Thermodynamic Profiling Reveals DNA Polymerase Template Binding, Substrate Incorporation, and Exonuclease Function
Abstract
1. Introduction
2. Results
2.1. Binding Affinity
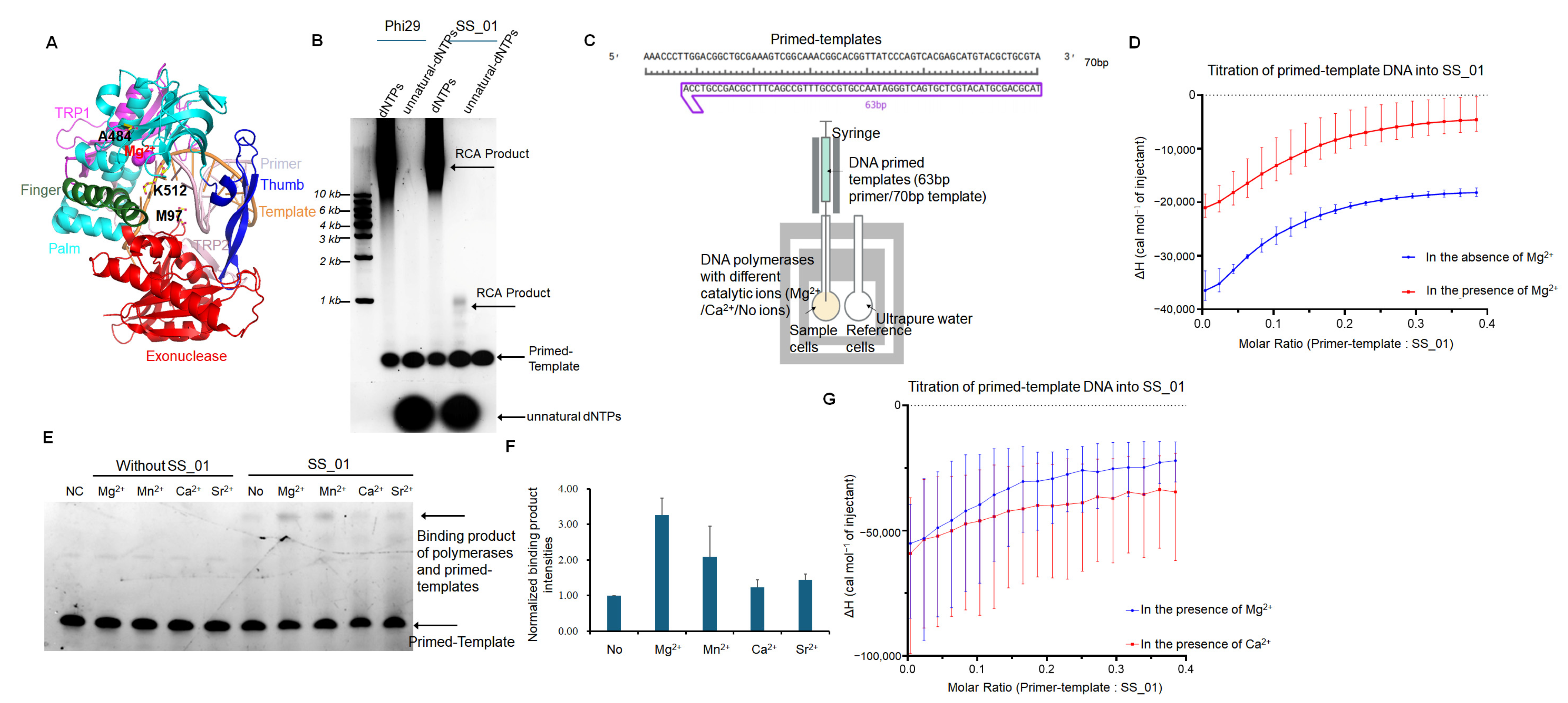
2.1.1. Binding Affinity Under Catalytic and Non-Catalytic Metal Ion Conditions
2.1.2. Binding Affinity Under Various Metal Ion Conditions
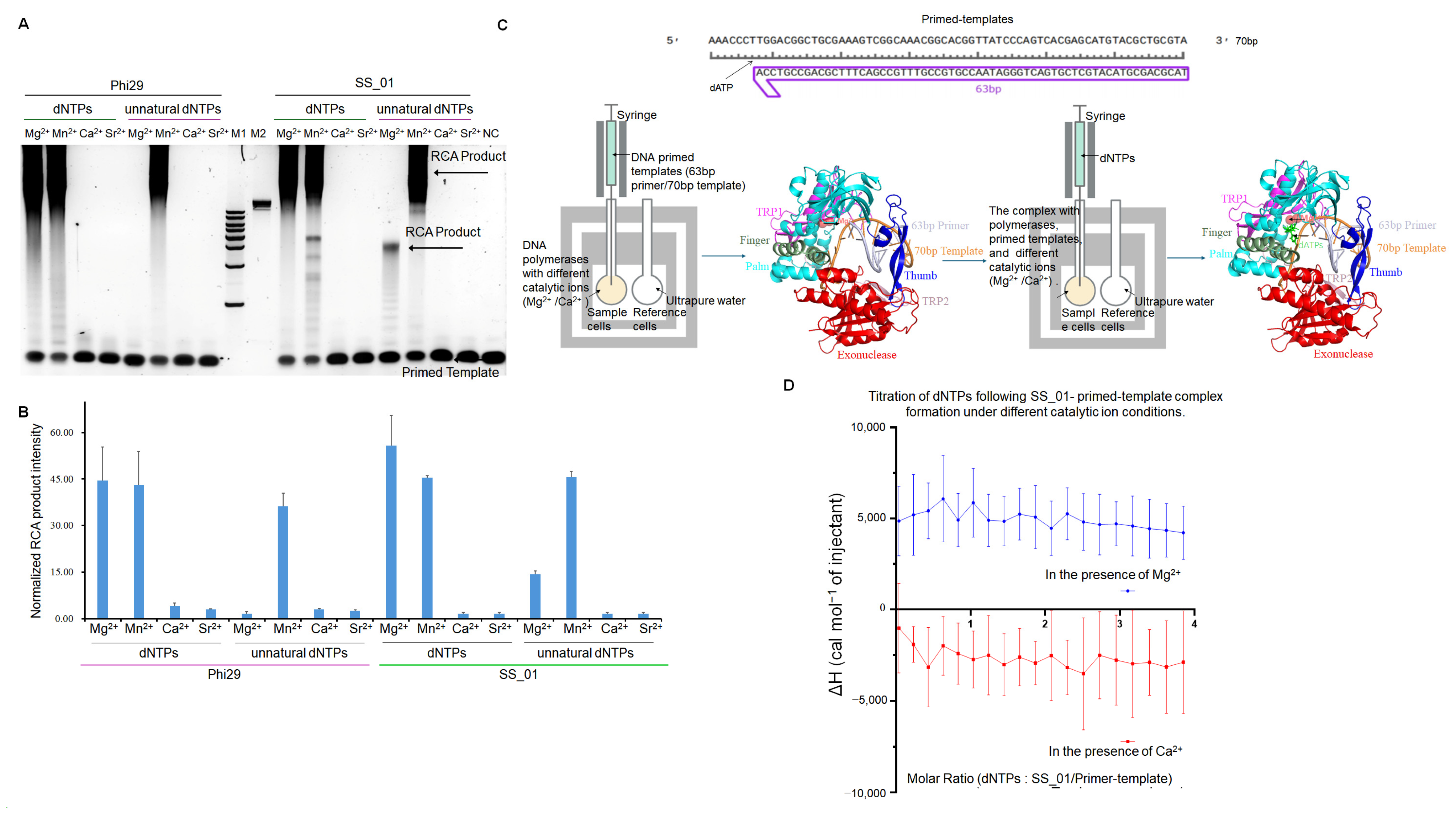
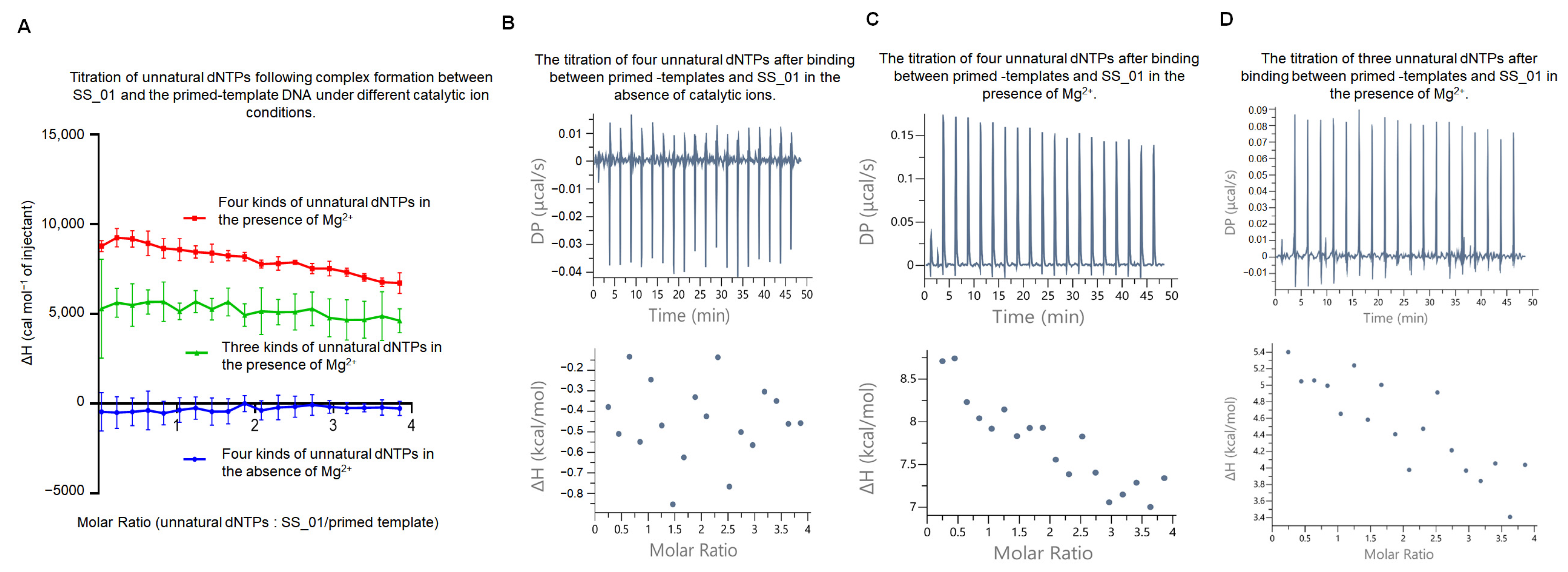
2.2. The Thermodynamic Changes During the Incorporation of SS_01 Using dNTPs
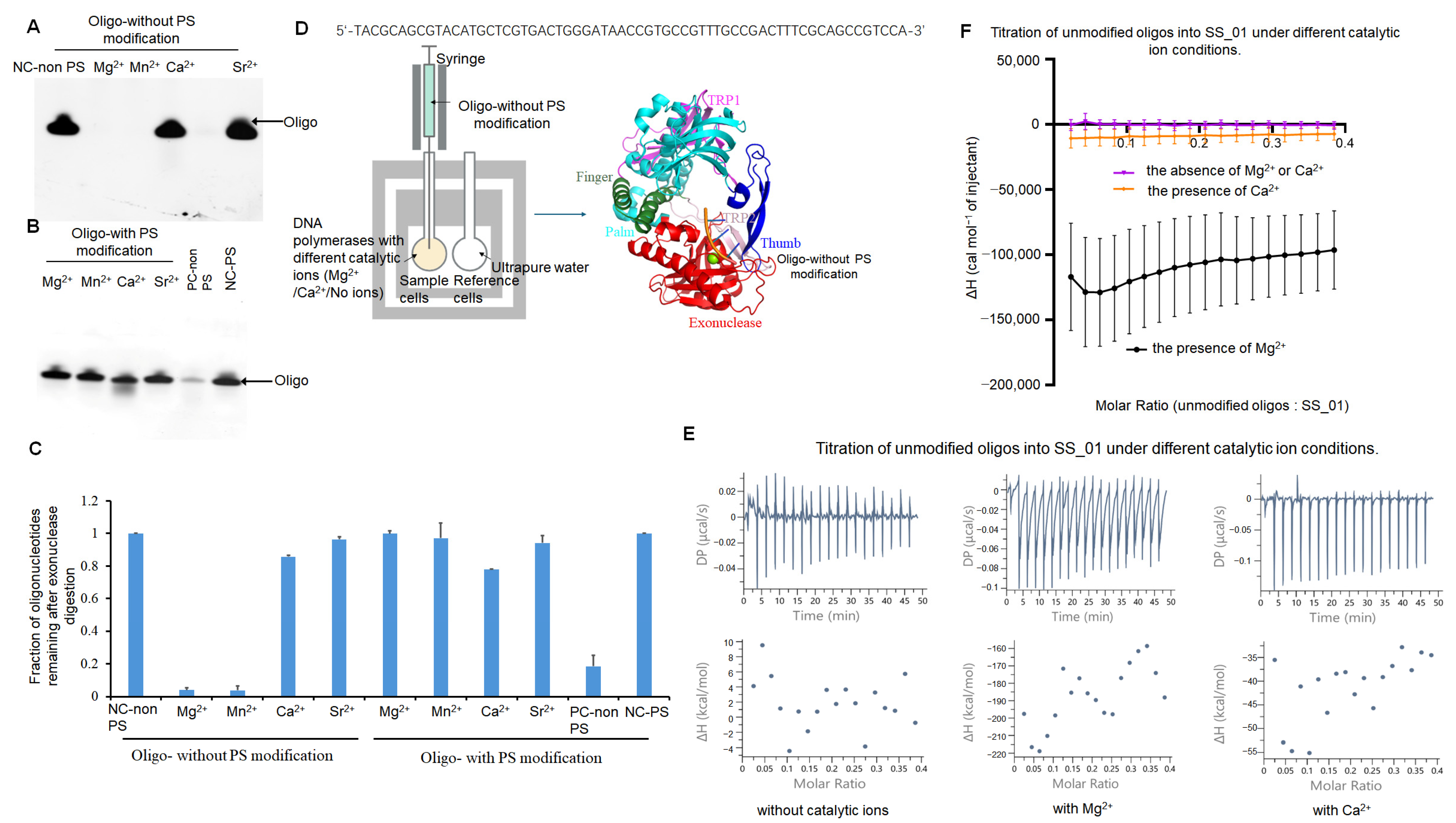
2.3. The Thermodynamic Changes During the Incorporation of SS_01 Using Oligonucleotide-Tagged dNTP Analogs
2.3.1. Thermodynamic Characterization of SS_01 Binding to Oligonucleotide-Tagged dNTPs Under Catalytic and Non-Catalytic Metal Ion Conditions
2.3.2. Thermodynamic Characterization of SS_01 Degrading the Oligonucleotides in the Presence of Mg2+
2.3.3. Thermodynamic Characterization of the Incorporation of SS_01 Using Oligonucleotide-Tagged dNTPs
3. Discussion
3.1. Metal Ion Dependence of Polymerase-Primed Templates Binding
3.2. Polymerization Activity and Thermodynamic Profiles
3.3. Oligonucleotide-Tagged dNTP or Single-Stranded DNA Binding with Polymerases
4. Materials and Methods
4.1. Plasmid Construction
4.2. Protein Expression
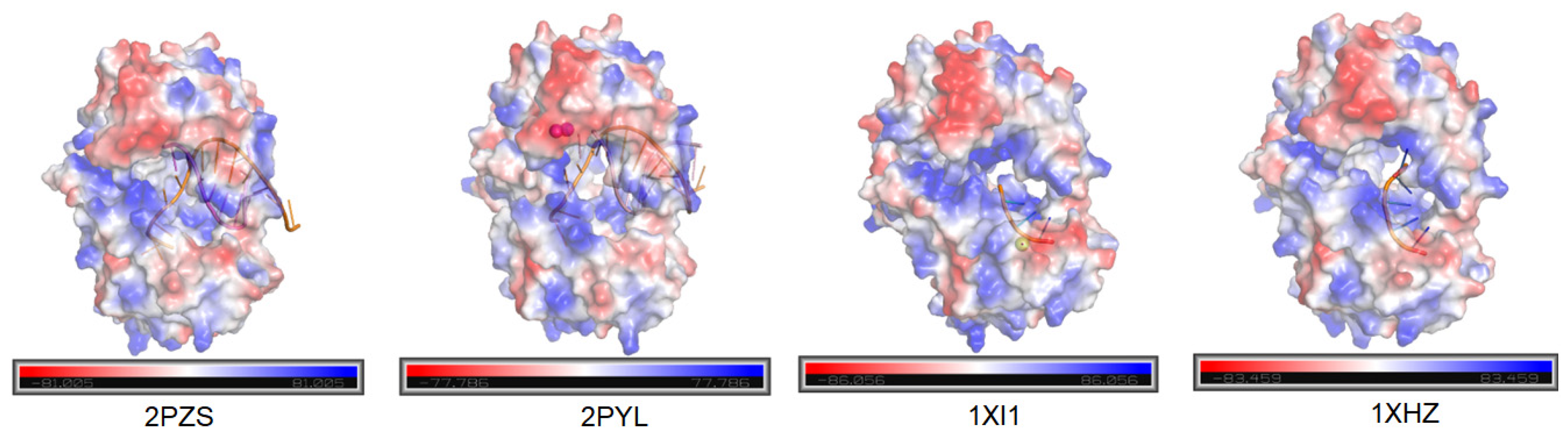
4.3. Structural Analysis
4.4. Exonuclease Activity
4.5. The Preparation of Single-Strand Circular (SSC) Templates
4.6. Rolling Circle Amplification (RCA)
4.7. Template Binding Affinity of DNA Polymerases in Various Metal Ion Conditions
4.8. A Pre-Annealed 63/70-mer DNA
4.9. Isothermal Titration Calorimetry (ITC)
4.9.1. Binding of SS_01 to 63/70-mer DNA Under Different Metal Ion Conditions
4.9.2. Two-Step ITC Assay for SS_01-Primed-Template Binding Followed by dNTPs or Unnatural Substrate Incorporation
4.9.3. Binding Affinity of SS_01 to Unmodified Oligonucleotides or Unnatural Substrates
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fuller, C.W.; Kumar, S.; Porel, M.; Chien, M.; Bibillo, A.; Stranges, P.B.; Dorwart, M.; Tao, C.; Li, Z.; Guo, W.; et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl. Acad. Sci. USA 2016, 113, 5233–5238. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; Shah, F.I.; Ibrar, R.; Fatima, T.; Haq, I.U.; Naseem, W.; Gul, M.A.; Tehreem, L.; Haider, G. Bacterial thermophilic DNA polymerases: A focus on prominent biotechnological applications. Anal. Biochem. 2023, 671, 115150. [Google Scholar] [CrossRef]
- Sun, Y.; Ko, D.H.; Gao, J.; Fu, K.; Gao, Y.; Zhang, Q.; Baldi, S.; Hong, T.; Ivanov, I.; He, Y.; et al. Unraveling the salt tolerance of Phi29 DNA polymerase using compartmentalized self-replication and microfluidics platform. Front. Microbiol. 2023, 14, 1267196. [Google Scholar] [CrossRef]
- Zhang, J.; Su, X.; Wang, Y.; Wang, X.; Zhou, S.; Jia, H.; Jing, X.; Gong, Y.; Wang, J.; Xu, J. Improved single-cell genome amplification by a high-efficiency phi29 DNA polymerase. Front. Bioeng. Biotechnol. 2023, 11, 1233856, Correction in Front. Bioeng. Biotechnol. 2023, 11, 1263634. https://doi.org/10.3389/fbioe.2023.1263634. [Google Scholar] [CrossRef]
- Gutfreund, C.; Betz, K.; Abramov, M.; Coosemans, F.; Holliger, P.; Herdewijn, P.; Marx, A. Structural insights into a DNA polymerase reading the xeno nucleic acid HNA. Nucleic Acids Res. 2025, 53, gkae1156. [Google Scholar] [CrossRef]
- Hottin, A.; Marx, A. Structural Insights into the Processing of Nucleobase-Modified Nucleotides by DNA Polymerases. Acc. Chem. Res. 2016, 49, 418–427. [Google Scholar] [CrossRef]
- Bermek, O.; Grindley, N.D.; Joyce, C.M. Distinct roles of the active-site Mg2+ ligands, Asp882 and Asp705, of DNA polymerase I (Klenow fragment) during the prechemistry conformational transitions. J. Biol. Chem. 2011, 286, 3755–3766. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.C.; Kool, E.T. Minor Groove Interactions between Polymerase and DNA: More Essential to Replication than Watson-Crick Hydrogen Bonds? J. Am. Chem. Soc. 1999, 121, 2323–2324. [Google Scholar] [CrossRef]
- Joyce, C.M.; Steitz, T.A. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994, 63, 777–822. [Google Scholar] [CrossRef] [PubMed]
- Kamtekar, S.; Berman, A.J.; Wang, J.; Lazaro, J.M.; de Vega, M.; Blanco, L.; Salas, M.; Steitz, T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell 2004, 16, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.J.; Kamtekar, S.; Goodman, J.L.; Lazaro, J.M.; de Vega, M.; Blanco, L.; Salas, M.; Steitz, T.A. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007, 26, 3494–3505. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Campoy, A.; Leavitt, S.A.; Freire, E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 2015, 1278, 183–204. [Google Scholar] [CrossRef]
- Upadhyay, V.; Lucas, A.; Patrick, C.; Mallela, K.M.G. Isothermal titration calorimetry and surface plasmon resonance methods to probe protein-protein interactions. Methods 2024, 225, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Dam, T.K.; Brewer, C.F. Thermodynamic studies of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem. Rev. 2002, 102, 387–429. [Google Scholar] [CrossRef]
- Datta, K.; LiCata, V.J. Thermodynamics of the binding of Thermus aquaticus DNA polymerase to primed-template DNA. Nucleic Acids Res. 2003, 31, 5590–5597. [Google Scholar] [CrossRef]
- Yang, W.; Weng, P.J.; Gao, Y. A new paradigm of DNA synthesis: Three-metal-ion catalysis. Cell Biosci. 2016, 6, 51, Erratum in Cell Biosci. 2017, 7, 32. [Google Scholar] [CrossRef]
- Chang, C.; Lee Luo, C.; Gao, Y. In crystallo observation of three metal ion promoted DNA polymerase misincorporation. Nat. Commun. 2022, 13, 2346. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Cryst. 1976, A32, 751–767. [Google Scholar] [CrossRef]
- Nikiforov, T.T. Oligonucleotides labeled with single fluorophores as sensors for deoxynucleotide triphosphate binding by DNA polymerases. Anal. Biochem. 2014, 444, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Stranges, P.B.; Palla, M.; Kalachikov, S.; Nivala, J.; Dorwart, M.; Trans, A.; Kumar, S.; Porel, M.; Chien, M.; Tao, C.; et al. Design and characterization of a nanopore-coupled polymerase for single-molecule DNA sequencing by synthesis on an electrode array. Proc. Natl. Acad. Sci. USA 2016, 113, E6749–E6756. [Google Scholar] [CrossRef]
- Wang, J.; Kamtekar, S.; Berman, A.J.; Steitz, T.A. Correction of X-ray intensities from single crystals containing lattice-translocation defects. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; He, Y.; Ivanov, I.; Yang, X.; Tian, H.; Liu, X. Expression and functional study of VpV262 Pol, a moderately halophilic DNA polymerase from the Vibrio parahaemolyticus phage VpV262. Enzym. Microb. Technol. 2020, 139, 109588. [Google Scholar] [CrossRef]
- Sun, Y.; Ko, D.H.; Gao, J.; Fu, K.; Mao, Y.; He, Y.; Tian, H. Engineering psychrophilic polymerase for nanopore long-read sequencing. Front. Bioeng. Biotechnol. 2024, 12, 1406722. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ko, D.H.; Gao, J.; Fu, K.; Mao, Y.; He, Y.; Tian, H. Expression and functional study of DNA polymerases from Psychrobacillus sp. BL-248-WT-3 and FJAT-21963. Front. Microbiol. 2024, 15, 1501020. [Google Scholar] [CrossRef]
- Gao, Y.; He, Y.; Chen, L.; Liu, X.; Ivanov, I.; Yang, X.; Tian, H. Chimeric Phi29 DNA polymerase with helix-hairpin-helix motifs shows enhanced salt tolerance and replication performance. Microb. Biotechnol. 2021, 14, 1642–1656. [Google Scholar] [CrossRef]
- Sun, Y.; Fu, K.; Lin, W.; Gao, J.; Zhao, X.; He, Y.; Tian, H. Characterization and Engineering of Two Novel Strand-Displacing B Family DNA Polymerases from Bacillus Phage SRT01hs and BeachBum. Biomolecules 2025, 15, 1126. [Google Scholar] [CrossRef]
- Blanco, L.; Bernad, A.; Lazaro, J.M.; Martin, G.; Garmendia, C.; Salas, M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989, 264, 8935–8940. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.K.; Chase, S.F.; Laue, T.M.; Jen-Jacobson, L.; Trakselis, M.A. Differential temperature-dependent multimeric assemblies of replication and repair polymerases on DNA increase processivity. Biochemistry 2012, 51, 7367–7382. [Google Scholar] [CrossRef]
- Weaver, T.M.; Washington, M.T.; Freudenthal, B.D. New insights into DNA polymerase mechanisms provided by time-lapse crystallography. Curr. Opin. Struct. Biol. 2022, 77, 102465. [Google Scholar] [CrossRef]
- Gong, S.; Kirmizialtin, S.; Chang, A.; Mayfield, J.E.; Zhang, Y.J.; Johnson, K.A. Kinetic and thermodynamic analysis defines roles for two metal ions in DNA polymerase specificity and catalysis. J. Biol. Chem. 2021, 296, 100184. [Google Scholar] [CrossRef]
- Dahl, J.M.; Lieberman, K.R.; Wang, H. Modulation of DNA Polymerase Noncovalent Kinetic Transitions by Divalent Cations. J. Biol. Chem. 2016, 291, 6456–6470. [Google Scholar] [CrossRef]
- Garcia-Diaz, M.; Bebenek, K.; Krahn, J.M.; Pedersen, L.C.; Kunkel, T.A. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair 2007, 6, 1333–1340. [Google Scholar] [CrossRef]
- Minetti, C.A.; Remeta, D.P.; Miller, H.; Gelfand, C.A.; Plum, G.E.; Grollman, A.P.; Breslauer, K.J. The thermodynamics of template-directed DNA synthesis: Base insertion and extension enthalpies. Proc. Natl. Acad. Sci. USA 2003, 100, 14719–14724. [Google Scholar] [CrossRef]
- Ralec, C.; Henry, E.; Lemor, M.; Killelea, T.; Henneke, G. Calcium-driven DNA synthesis by a high-fidelity DNA polymerase. Nucleic Acids Res. 2017, 45, 12425–12440. [Google Scholar] [CrossRef] [PubMed]
- Elisseeva, E.; Mandal, S.S.; Reha-Krantz, L.J. Mutational and pH studies of the 3′ → 5′ exonuclease activity of bacteriophage T4 DNA polymerase. J. Biol. Chem. 1999, 274, 25151–25158. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 2022, 31, e4482. [Google Scholar] [CrossRef] [PubMed]
- Kagami, L.P.; das Neves, G.M.; Timmers, L.; Caceres, R.A.; Eifler-Lima, V.L. Geo-Measures: A PyMOL plugin for protein structure ensembles analysis. Comput. Biol. Chem. 2020, 87, 107322. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Wowor, A.J.; Richard, A.J.; LiCata, V.J. Temperature dependence and thermodynamics of Klenow polymerase binding to primed-template DNA. Biophys. J. 2006, 90, 1739–1751. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Lin, W.; Fu, K.; Gao, J.; Zhao, X.; He, Y.; Tian, H. Thermodynamic Profiling Reveals DNA Polymerase Template Binding, Substrate Incorporation, and Exonuclease Function. Int. J. Mol. Sci. 2025, 26, 11909. https://doi.org/10.3390/ijms262411909
Sun Y, Lin W, Fu K, Gao J, Zhao X, He Y, Tian H. Thermodynamic Profiling Reveals DNA Polymerase Template Binding, Substrate Incorporation, and Exonuclease Function. International Journal of Molecular Sciences. 2025; 26(24):11909. https://doi.org/10.3390/ijms262411909
Chicago/Turabian StyleSun, Yaping, Wu Lin, Kang Fu, Jie Gao, Xianhui Zhao, Yun He, and Hui Tian. 2025. "Thermodynamic Profiling Reveals DNA Polymerase Template Binding, Substrate Incorporation, and Exonuclease Function" International Journal of Molecular Sciences 26, no. 24: 11909. https://doi.org/10.3390/ijms262411909
APA StyleSun, Y., Lin, W., Fu, K., Gao, J., Zhao, X., He, Y., & Tian, H. (2025). Thermodynamic Profiling Reveals DNA Polymerase Template Binding, Substrate Incorporation, and Exonuclease Function. International Journal of Molecular Sciences, 26(24), 11909. https://doi.org/10.3390/ijms262411909




