Comparative Tumor Microenvironment Analysis for HCC and PDAC Using KMplotter
Abstract
1. Introduction
2. Results
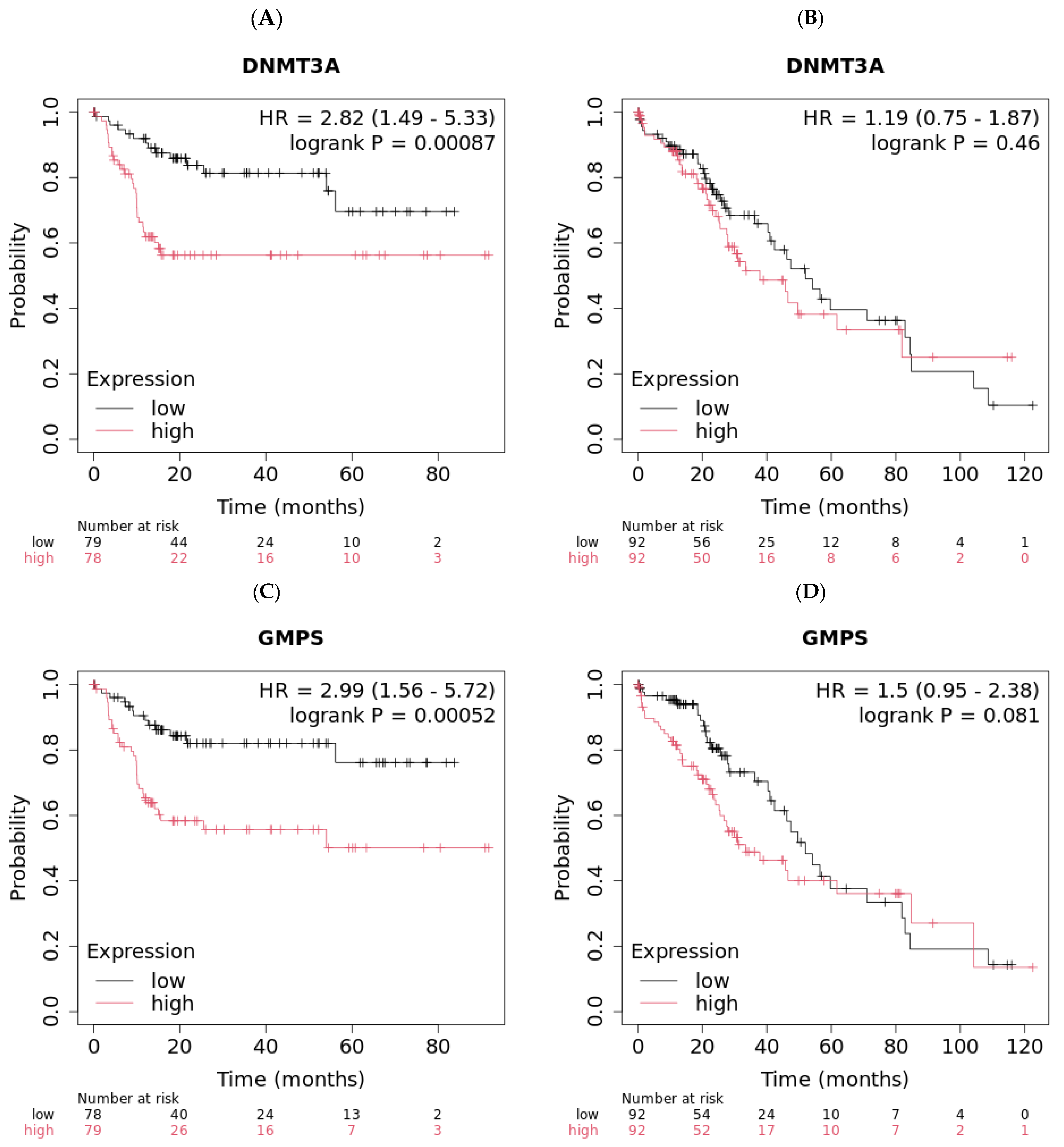
2.1. DNMT3A and GMPS’ Impact on OS in Liver Cancer
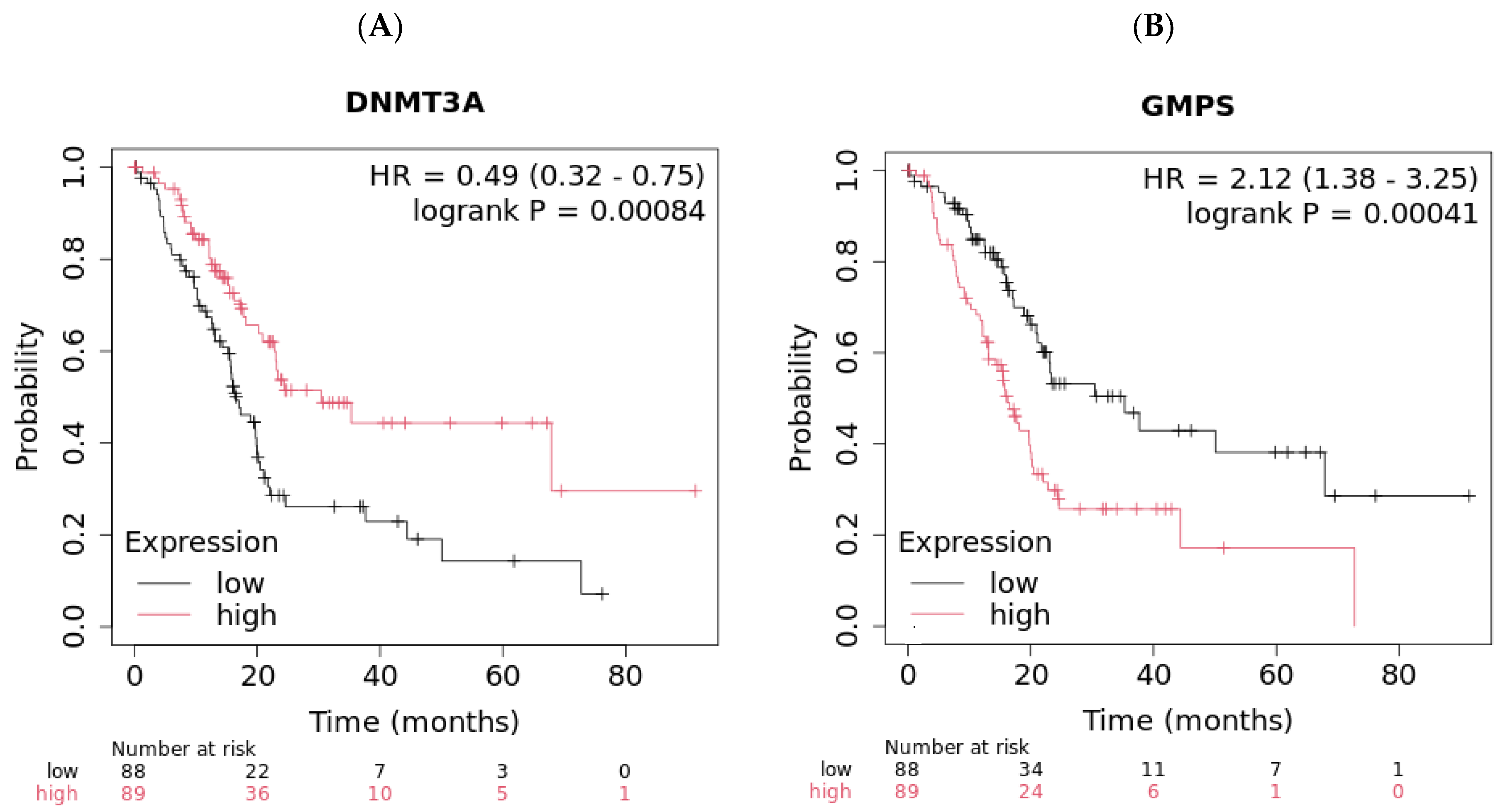
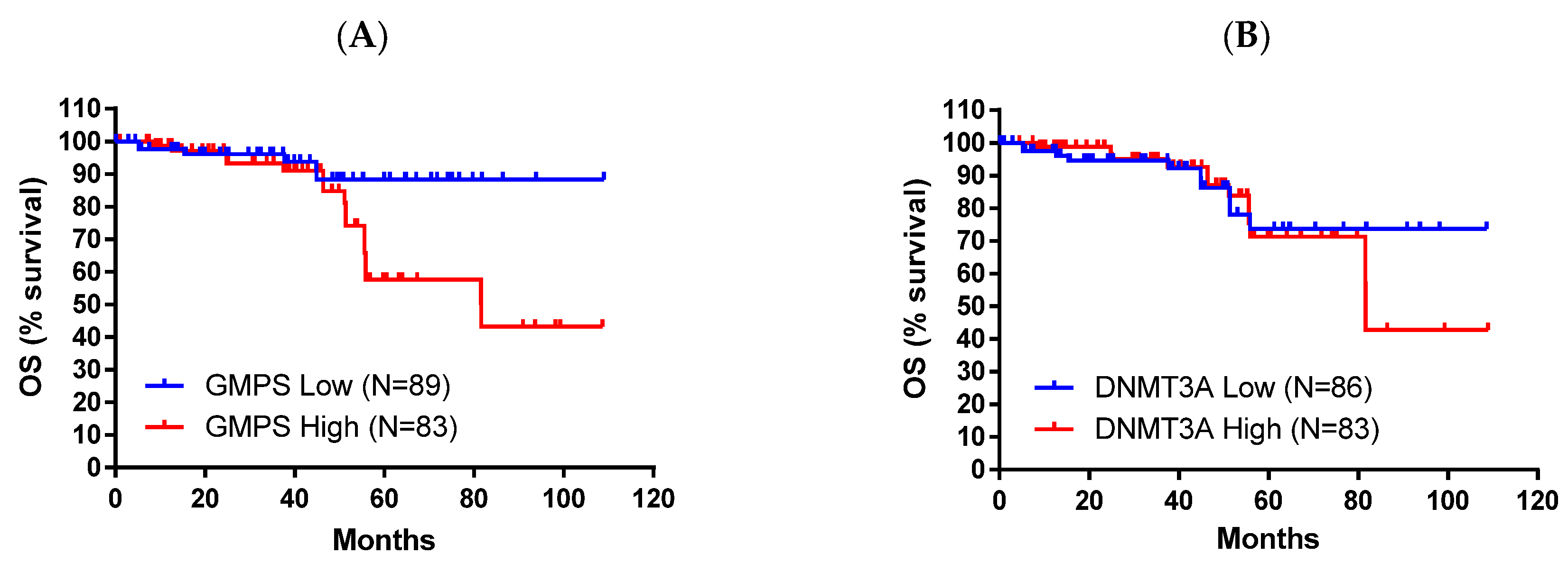
2.2. DNMT3A and GMPS’ Impact on OS in Pancreatic Cancer
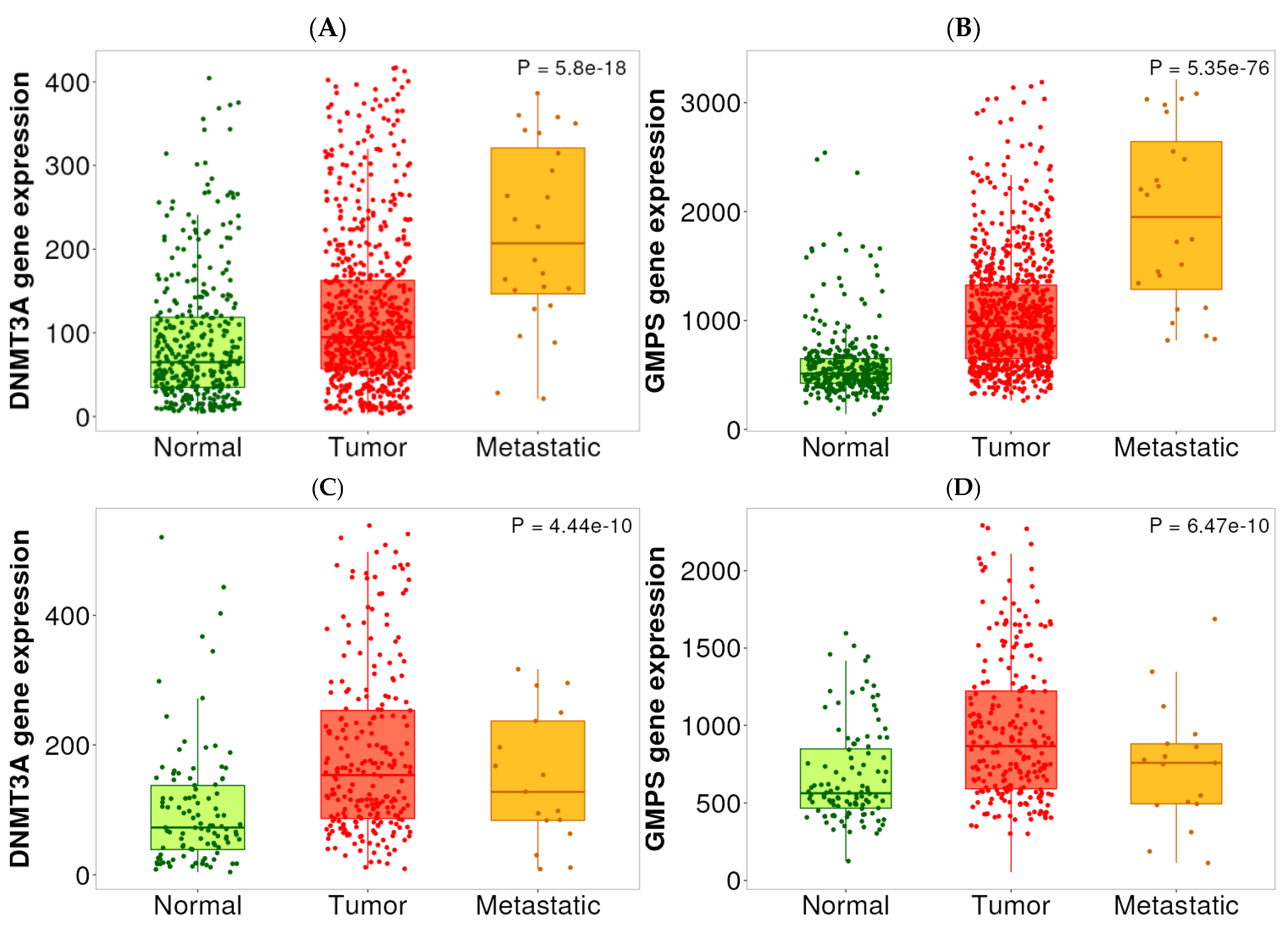
2.3. Expression Analysis Among Normal, Tumor, and Metastatic Tissues
2.4. Toll-like Receptors (TLRs) TME Analysis
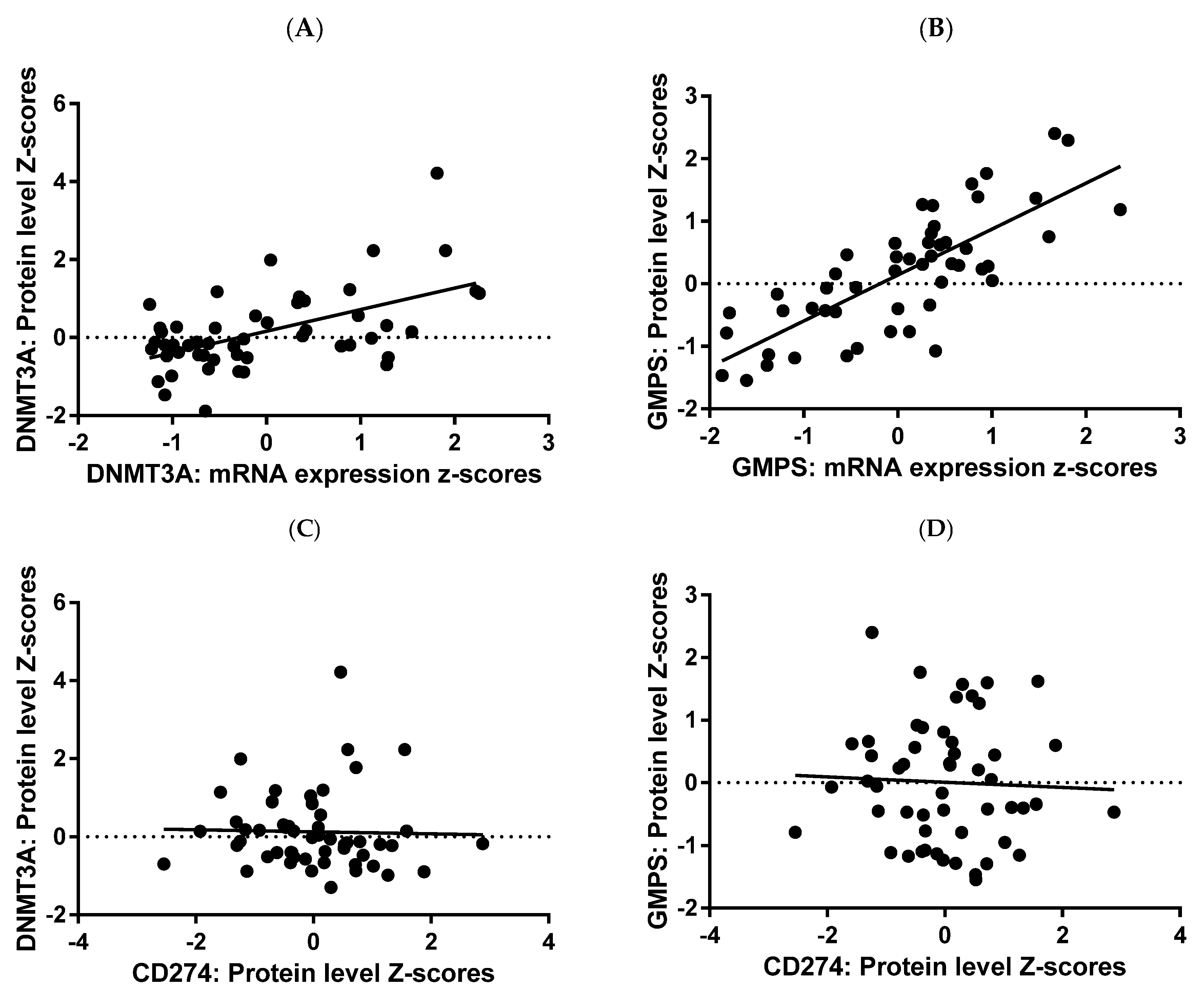
2.5. DNMT3A and GMPS Analyses at Protein Level
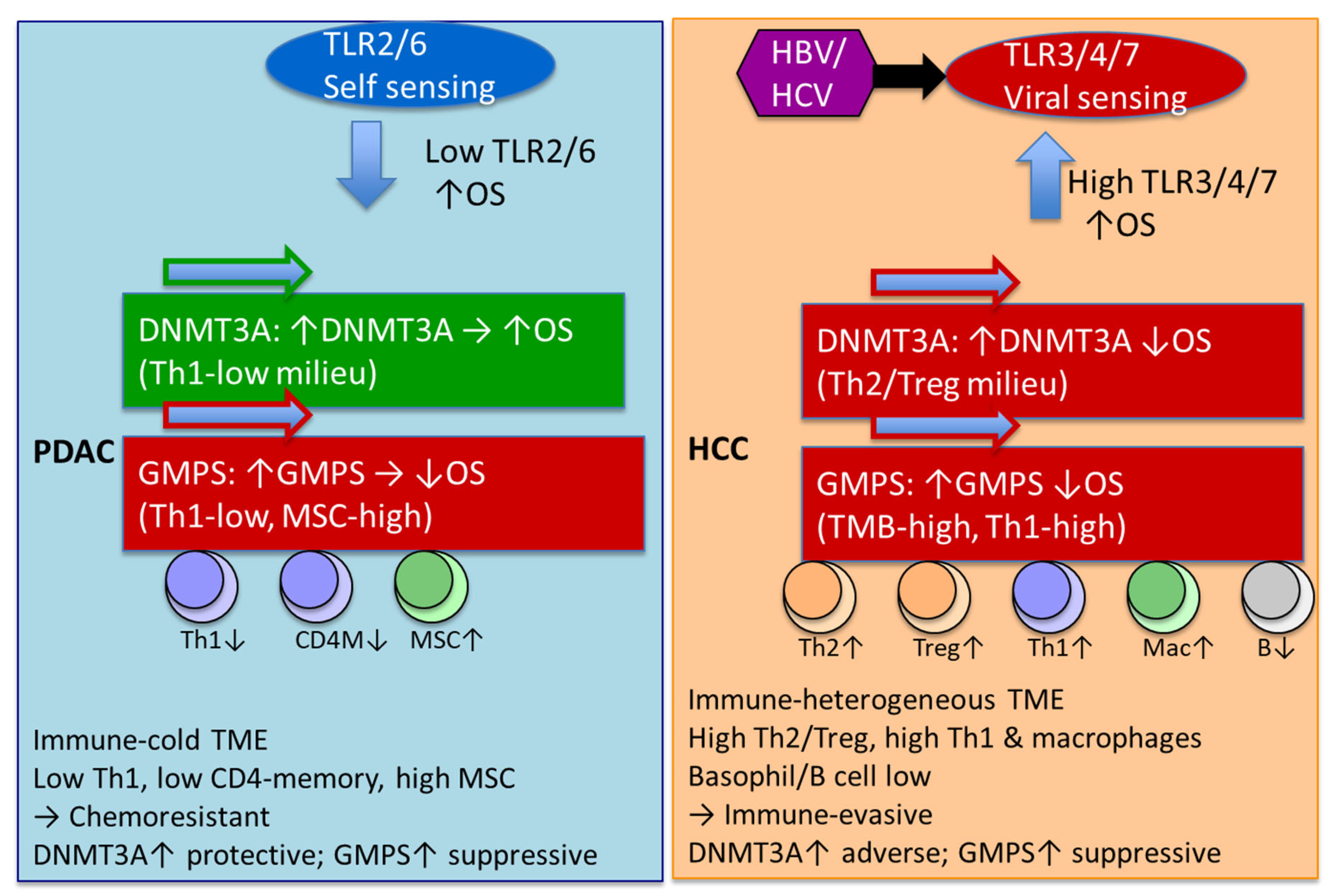
3. Discussion
4. Materials and Methods
4.1. Domain-Specific Identification of PubMed Articles Augmented by Artificial Intelligence
4.2. KMplotter and TNM Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, H.; Lu, F.; Lu, R.; Huang, M.; Li, X.; Yuan, J.; Wang, F. A novel tumor 4-driver gene signature for the prognosis of hepatocellular carcinoma. Heliyon 2023, 9, e17054. [Google Scholar] [CrossRef]
- Parpart, S.; Roessler, S.; Dong, F.; Rao, V.; Takai, A.; Ji, J.; Qin, L.X.; Ye, Q.H.; Jia, H.L.; Tang, Z.Y.; et al. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology 2014, 60, 872–883. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xie, Z. Therapeutic potential of tLyp-1-EV-shCTCF in inhibiting liver cancer stem cell self-renewal and immune escape via SALL3 modulation in hepatocellular carcinoma. Transl. Oncol. 2024, 49, 102048. [Google Scholar] [CrossRef]
- Schulze, K.; Rose, T.D.; Adlung, L.; Peschka, M.; Pagani, F.; Gorgulho, J.; Frundt, T.W.; Labgaa, I.; Haber, P.K.; Zimpel, C.; et al. Metabolomic liquid biopsy dynamics predict early-stage HCC and actionable candidates of human hepatocarcinogenesis. JHEP Rep. 2025, 7, 101340. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Hsieh, S.Y.; Sheen, I.S.; Lee, W.C.; Chen, T.C.; Shyu, W.C.; Liaw, Y.F. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001, 61, 4238–4243. [Google Scholar] [PubMed]
- Ohni, S.; Yamaguchi, H.; Hirotani, Y.; Nakanishi, Y.; Midorikawa, Y.; Sugitani, M.; Naruse, H.; Nakayama, T.; Makishima, M.; Esumi, M. Direct molecular evidence for both multicentric and monoclonal carcinogenesis followed by transdifferentiation from hepatocellular carcinoma to cholangiocarcinoma in a case of metachronous liver cancer. Oncol. Lett. 2022, 23, 22. [Google Scholar] [CrossRef]
- Deng, L.; Dou, L.; Huang, X.; Wang, P.; Shen, N. Machine Learning-based Gene Biomarker Identification for Improving Prognosis and Therapy in Hepatocellular Carcinoma. Curr. Med. Chem. 2025, 32, 8975–8996. [Google Scholar] [CrossRef]
- Zhang, C.; Peng, L.; Zhang, Y.; Liu, Z.; Li, W.; Chen, S.; Li, G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 2017, 34, 101. [Google Scholar] [CrossRef]
- Tan, Y.J. Hepatitis B virus infection and the risk of hepatocellular carcinoma. World J. Gastroenterol. 2011, 17, 4853–4857. [Google Scholar] [CrossRef]
- Tarocchi, M.; Polvani, S.; Marroncini, G.; Galli, A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J. Gastroenterol. 2014, 20, 11630–11640. [Google Scholar] [CrossRef]
- Tian, Z.; Xu, C.; Yang, P.; Lin, Z.; Wu, W.; Zhang, W.; Ding, J.; Ding, R.; Zhang, X.; Dou, K. Molecular pathogenesis: Connections between viral hepatitis-induced and non-alcoholic steatohepatitis-induced hepatocellular carcinoma. Front. Immunol. 2022, 13, 984728. [Google Scholar] [CrossRef]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273.e1, Correction in Gastroenterology 2012, 143, 269. [Google Scholar] [CrossRef]
- Coleman, W.B. Mechanisms of human hepatocarcinogenesis. Curr. Mol. Med. 2003, 3, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Ozen, C.; Yildiz, G.; Dagcan, A.T.; Cevik, D.; Ors, A.; Keles, U.; Topel, H.; Ozturk, M. Genetics and epigenetics of liver cancer. N. Biotechnol. 2013, 30, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Pourhoseingholi, M.A.; Vahedi, M.; Baghestani, A.R. Burden of gastrointestinal cancer in Asia; an overview. Gastroenterol. Hepatol. Bed Bench 2015, 8, 19–27. [Google Scholar]
- Cha, C.; DeMatteo, R.P. Molecular mechanisms in hepatocellular carcinoma development. Best Pract. Res. Clin. Gastroenterol. 2005, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Kanai, Y.; Sakamoto, M.; Saito, H.; Ishii, H.; Hirohashi, S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 2001, 33, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.K.; Kim, H.; Park, H.J.; Shim, Y.H.; Choi, J.; Park, C.; Park, Y.N. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int. J. Mol. Med. 2007, 20, 65–73. [Google Scholar] [CrossRef]
- Hassouna, M.M.; Naguib, M.; Radwan, E.M.; Abdel-Samiee, M.; Estaphan, S.; Abdelsameea, E. DNA Methyltransferases as Potential Biomarkers for HCV Related Hepatocellular Carcinoma. Asian Pac. J. Cancer Prev. 2020, 21, 3357–3363. [Google Scholar] [CrossRef]
- Choi, M.S.; Shim, Y.H.; Hwa, J.Y.; Lee, S.K.; Ro, J.Y.; Kim, J.S.; Yu, E. Expression of DNA methyltransferases in multistep hepatocarcinogenesis. Hum. Pathol. 2003, 34, 11–17. [Google Scholar] [CrossRef]
- Zhao, Z.; Wu, Q.; Cheng, J.; Qiu, X.; Zhang, J.; Fan, H. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. J. Biomed. Biotechnol. 2010, 2010, 737535. [Google Scholar] [CrossRef]
- Hu, S.; Luo, X.; Qian, J.; Hou, Y.; Shi, W. High expression of DNMT3A and DNMT3B regulatory factors of TGFB in non-neoplastic liver tissues of HCC. Cell. Mol. Biol. 2023, 69, 52–61. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, Y.; Ma, P.; Xu, Q.; Zeng, L.; Song, X.; Yu, F. Integrated GMPS and RAMP3 as a signature to predict prognosis and immune heterogeneity in hepatocellular carcinoma. Gene 2025, 933, 148958. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.N.; Ul Haq, A.; Siddiqui, O.A.; Khan, R. DNA methyltransferase 1, 3a, and 3b expression in hepatitis C associated human hepatocellular carcinoma and their clinicopathological association. Tumour Biol. 2016, 37, 10487–10497. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Song, N.; Liu, Y.P.; Qu, X.J.; Qi, Y.F.; Li, C.; Hou, K.Z.; Che, X.F.; Yang, X.H. DNMT3a promotes proliferation by activating the STAT3 signaling pathway and depressing apoptosis in pancreatic cancer. Cancer Manag. Res. 2019, 11, 6379–6396. [Google Scholar] [CrossRef] [PubMed]
- Anurag, M.; Jaehnig, E.J.; Krug, K.; Lei, J.T.; Bergstrom, E.J.; Kim, B.J.; Vashist, T.D.; Huynh, A.M.T.; Dou, Y.; Gou, X.; et al. Proteogenomic Markers of Chemotherapy Resistance and Response in Triple-Negative Breast Cancer. Cancer Discov. 2022, 12, 2586–2605. [Google Scholar] [CrossRef]
- Mba Medie, F.; Sharma-Kuinkel, B.K.; Ruffin, F.; Chan, L.C.; Rossetti, M.; Chang, Y.L.; Park, L.P.; Bayer, A.S.; Filler, S.G.; Ahn, R.; et al. Genetic variation of DNA methyltransferase-3A contributes to protection against persistent MRSA bacteremia in patients. Proc. Natl. Acad. Sci. USA 2019, 116, 20087–20096. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, X.; Fujiwara, K.; Jurcak, N.; Muth, S.; Zhou, J.; Xiao, Q.; Li, A.; Che, X.; Li, Z.; et al. Pancreatic cancer cells render tumor-associated macrophages metabolically reprogrammed by a GARP and DNA methylation-mediated mechanism. Signal Transduct. Target. Ther. 2021, 6, 366. [Google Scholar] [CrossRef]
- Park, H.J.; Yu, E.; Shim, Y.H. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006, 233, 271–278. [Google Scholar] [CrossRef]
- Guo, X.; Cui, T.; Sun, L.; Fu, Y.; Cheng, C.; Wu, C.; Zhu, Y.; Liang, S.; Liu, Y.; Zhou, S.; et al. A STT3A-dependent PD-L1 glycosylation modification mediated by GMPS drives tumor immune evasion in hepatocellular carcinoma. Cell Death Differ. 2025, 32, 944–958. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Wu, R.; Dong, C.S. Unveiling purine metabolism dysregulation orchestrated immunosuppression in advanced pancreatic cancer and concentrating on the central role of NT5E. Front. Immunol. 2025, 16, 1569088. [Google Scholar] [CrossRef] [PubMed]
- Hughey, C.C.; James, F.D.; Wang, Z.; Goelzer, M.; Wasserman, D.H. Dysregulated transmethylation leading to hepatocellular carcinoma compromises redox homeostasis and glucose formation. Mol. Metab. 2019, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.; Li, X.L.; Mo, Y.Z.; Fan, C.M.; Tang, L.; Xiong, F.; Guo, C.; Xiang, B.; Zhou, M.; Ma, J.; et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol. Cancer 2018, 17, 168. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Huang, L.; Liu, Y.; Yi, M.; Chu, Q.; Jiao, D.; Wu, K. Metabolic profiles of regulatory T cells and their adaptations to the tumor microenvironment: Implications for antitumor immunity. J. Hematol. Oncol. 2022, 15, 104. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Yang, Y.; Li, Z.; Chen, Y.; Shang, S.; Wang, Y. DNMT3A mutation promotes leukemia development through NAM-NAD metabolic reprogramming. J. Transl. Med. 2023, 21, 481. [Google Scholar] [CrossRef]
- Song, W.; Yu, Y.; Wang, S.; Cui, Z.; Zhu, Q.; Liu, W.; Wei, S.; Chi, J. Metabolic reprogramming shapes the immune microenvironment in pancreatic adenocarcinoma: Prognostic implications and therapeutic targets. Front. Immunol. 2025, 16, 1555287. [Google Scholar] [CrossRef]
- Delitto, D.; Delitto, A.E.; DiVita, B.B.; Pham, K.; Han, S.; Hartlage, E.R.; Newby, B.N.; Gerber, M.H.; Behrns, K.E.; Moldawer, L.L.; et al. Human Pancreatic Cancer Cells Induce a MyD88-Dependent Stromal Response to Promote a Tumor-Tolerant Immune Microenvironment. Cancer Res. 2017, 77, 672–683. [Google Scholar] [CrossRef]
- Ochi, A.; Graffeo, C.S.; Zambirinis, C.P.; Rehman, A.; Hackman, M.; Fallon, N.; Barilla, R.M.; Henning, J.R.; Jamal, M.; Rao, R.; et al. Toll-like receptor 7 regulates pancreatic carcinogenesis in mice and humans. J. Clin. Investig. 2012, 122, 4118–4129. [Google Scholar] [CrossRef]
- Farren, M.R.; Mace, T.A.; Geyer, S.; Mikhail, S.; Wu, C.; Ciombor, K.; Tahiri, S.; Ahn, D.; Noonan, A.M.; Villalona-Calero, M.; et al. Systemic Immune Activity Predicts Overall Survival in Treatment-Naive Patients with Metastatic Pancreatic Cancer. Clin. Cancer Res. 2016, 22, 2565–2574. [Google Scholar] [CrossRef]
- Tomasich, E.; Muhlbacher, J.; Woran, K.; Hatziioannou, T.; Herac, M.; Kleinberger, M.; Berger, J.M.; Dibon, L.K.; Berchtold, L.; Heller, G.; et al. Immune cell distribution and DNA methylation signatures differ between tumor and stroma enriched compartment in pancreatic ductal adenocarcinoma. Transl. Res. 2024, 271, 40–51. [Google Scholar] [CrossRef]
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; Castro de Moura, M.; Putra, J.; Camprecios, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017, 153, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Machida, K.; Feldman, D.E.; Tsukamoto, H. TLR4-dependent tumor-initiating stem cell-like cells (TICs) in alcohol-associated hepatocellular carcinogenesis. In Biological Basis of Alcohol-Induced Cancer; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2015; Volume 815, pp. 131–144. [Google Scholar] [CrossRef]
- An, Y.; Gao, S.; Zhao, W.C.; Qiu, B.A.; Xia, N.X.; Zhang, P.J.; Fan, Z.P. Transforming growth factor-beta and peripheral regulatory cells are negatively correlated with the overall survival of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, S.M.; Zhang, F.; Ding, C.; Montoya-Durango, D.E.; Hu, X.; Yang, C.; Wang, Z.; Yuan, F.; Fox, M.; Zhang, H.G.; et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021, 33, 2040–2058.e10. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, N.; Yan, T.; Wei, J.; Hao, L.; Sun, C.; Zhao, H.; Jiang, S. Lactate-mediated metabolic reprogramming of tumor-associated macrophages: Implications for tumor progression and therapeutic potential. Front. Immunol. 2025, 16, 1573039. [Google Scholar] [CrossRef]
- Wei, X.; Michelakos, T.; He, Q.; Wang, X.; Chen, Y.; Kontos, F.; Wang, H.; Liu, X.; Liu, H.; Zheng, W.; et al. Association of Tumor Cell Metabolic Subtype and Immune Response with the Clinical Course of Hepatocellular Carcinoma. Oncologist 2023, 28, e1031–e1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zeng, L.; Chen, R.; Zheng, S.; Zhou, Y.; Chen, R. Characterization of heterogeneous metabolism in hepatocellular carcinoma identifies new therapeutic target and treatment strategy. Front. Immunol. 2023, 14, 1076587. [Google Scholar] [CrossRef]
- Beilmann-Lehtonen, I.; Kasurinen, J.; Hagstrom, J.; Kaprio, T.; Bockelman, C.; Haglund, C. High tissue expression of TLRs combined with high density of tumor infiltrating lymphocytes predicts a better prognosis in colorectal cancer patients. PLoS ONE 2023, 18, e0280085. [Google Scholar] [CrossRef]
- Cammarota, R.; Bertolini, V.; Pennesi, G.; Bucci, E.O.; Gottardi, O.; Garlanda, C.; Laghi, L.; Barberis, M.C.; Sessa, F.; Noonan, D.M.; et al. The tumor microenvironment of colorectal cancer: Stromal TLR-4 expression as a potential prognostic marker. J. Transl. Med. 2010, 8, 112. [Google Scholar] [CrossRef]
- Kasurinen, A.; Hagstrom, J.; Laitinen, A.; Kokkola, A.; Bockelman, C.; Haglund, C. Evaluation of toll-like receptors as prognostic biomarkers in gastric cancer: High tissue TLR5 predicts a better outcome. Sci. Rep. 2019, 9, 12553. [Google Scholar] [CrossRef]
- Leppanen, J.; Helminen, O.; Huhta, H.; Kauppila, J.H.; Isohookana, J.; Haapasaari, K.M.; Lehenkari, P.; Saarnio, J.; Karttunen, T.J. High toll-like receptor (TLR) 9 expression is associated with better prognosis in surgically treated pancreatic cancer patients. Virchows Arch. 2017, 470, 401–410. [Google Scholar] [CrossRef]
- Ridnour, L.A.; Cheng, R.Y.; Switzer, C.H.; Heinecke, J.L.; Ambs, S.; Glynn, S.; Young, H.A.; Trinchieri, G.; Wink, D.A. Molecular pathways: Toll-like receptors in the tumor microenvironment--poor prognosis or new therapeutic opportunity. Clin. Cancer Res. 2013, 19, 1340–1346. [Google Scholar] [CrossRef]
- Matijevic, T.; Pavelic, J. Toll-like receptors: Cost or benefit for cancer? Curr. Pharm. Des. 2010, 16, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Cao, Y.; Wan, L.; Ren, Z.; Wang, X.; Huang, M.; Guo, Y. Comparison of Helicobacter pylori positive and negative gastric cancer via multi-omics analysis. mBio 2023, 14, e0153123. [Google Scholar] [CrossRef] [PubMed]
- Bednarz-Misa, I.; Fortuna, P.; Diakowska, D.; Jamrozik, N.; Krzystek-Korpacka, M. Distinct Local and Systemic Molecular Signatures in the Esophageal and Gastric Cancers: Possible Therapy Targets and Biomarkers for Gastric Cancer. Int. J. Mol. Sci. 2020, 21, 4509. [Google Scholar] [CrossRef] [PubMed]
- Greten, T.F.; Schwabe, R.; Bardeesy, N.; Ma, L.; Goyal, L.; Kelley, R.K.; Wang, X.W. Immunology and immunotherapy of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 349–365. [Google Scholar] [CrossRef]
- Gyorffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Posta, M.; Gyorffy, B. Pathway-level mutational signatures predict breast cancer outcomes and reveal therapeutic targets. Br. J. Pharmacol. 2025, 182, 5734–5747. [Google Scholar] [CrossRef]
- Gyorffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. GeroScience 2023, 45, 1889–1898. [Google Scholar] [CrossRef]
- Gyorffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 2024, 181, 362–374. [Google Scholar] [CrossRef]
- Menyhart, O.; Nagy, A.; Gyorffy, B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef]
- Posta, M.; Gyorffy, B. Analysis of a large cohort of pancreatic cancer transcriptomic profiles to reveal the strongest prognostic factors. Clin. Transl. Sci. 2023, 16, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets with tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Aronow, R.A.; Kirshtein, A.; Shahriyari, L. A review of digital cytometry methods: Estimating the relative abundance of cell types in a bulk of cells. Brief. Bioinform. 2021, 22, bbaa219. [Google Scholar] [CrossRef] [PubMed]
| Hepatocellular Carcinoma | |||
|---|---|---|---|
| N | % | ||
| Total | 370 | 100.00% | |
| Gender | Male | 249 | 67.30% |
| Female | 121 | 32.70% | |
| Race | Caucasian | 184 | 49.73% |
| Asian | 157 | 42.43% | |
| African | 17 | 4.59% | |
| Stage | 1 | 171 | 46.22% |
| 2 | 85 | 22.97% | |
| 3 | 85 | 22.97% | |
| 4 | 5 | 1.35% | |
| Grade | 1 | 55 | 14.86% |
| 2 | 177 | 47.84% | |
| 3 | 121 | 32.70% | |
| 4 | 12 | 3.24% | |
| Pancreatic Ductal Adenocarcinoma | |||
|---|---|---|---|
| N | % | ||
| Total | 177 | 100.00% | |
| Gender | Male | 97 | 54.80% |
| Female | 80 | 45.20% | |
| Race | Caucasian | 156 | 88.14% |
| Asian | 11 | 6.21% | |
| African | 6 | 3.39% | |
| Stage | 1 | 21 | 11.86% |
| 2 | 146 | 82.49% | |
| 3 | 3 | 1.69% | |
| 4 | 4 | 2.26% | |
| Grade | 1 | 31 | 17.51% |
| 2 | 94 | 53.11% | |
| 3 | 48 | 27.12% | |
| 4 | 2 | 1.13% | |
| HCC (n = 157) | PDAC (n = 177) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNMT3A—Asian | GMPS—Asian | DNMT3A | GMPS | |||||||||||
| p-Value | OS Low | OS High | p-Value | OS Low | OS High | p-Value | OS Low | OS High | p-Value | OS Low | OS High | |||
| All patients | 0.00087 | 56.17 | 9.97 | 0.00052 | >57 | 12.5 | 0.00084 | 16.6 | 30.43 | 0.00041 | 35.3 | 16.2 | ||
| Basophils | ↓ | 0.00081 | 56.17 | 9.97 | 0.00057 | >57 | 13.0 | ↓ | 0.0011 | 16.03 | 23.4 | 0.001 | 23.4 | 15.57 |
| B cells | ↓ | 0.00054 | >57 | 10.5 | 0.0014 | >57 | 13.0 | ↓ | 0.0029 | 15.77 | 24.6 | 0.0043 | 37.67 | 15.77 |
| CD4+ Tmem | ↓ | 0.0012 | >57 | 12.5 | 0.0027 | >57 | 12.5 | ↓ | 0.011 | 16.17 | 24.3 | 0.00022 | 37.67 | 15.53 |
| CD8+ T cells | ↓ | 0.0021 | >57 | 12.5 | 0.0022 | >57 | 13.0 | ↓ | 0.0032 | 15.57 | 23.17 | 0.0048 | 23.17 | 15.33 |
| Eosinophils | ↓ | 0.0012 | 56.17 | 10 | 0.001 | >57 | 14.0 | ↑ | 0.0043 | 17.27 | 35.3 | 0.0039 | 35.3 | 17.27 |
| MSC | ↓ | 0.00049 | >57 | 11.0 | 0.0015 | >57 | 10.5 | ↑ | 0.014 | 16.6 | 23.4 | 0.00068 | 23.4 | 15.57 |
| Macrophages | ↑ | 0.0081 | 56.17 | 9.97 | 0.00056 | >57 | 7.5 | ↓ | 0.045 | 8.13 | 13.1 | 0.00079 | 16.17 | 8.33 |
| NK T cells | ↑ | 0.0022 | 54.07 | 9.3 | 0.0026 | 56.17 | 9.87 | n.s. | -- | -- | n.s. | -- | -- | |
| Reg T cells | ↑ | 1.60 × 10−5 | >57 | 12.0 | 0.0014 | >57 | 14.0 | n.s. | -- | -- | n.s. | -- | -- | |
| Th1 cells | ↑ | 0.0047 | 54.07 | 9.3 | 0.00038 | 56.17 | 9.3 | ↓ | 0.00085 | 15.87 | 24.4 | 0.00074 | 30.43 | 15.77 |
| Th2 cells | ↑ | 0.00055 | 25.6 | 5.7 | 0.025 | 15.63 | 6.5 | n.s. | -- | -- | n.s. | -- | -- | |
| Mutation Burden | ↑ | 0.0039 | 54.07 | 8.73 | 5.8 × 10−5 | 56.17 | 8.73 | n.s. | -- | -- | n.s. | -- | -- | |
| HCC (n = 157) | PDAC (n = 177) | |||
|---|---|---|---|---|
| DNMT3A | GMPS | DNMT3A | GMPS | |
| Dunn.test.P | ||||
| Normal-Tumor | 6.09 × 10−16 | 2.25 × 10−69 | 2.75 × 10−11 | 7.45 × 10−11 |
| Normal-Metastatic | 2.57 × 10−8 | 4.84 × 10−22 | 4.04 × 10−2 | 1.85 × 10−1 |
| Tumor-Metastatic | 8.79 × 10−4 | 4.23 × 10−6 | 1.16 × 10−1 | 2.21 × 10−2 |
| PDAC (n = 177) | HCC—Asian (n = 157) | HCC—Caucasian (n = 184) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | OS Low | OS High | p-Value | OS Low | OS High | p-Value | OS Low | OS High | |||
| DNMT3A | TLR1 | High | n.s. | -- | -- | n.s. | -- | -- | n.s. | -- | -- |
| Low | 0.0024 | 14.33 | 67.87 | 0.042 | 54.07 | 11.47 | n.s. | -- | -- | ||
| TLR2 | High | n.s. | -- | -- | 7.9 × 10−5 | >57 | 6.0 | n.s. | -- | -- | |
| Low | 0.00014 | 15.33 | 67.87 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR3 | high | 0.0019 | 12.7 | 23.17 | 0.00072 | >57 | 10.0 | n.s. | -- | -- | |
| Low | n.s. | -- | -- | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR4 | high | 0.038 | 18.93 | 23.17 | 0.00014 | >57 | 8.0 | n.s. | -- | -- | |
| low | 0.019 | 16.6 | 35.3 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR5 | high | n.s. | -- | -- | 0.0023 | 56.17 | 5.7 | n.s. | -- | -- | |
| low | 0.0015 | 15.67 | 67.87 | n.s. | -- | -- | 0.0015 | 59.7 | 31.03 | ||
| TLR6 | high | n.s. | -- | -- | 0.0081 | >57 | 7.0 | n.s. | -- | -- | |
| low | 0.00035 | 15.77 | 72.73 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR7 | high | n.s. | -- | -- | 0.0055 | 54.07 | 4.3 | n.s. | -- | -- | |
| Low | 0.013 | 15.77 | 67.87 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR8 | High | n.s. | -- | -- | 0.00033 | >57 | 5.0 | n.s. | -- | -- | |
| Low | 0.0045 | 15.87 | 67.87 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR9 | High | n.s. | -- | -- | n.s. | -- | -- | n.s. | -- | -- | |
| Low | 0.024 | 15.87 | 24.6 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR10 | High | n.s. | -- | -- | 0.0022 | >57 | 5.0 | n.s. | -- | -- | |
| Low | 0.00035 | 15.67 | 67.87 | n.s. | -- | -- | 0.036 | 52.0 | 57.57 | ||
| GMPS | TLR1 | High | n.s. | -- | -- | n.s. | -- | -- | n.s. | -- | -- |
| Low | 0.00024 | 67.87 | 13.1 | n.s. | -- | -- | 0.018 | 71.03 | 27.57 | ||
| TLR2 | High | n.s. | -- | -- | 0.00013 | >57 | 5.0 | n.s. | |||
| Low | 0.000017 | 67.87 | 15.33 | n.s. | -- | -- | 0.0036 | 71.03 | 29.97 | ||
| TLR3 | high | n.s. | -- | -- | 0.011 | >57 | 15.0 | n.s. | -- | -- | |
| Low | 0.00062 | 67.87 | 19.93 | 0.037 | 21.63 | 9.97 | n.s. | -- | -- | ||
| TLR4 | high | 0.0093 | 23.17 | 18.17 | 1.3 × 10−5 | >57 | 6.0 | n.s. | -- | -- | |
| low | 0.012 | 37.67 | 15.53 | n.s. | -- | -- | 0.0077 | 56.47 | 27.57 | ||
| TLR5 | high | n.s. | -- | -- | 0.0045 | 56.17 | 6.5 | n.s. | -- | -- | |
| low | 0.00036 | 37.67 | 13.13 | 0.044 | >57 | 15.0 | 0.036 | 59.7 | 31.03 | ||
| TLR6 | high | 0.013 | 23.4 | 15.57 | 0.00047 | >57 | 6.0 | n.s. | -- | -- | |
| low | 0.0077 | 17.03 | 9.77 | n.s. | -- | -- | 0.016 | 59.7 | 31.03 | ||
| TLR7 | high | n.s. | -- | -- | 0.0029 | 54.07 | 4.67 | n.s. | -- | -- | |
| Low | 0.0016 | 67.87 | 13.13 | n.s. | -- | -- | 0.021 | 59.7 | 27.57 | ||
| TLR8 | High | n.s. | -- | -- | 0.00044 | >57 | 5.0 | n.s. | -- | -- | |
| Low | 0.0012 | 67.87 | 15.53 | n.s. | -- | -- | 0.004 | 59.7 | 29.97 | ||
| TLR9 | High | n.s. | -- | -- | 0.01 | NA | NA | n.s. | -- | -- | |
| Low | 0.032 | 30.43 | 15.87 | n.s. | -- | -- | n.s. | -- | -- | ||
| TLR10 | High | n.s. | -- | -- | 0.0048 | >57 | 7.0 | n.s. | -- | -- | |
| Low | 0.034 | 37.67 | 14.33 | 0.012 | NA | NA | n.s. | -- | -- | ||
| Low Expression | TLR1 | TLR2 | TLR3 | TLR4 | TLR5 | TLR6 | TLR7 | TLR8 | TLR9 | TLR10 | Sum |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDAC | 1 | 1 | |||||||||
| PDAC-Seq | 1 | 1 | 1 | 1 | 4 | ||||||
| Breast | 1 | 1 | |||||||||
| Breast-TCGA | 0 | ||||||||||
| AML | 1 | 1 | 1 | 1 | 4 | ||||||
| Gastric | 1 | 1 | 2 | ||||||||
| Lung | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | ||
| Myeloma | 0 | ||||||||||
| Ovarian | 1 | 1 | 1 | 1 | 1 | 5 | |||||
| HCC-Seq | 0 | ||||||||||
| HCC-Seq-Caucasian | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | |||
| Total | 4 | 3 | 4 | 3 | 3 | 3 | 2 | 3 | 5 | 2 | |
| High Expression | TLR1 | TLR2 | TLR3 | TLR4 | TLR5 | TLR6 | TLR7 | TLR8 | TLR9 | TLR10 | Sum |
| PDAC | 1 | 1 | |||||||||
| PDAC-Seq | 0 | ||||||||||
| Breast | 0 | ||||||||||
| Breast-TCGA | 1 | 1 | 1 | 1 | 4 | ||||||
| AML | 0 | ||||||||||
| Gastric | 1 | 1 | 1 | 1 | 1 | 5 | |||||
| Lung | 0 | ||||||||||
| Myeloma | 1 | 1 | |||||||||
| Ovarian | 0 | ||||||||||
| HCC-Seq-Asian | 1 | 1 | 1 | 1 | 4 | ||||||
| Total | 2 | 3 | 1 | 2 | 0 | 2 | 2 | 2 | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, W.-H.; Shah, D.; Myers, S.; Potts, M.; Qazi, S.; Trieu, V. Comparative Tumor Microenvironment Analysis for HCC and PDAC Using KMplotter. Int. J. Mol. Sci. 2025, 26, 11920. https://doi.org/10.3390/ijms262411920
Chang W-H, Shah D, Myers S, Potts M, Qazi S, Trieu V. Comparative Tumor Microenvironment Analysis for HCC and PDAC Using KMplotter. International Journal of Molecular Sciences. 2025; 26(24):11920. https://doi.org/10.3390/ijms262411920
Chicago/Turabian StyleChang, Wen-Han, Drashya Shah, Scott Myers, Michael Potts, Sanjive Qazi, and Vuong Trieu. 2025. "Comparative Tumor Microenvironment Analysis for HCC and PDAC Using KMplotter" International Journal of Molecular Sciences 26, no. 24: 11920. https://doi.org/10.3390/ijms262411920
APA StyleChang, W.-H., Shah, D., Myers, S., Potts, M., Qazi, S., & Trieu, V. (2025). Comparative Tumor Microenvironment Analysis for HCC and PDAC Using KMplotter. International Journal of Molecular Sciences, 26(24), 11920. https://doi.org/10.3390/ijms262411920






