer, 12th Edition: Part I. Esophagus 2024, 21, 179–215. [Google Scholar] [CrossRef]
- Doki, Y.; Tanaka, K.; Kawachi, H.; Shirakawa, Y.; Kitagawa, Y.; Toh, Y.; Yasuda, T.; Watanabe, M.; Kamei, T.; Oyama, T.; et al. Japanese Classification of Esophageal Cancer, 12th Edition: Part II. Esophagus 2024, 21, 216–269. [Google Scholar] [CrossRef] [PubMed]
- Brierley, J.D.; Giuliani, M.; O’Sullivan, B.; Rous, B.; Van Eycken, E. (Eds.) TNM Classification of Malignant Tumours, 9th ed.; Wiley-Blackwell: Oxford, UK, 2025. [Google Scholar]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Tang, C.M.; Banerjee, S.; Yebra, M.; Noh, S.; Burgoyne, A.M.; Torre, J.; Siena, M.; Liu, M.; Klug, L.R.; et al. Cancer-associated fibroblast secretion of PDGFC promotes gastrointestinal stromal tumor growth and metastasis. Oncogene 2021, 40, 1957–1973. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Du, R.; Dong, J.; Sun, Y.; Zhou, F.; Feng, F.; Feng, B.; Han, Y.; Shang, Y. Cancer associated fibroblast derived SLIT2 drives gastric cancer cell metastasis by activating NEK9. Cell Death Dis. 2023, 14, 421. [Google Scholar] [CrossRef]
- Hu, S.; Xiao, Q.; Gao, R.; Qin, J.; Nie, J.; Chen, Y.; Lou, J.; Ding, M.; Pan, Y.; Wang, S. Identification of BGN positive fibroblasts as a driving factor for colorectal cancer and development of its related prognostic model combined with machine learning. BMC Cancer 2024, 24, 516. [Google Scholar] [CrossRef]
- Zheng, S.; Zou, Y.; Tang, Y.; Yang, A.; Liang, J.Y.; Wu, L.; Tian, W.; Xiao, W.; Xie, X.; Yang, L.; et al. Landscape of cancer-associated fibroblasts identifies the secreted biglycan as a protumor and immunosuppressive factor in triple-negative breast cancer. Oncoimmunology 2022, 11, 2020984. [Google Scholar] [CrossRef]
- Schönherr, E.; Witsch-Prehm, P.; Harrach, B.; Robenek, H.; Rauterberg, J.; Kresse, H. Interaction of biglycan with type I collagen. J. Biol. Chem. 1995, 270, 2776–2783. [Google Scholar] [CrossRef]
- Hua, R.; Han, Y.; Ni, Q.; Fajardo, R.J.; Iozzo, R.V.; Ahmed, R.; Nyman, J.S.; Wang, X.; Jiang, J.X. Pivotal roles of biglycan and decorin in regulating bone mass, water retention, and bone toughness. Bone Res. 2025, 13, 2. [Google Scholar] [CrossRef]
- Beach, Z.M.; Bonilla, K.A.; Dekhne, M.S.; Sun, M.; Adams, T.H.; Adams, S.M.; Weiss, S.N.; Rodriguez, A.B.; Shetye, S.S.; Birk, D.E.; et al. Biglycan has a major role in maintenance of mature tendon mechanics. J. Orthop. Res. 2022, 40, 2546–2556. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.K.; Sommer, G.; Michl, P.; Fensterer, H.; Weimer, M.; Gansauge, F.; Leder, G.; Adler, G.; Gress, T.M. Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology 2001, 121, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Maishi, N.; Annan, D.A.; Young, M.F.; Morimoto, H.; Morimoto, M.; Nam, J.M.; Hida, Y.; Hida, K. Inhibition of stromal biglycan promotes normalization of the tumor microenvironment and enhances chemotherapeutic efficacy. Breast Cancer Res. 2021, 23, 51. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Zhu, Y.; Li, G.; Xu, Y.; Wen, C.; Ye, W. Biglycan promotes tumour proliferation and invasion in human colon cancers. J. Int. Med. Res. 2024, 52, 3000605241300067. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, R.; Feng, L.; Ma, H.; Fang, J. LINC00460 Promotes Cell Proliferation, Migration, Invasion, and Epithelial-Mesenchymal Transition of Head and Neck Squamous Cell Carcinoma via miR-320a/BGN Axis. Onco Targets Ther. 2021, 14, 2279–2291. [Google Scholar] [CrossRef] [PubMed]
- Giatagana, E.M.; Berdiaki, A.; Gaardløs, M.; Samsonov, S.A.; Tzanakakis, G.N.; Nikitovic, D. Biglycan Interacts with Type I Insulin-like Receptor (IGF-IR) Signaling Pathway to Regulate Osteosarcoma Cell Growth and Response to Chemotherapy. Cancers 2022, 14, 1196. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, A.D.; Fisher, L.W.; Kilts, T.M.; Owens, R.T.; Robey, P.G.; Gutkind, J.S.; Young, M.F. Modulation of canonical Wnt signaling by the extracellular matrix component biglycan. Proc. Natl. Acad. Sci. USA 2011, 108, 17022–17027. [Google Scholar] [CrossRef]
- Tao, M.; Ruan, X.; Yu, J.; Dong, Q.; Luo, W.; Zhang, W.; Tian, M.; Hou, X.; Hu, L.; Zhao, J.; et al. Enhancer-mediated NR2F2 recruitment activates BGN to promote tumor growth and shape tumor microenvironment in papillary thyroid cancer. Theranostics 2026, 16, 298–324. [Google Scholar] [CrossRef]
- Inkson, C.A.; Ono, M.; Bi, Y.; Kuznetsov, S.A.; Fisher, L.W.; Young, M.F. The potential functional interaction of biglycan and WISP-1 in controlling differentiation and proliferation of osteogenic cells. Cells Tissues Organs 2009, 189, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Acar, A.; Eaton, E.N.; Mellody, K.T.; Scheel, C.; Ben-Porath, I.; Onder, T.T.; Wang, Z.C.; Richardson, A.L.; Weinberg, R.A.; et al. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. USA 2010, 107, 20009–20014. [Google Scholar] [CrossRef]
- Sjöberg, E.; Meyrath, M.; Milde, L.; Herrera, M.; Lövrot, J.; Hägerstrand, D.; Frings, O.; Bartish, M.; Rolny, C.; Sonnhammer, E.; et al. A Novel ACKR2-Dependent Role of Fibroblast-Derived CXCL14 in Epithelial-to-Mesenchymal Transition and Metastasis of Breast Cancer. Clin. Cancer Res. 2019, 25, 3702–3717. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, H.; Liu, J.; Zhao, Z.; Wang, F.; Qiao, L.; Yin, S.; Zhang, C.; Huo, M. Exosomal Biglycan promotes gastric cancer progression via M2 polarization and CXCL10-mediated JAK/STAT1 activation. Cancer Lett. 2025, 626, 217758. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Liang, J.Y.; Tang, Y.; Xie, J.; Zou, Y.; Yang, A.; Shao, N.; Kuang, X.; Ji, F.; Liu, X.; et al. Dissecting the role of cancer-associated fibroblast-derived biglycan as a potential therapeutic target in immunotherapy resistance: A tumor bulk and single-cell transcriptomic study. Clin. Transl. Med. 2023, 13, e1189. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Bianco, P.; Fisher, L.W.; Longenecker, G.; Smith, E.; Goldstein, S.; Bonadio, J.; Boskey, A.; Heegaard, A.M.; Sommer, B.; et al. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat. Genet. 1998, 20, 78–82. [Google Scholar] [CrossRef]
- Nielsen, M.C.; Andersen, M.N.; Møller, H.J. Monocyte isolation techniques significantly impact the phenotype of both isolated monocytes and derived macrophages in vitro. Immunology 2020, 159, 63–74. [Google Scholar] [CrossRef]
- Urakami, S.; Koma, Y.I.; Tsukamoto, S.; Azumi, Y.; Miyako, S.; Kitamura, Y.; Kodama, T.; Nishio, M.; Shigeoka, M.; Abe, H.; et al. Biological and clinical significance of the YKL-40/osteopontin-integrin β4-p70S6K axis induced by macrophages in early oesophageal squamous cell carcinoma. J. Pathol. 2023, 261, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Peng, L.; Luo, Y.; Zhang, S.; Pu, Y.; Chen, Y.; Guo, W.; Yao, J.; Shao, M.; Fan, W.; et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 2021, 12, 5291. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
| Accession Number | Symbol | Description | Signal Intensity (Global Normalization) | Ratio | ||||
|---|---|---|---|---|---|---|---|---|
| MSC mono | CAF9 | TE-9 mono | TE-9 co | CAF9 /MSC mono | TE-9 co/TE-9 mono | |||
| NM_001711.5 | BGN | Biglycan | 2750 | 6702 | 40 | 76 | 2.44 | 1.92 |
| NM_001164098.1 | VCAN | Versican | 1034 | 2355 | 45 | 62 | 2.28 | 1.37 |
| NM_197955.2 | C15orf48 | chromosome 15 open reading frame 48 | 79 | 2346 | 36 | 52 | 29.88 | 1.48 |
| NM_001856.3 | COL16A1 | collagen type XVI alpha 1 | 885 | 2139 | 30 | 50 | 2.42 | 1.67 |
| XM_005271110.2 | P3H1 | prolyl 3-hydroxylase 1 | 494 | 1221 | 72 | 82 | 2.47 | 1.14 |
| XM_005269005.2 | DRAM1 | DNA damage regulated autophagy modulator 1 | 375 | 1189 | 31 | 41 | 3.17 | 1.33 |
| NM_000064.3 | C3 | complement component 3 | 28 | 1142 | 20 | 25 | 41.42 | 1.27 |
| NM_001206.2 | KLF9 | Kruppel-like factor 9 | 358 | 1013 | 11 | 13 | 2.83 | 1.19 |
| Case Number | Expression of BGN | p-Value | ||
|---|---|---|---|---|
| Low (n = 31) | High (n = 35) | |||
| Age (years) | 0.632 | |||
| <65 | 32 | 16 | 16 | |
| ≥65 | 34 | 15 | 19 | |
| Sex | 0.798 | |||
| Male | 52 | 24 | 28 | |
| Female | 14 | 7 | 7 | |
| Histological grade a | 0.575 | |||
| SCCIS + WDSCC | 15 | 8 | 7 | |
| MDSCC + PDSCC | 51 | 23 | 28 | |
| Depth of tumor invasion a | <0.001 * | |||
| T1 | 45 | 29 | 16 | |
| T2 + T3 | 21 | 2 | 19 | |
| Lymphatic vessel invasion a | 0.001 * | |||
| Negative | 35 | 23 | 12 | |
| Positive | 31 | 8 | 23 | |
| Blood vessel invasion a | 0.057 | |||
| Negative | 41 | 23 | 18 | |
| Positive | 25 | 8 | 17 | |
| Lymphatic node metastasis a | 0.057 | |||
| Negative | 41 | 23 | 18 | |
| Positive | 25 | 8 | 17 | |
| Stage b | 0.070 | |||
| 0 + I | 37 | 21 | 16 | |
| II + III + IV | 29 | 10 | 19 | |
| Expression of αSMA c | 0.003 * | |||
| Low | 34 | 22 | 12 | |
| High | 32 | 9 | 23 | |
| Expression of FAP c | 0.022 * | |||
| Low | 37 | 22 | 15 | |
| High | 29 | 9 | 20 | |
| Expression of CD68 c | 0.049 * | |||
| Low | 32 | 19 | 13 | |
| High | 34 | 12 | 22 | |
| Expression of CD163 c | 0.007 * | |||
| Low | 31 | 20 | 11 | |
| High | 35 | 11 | 24 | |
| Expression of CD204 c | 0.141 | |||
| Low | 32 | 18 | 14 | |
| High | 34 | 13 | 21 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokoo, H.; Koma, Y.-i.; Nomura, N.; Torigoe, R.; Omori, M.; Nakanishi, T.; Miyako, S.; Nakanishi, T.; Kodama, T.; Shigeoka, M.; et al. BGN Secreted by Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression via Activation of TLR4-Mediated Erk and NF-κB Signaling Pathways. Int. J. Mol. Sci. 2025, 26, 12024. https://doi.org/10.3390/ijms262412024
Yokoo H, Koma Y-i, Nomura N, Torigoe R, Omori M, Nakanishi T, Miyako S, Nakanishi T, Kodama T, Shigeoka M, et al. BGN Secreted by Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression via Activation of TLR4-Mediated Erk and NF-κB Signaling Pathways. International Journal of Molecular Sciences. 2025; 26(24):12024. https://doi.org/10.3390/ijms262412024
Chicago/Turabian StyleYokoo, Hiroki, Yu-ichiro Koma, Naozane Nomura, Rikuya Torigoe, Masaki Omori, Takashi Nakanishi, Shoji Miyako, Takaaki Nakanishi, Takayuki Kodama, Manabu Shigeoka, and et al. 2025. "BGN Secreted by Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression via Activation of TLR4-Mediated Erk and NF-κB Signaling Pathways" International Journal of Molecular Sciences 26, no. 24: 12024. https://doi.org/10.3390/ijms262412024
APA StyleYokoo, H., Koma, Y.-i., Nomura, N., Torigoe, R., Omori, M., Nakanishi, T., Miyako, S., Nakanishi, T., Kodama, T., Shigeoka, M., Kakeji, Y., & Horie, M. (2025). BGN Secreted by Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression via Activation of TLR4-Mediated Erk and NF-κB Signaling Pathways. International Journal of Molecular Sciences, 26(24), 12024. https://doi.org/10.3390/ijms262412024






