Comparative Transcriptomic Analysis Underlies the Differential Virulence of Vibrio harveyi and Vibrio vulnificus in American Eels (Anguilla rostrata)
Abstract
1. Introduction
2. Results
2.1. Comparative Histopathology of Liver, Kidney, and Spleen
2.2. Sequencing Data and Mapping Rate
2.3. Gene Expression, Venn Analysis, Correlation and Clustering Between Samples
2.4. Differential Expressed Genes (DEGs): Venn and Heatmap Analysis of DEGs
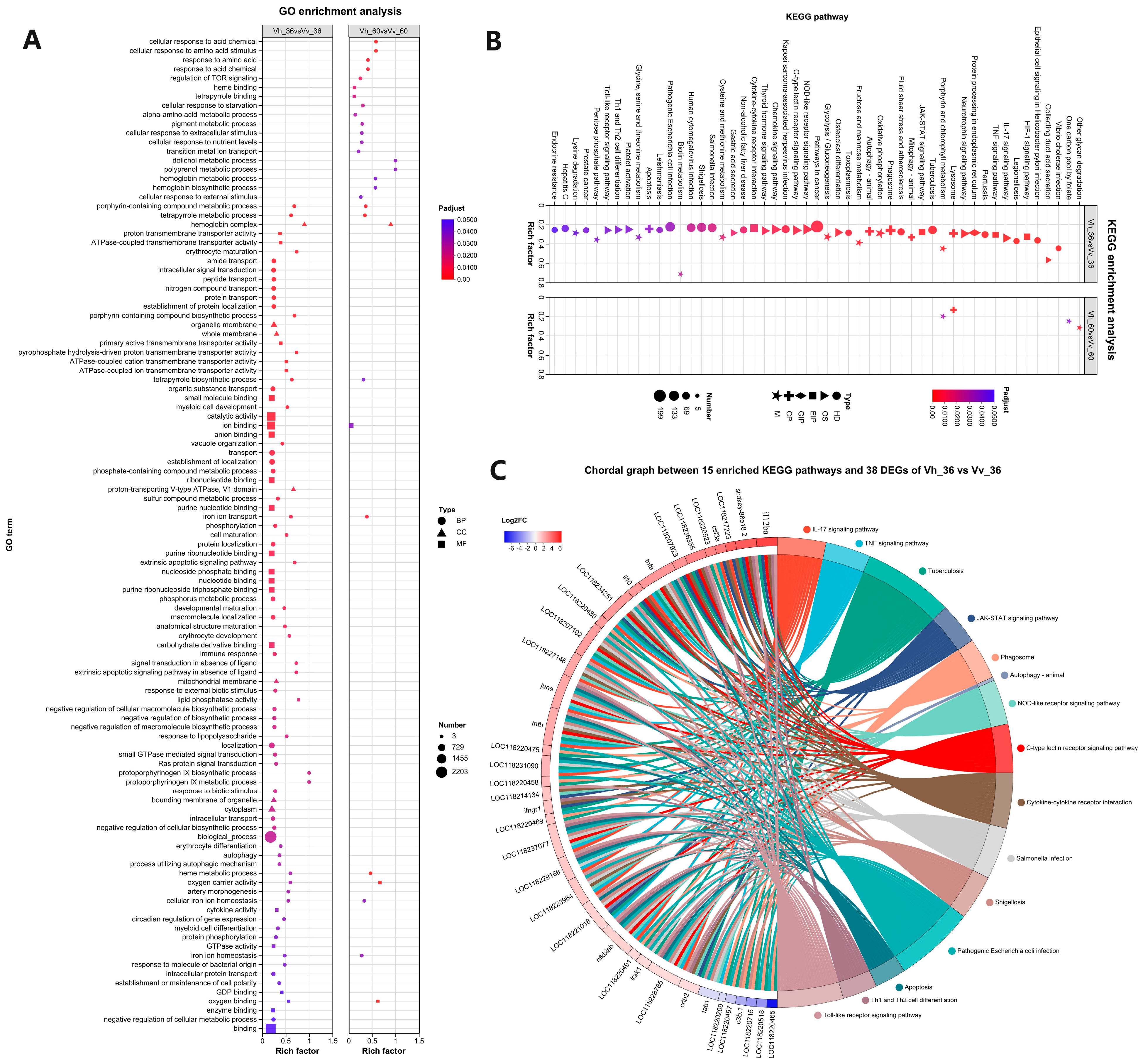
2.5. GO and KEGG Enrichment Analysis
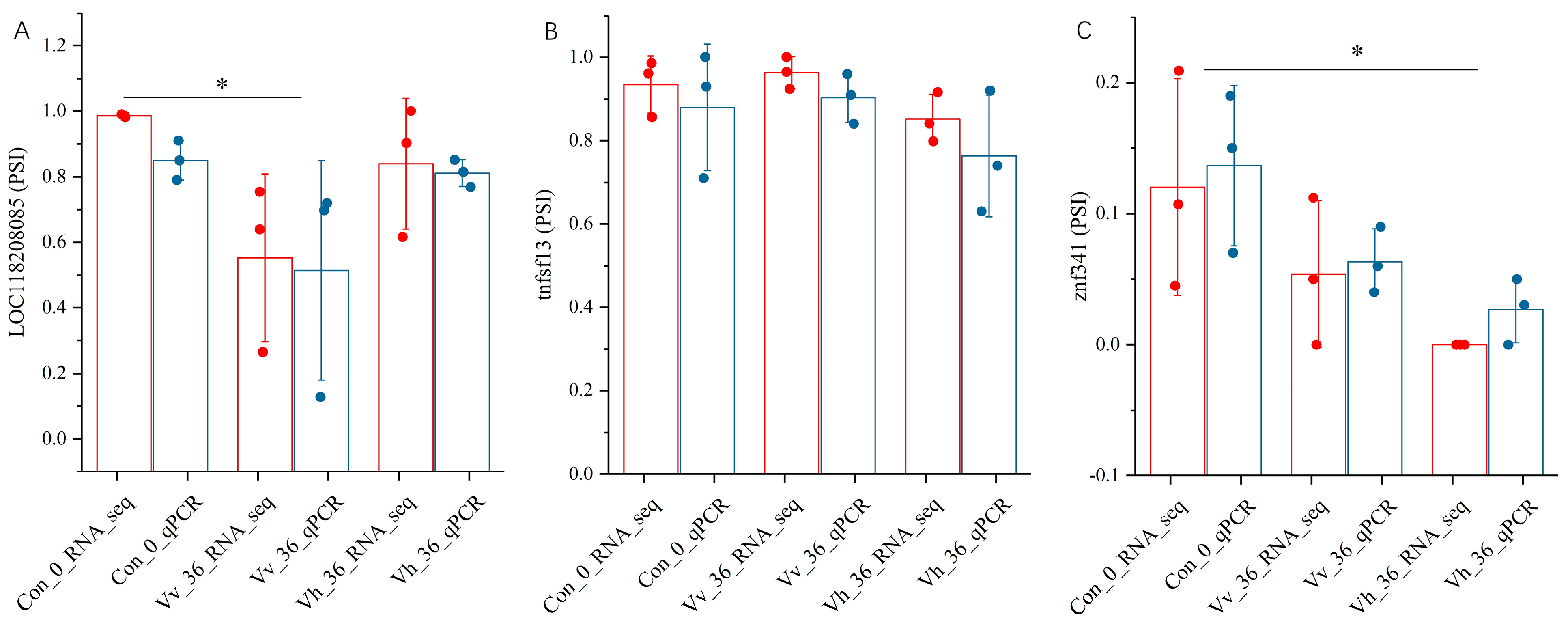
2.6. Validation of qRT-PCR for RNA_Seq
2.7. The Differential Alternative Splicing Genes (DASs) Analysis
2.8. The Results of the AS Verification
2.9. Annotation and Enrichment Analysis of DASs, DEGs, and Overlapping DAS-DEGs
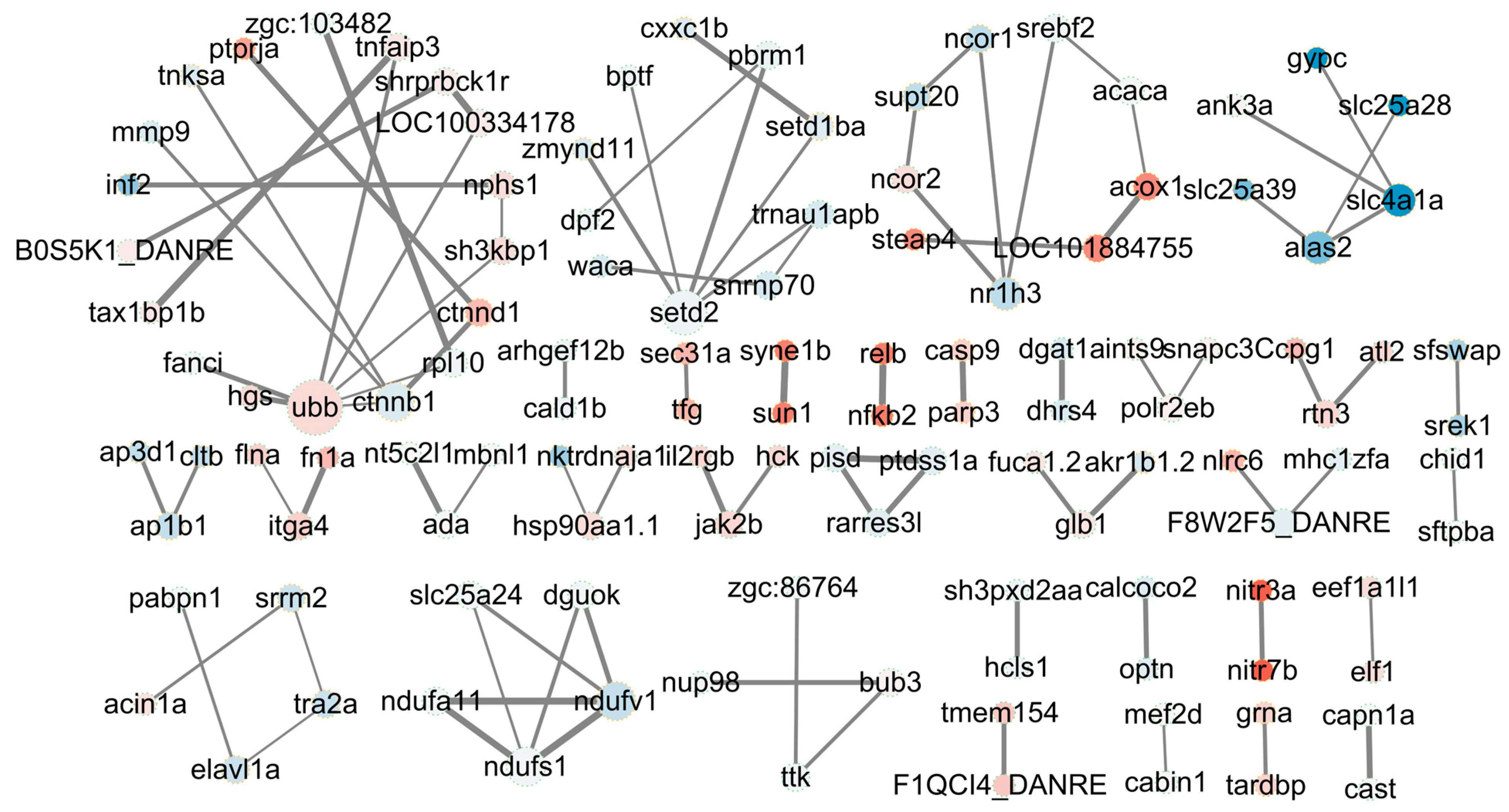
2.10. Protein–Protein Interaction Network of DAS Gene-Encoded Proteins
3. Discussion
3.1. Pathological Changes and Differences in Virulence
3.2. Analysis of DEGs and Virulence Differences
3.3. Analysis of DAS Genes and Virulence Differences
4. Materials and Methods
4.1. Bacteria Strain and American Eels
4.2. Experimental Infection and Sampling
4.3. Preparation of Paraffin Sections
4.4. Transcriptome Analysis Via RNA-Seq
4.5. Analysis of Gene Expression and Differentially Expressed Genes
4.6. QRT-PCR Validation
4.7. Alternative Splicing Events Analysis
4.8. QPCR Verification of Alternative Splicing Events
4.9. Protein–Protein Interaction Network of Proteins Encoded by DAS Genes
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Noorian, P.; Hoque, M.M.; Espinoza-Vergara, G.; McDougald, D. Environmental reservoirs of pathogenic Vibrio spp. and their role in disease: The list keeps expanding. Adv. Exp. Med. Biol. 2023, 1404, 99–126. [Google Scholar]
- Ina-Salwany, M.Y.; Al-Saari, N.; Mohamad, A.; Mursidi, F.A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in fish: A review on disease development and prevention. J. Aquat. Anim. Health. 2019, 31, 3–22. [Google Scholar] [CrossRef]
- Wan, Q.; Wu, L.; Yang, Q.; Lin, M.; Guo, S. First identification and pathogenicity study of Vibrio harveyi isolated from diseased American eel (Anguilla rostrata) cultivated in freshwater. Aquac. Res. 2022, 53, 1240–1253. [Google Scholar] [CrossRef]
- He, L.; Duan, L.; Feng, J.; Lin, P.; Guo, S. Immunogenicity study of an expressed outer membrane protein U of Vibrio vulnificus in Japanese eel (Anguilla japonica). J. Appl. Microbiol. 2018, 125, 1642–1654. [Google Scholar]
- Almagro-Moreno, S.; Martinez-Urtaza, J.; Pukatzki, S. Vibrio infections and the twenty-first century. Adv. Exp. Med. Biol. 2023, 1404, 1–16. [Google Scholar] [PubMed]
- Amaro, C.; Carmona-Salido, H. Vibrio vulnificus, an underestimated zoonotic pathogen. Adv. Exp. Med. Biol. 2023, 1404, 175–194. [Google Scholar] [PubMed]
- Chen, M.; Wan, Q.; Xu, M.; Chen, Z.; Guo, S. Transcriptome analysis of host anti-Vibrio harveyi infection revealed the pathogenicity of V. harveyi to American eel (Anguilla rostrata). Mar. Biotechnol. 2024, 26, 306–323. [Google Scholar] [CrossRef] [PubMed]
- De Souza Valente, C.; Wan, A.H.L. Vibrio and major commercially important vibriosis diseases in decapod crustaceans. J. Invertebr. Pathol. 2021, 181, 107527. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, S.; Chen, M.; Xu, M.; Guo, S. Comparative phenotype and transcriptome analysis revealed the role of ferric uptake regulator (Fur) in the virulence of Vibrio harveyi isolated from diseased American eel (Anguilla rostrata). J. Fish Dis. 2024, 47, e13931. [Google Scholar] [CrossRef]
- Hernández-Cabanyero, C.; Lee, C.T.; Tolosa-Enguis, V.; Sanjuán, E.; Pajuelo, D.; Reyes-López, F.; Tort, L.; Amaro, C. Adaptation to host in Vibrio vulnificus, a zoonotic pathogen that causes septicemia in fish and humans. Environ. Microbiol. 2019, 21, 3118–3139. [Google Scholar] [CrossRef]
- Davidson, N.; Edwards, F.; Harris, P.N.A.; Laupland, K.B. Vibrio species bloodstream infections in Queensland Australia. Intern. Med. J. 2024, 54, 157–163. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Wan, Q.; Chen, M.; Guo, S. RNA-seq analysis revealed the pathogenicity of Vibrio vulnificus to American eel (Anguilla rostrata) and the strategy of host anti-V. vulnificus infection. Microb. Pathog. 2024, 186, 106498. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wu, L.; Ling, P.; Zhai, S.; Guo, S.; Xiao, Y.; Wan, Q. First expression and immunogenicity study of a novel trivalent outer membrane protein (OmpII-U-A) from Aeromonas hydrophila, Vibrio vulnificus and Edwardsiella anguillarum. Aquaculture 2020, 519, 734932. [Google Scholar] [CrossRef]
- Guo, S.; He, L.; Wu, L.; Xiao, Y.; Zhai, S.; Yan, Q. Immunization of a novel bivalent outer membrane protein simultaneously resisting Aeromonas hydrophila, Edwardsiella anguillarum and Vibrio vulnificus infection in European eels (Anguilla anguilla). Fish Shellfish Immunol. 2020, 97, 46–57. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, S.; Chen, M.; Xu, M.; Guo, S. Δfur mutant as a potential live attenuated vaccine (LAV) candidate protects American eels (Anguilla rostrata) from Vibrio harveyi infection. Microb. Pathog. 2024, 189, 106591. [Google Scholar] [CrossRef]
- Choi, G.; Choi, S.H. Complex regulatory networks of virulence factors in Vibrio vulnificus. Trends Microbiol. 2022, 30, 1205–1216. [Google Scholar] [CrossRef]
- Li, G.; Wang, M.Y. The role of Vibrio vulnificus virulence factors and regulators in its infection-induced sepsis. Folia Microbiol 2020, 65, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Ni, P.; Yang, Q.; Hu, H.; Wang, Q.; Ye, S.; Liu, Y. Delineating the key virulence factors and intraspecies divergence of Vibrio harveyi via whole-genome sequencing. Can. J. Microbiol. 2021, 67, 231–248. [Google Scholar] [CrossRef]
- Church, S.R.; Lux, T.; Baker-Austin, C.; Buddington, S.P.; Michell, S.L. Vibrio vulnificus type 6 secretion system 1 contains anti-bacterial properties. PLoS ONE 2016, 11, e0165500. [Google Scholar] [CrossRef]
- Ruwandeepika, H.A.; Karunasagar, I.; Bossier, P.; Defoirdt, T. Expression and quorum sensing regulation of type iii secretion system genes of Vibrio harveyi during infection of gnotobiotic brine shrimp. PLoS ONE 2015, 10, e0143935. [Google Scholar] [CrossRef]
- Guo, S.; Hu, L.; Feng, J.; Lin, P.; He, L.; Yan, Q. Immunogenicity of a bivalent protein as a vaccine against Edwardsiella anguillarum and Vibrio vulnificus in Japanese eel (Anguilla japonica). Microbiologyopen 2019, 8, e00766. [Google Scholar] [CrossRef]
- Goo, S.Y.; Lee, H.J.; Kim, W.H.; Han, K.L.; Park, D.K.; Lee, H.J. Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 2006, 74, 5586–5594. [Google Scholar] [CrossRef]
- Guo, S.L.; Lu, P.P.; Feng JianJun Zhao, J.P.; Lin, P.; Duan, L.H. A novel recombinant bivalent outer membrane protein of Vibrio vulnificus and Aeromonas hydrophila as a vaccine antigen of American eel (Anguilla rostrata). Fish Shellfish Immunol. 2015, 43, 477–484. [Google Scholar]
- Wang, Y. Comparative Analysis of Antibacterial-Related Gene Expressions in American Eel (Anguilla rostrata) Infected with Different Bacterial Pathogens. Master’s Thesis, JiMei University, Xiamen, China, 2023. [Google Scholar]
- Forlenza, A.E.; Galbraith, H.S.; Blakeslee, C.J.; Glazier, D.S. Ontogenetic Changes in Body Shape and the Scaling of Metabolic Rate in the American Eel (Anguilla rostrata). Physiol. Biochem. Zool. 2022, 95, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Nie, P.; Liu, Y. Detection of Dermocystidium anguillae in imported elvers of American eel Anguilla rostrata in China. Folia Parasitol. 2022, 69, 2022.013. [Google Scholar] [CrossRef]
- Guo, R.; Zhang, Z.; He, T.; Li, M.; Zhuo, Y.; Yang, X.; Fan, H.; Chen, X. Isolation and Identification of a New Isolate of Anguillid Herpesvirus 1 from Farmed American Eels (Anguilla rostrata) in China. Viruses 2022, 14, 2722. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, G.; Guo, S. Differential expressed genes (DEGs) and differential alternative splicing genes (DASs) revealed the common pathologic mechanism of three bacterial pathogens to American eels (Anguilla rostrata). Aquac. Rep. 2025, 40, 102632. [Google Scholar] [CrossRef]
- Wann, S.R.; Lo, H.R.; Chang, Y.T.; Liao, J.B.; Wen, Z.H.; Chi, P.L. P2X7 receptor blockade reduces pyroptotic inflammation and promotes phagocytosis in Vibrio vulnificus infection. J. Cell Physiol. 2023, 238, 2316–2334. [Google Scholar] [CrossRef]
- Yang, A.; Li, W.; Tao, Z.; Ye, H.; Xu, Z.; Li, Y.; Gao, Y.; Yan, X. Vibrio harveyi isolated from marine aquaculture species in eastern China and virulence to the large yellow croaker (Larimichthys crocea). J. Appl. Microbiol. 2021, 131, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Mohi, M.M.; Kuratani, M.; Miyazaki, T.; Yoshida, T. Histopathological studies on Vibrio harveyi-infected tiger puffer, Takifugu rubripes (Temminck et Schlegel), cultured in Japan. J. Fish Dis. 2010, 33, 833–840. [Google Scholar] [CrossRef]
- Kim, I.H.; Kim, I.J.; Wen, Y.; Park, N.Y.; Park, J.; Lee, K.W.; Koh, A.; Lee, J.H.; Koo, S.H.; Kim, K.S. Vibrio vulnificus secretes an insulin-degrading enzyme that promotes bacterial proliferation in vivo. J. Biol. Chem. 2015, 290, 18708–18720. [Google Scholar] [CrossRef]
- Chen, C.L.; Chien, S.C.; Leu, T.H.; Harn, H.I.; Tang, M.J.; Hor, L.I. Vibrio vulnificus MARTX cytotoxin causes inactivation of phagocytosis-related signaling molecules in macrophages. J. Biomed. Sci. 2017, 24, 58. [Google Scholar] [CrossRef]
- Liu, K.D.; Altmann, C.; Smits, G.; Krawczeski, C.D.; Edelstein, C.L.; Devarajan, P.; Faubel, S. Serum interleukin-6 and interleukin-8 are early biomarkers of acute kidney injury and predict prolonged mechanical ventilation in children undergoing cardiac surgery: A case-control study. Crit. Care 2009, 13, R104. [Google Scholar] [CrossRef] [PubMed]
- Peerapornratana, S.; Manrique-Caballero, C.L.; Gómez, H.; Kellum, J.A. Acute kidney injury from sepsis: Current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int. 2019, 96, 1083–1099. [Google Scholar] [CrossRef]
- Senturk, S.; Ulusoy, S.; Bosgelmez-Tinaz, G.; Yagci, A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J. Infect. Dev. Ctries 2012, 6, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Yang, C. Renal tubular epithelial cells: The neglected mediator of tubulointerstitial fibrosis after injury. Cell Death Dis. 2018, 9, 1126. [Google Scholar] [CrossRef] [PubMed]
- Zarbock, A.; Gomez, H.; Kellum, J.A. Sepsis-induced acute kidney injury revisited: Pathophysiology, prevention and future therapies. Curr. Opin. Crit. Care 2014, 20, 588–595. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Y.; Li, L.; Liu, S.; Wang, C.; Yuan, Y.; Yang, G.; Chen, Y.; Cheng, J.; Lu, Y.; et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics 2021, 11, 1845–1863. [Google Scholar] [CrossRef]
- Pefanis, A.; Ierino, F.L.; Murphy, J.M.; Cowan, P.J. Regulated necrosis in kidney ischemia-reperfusion injury. Kidney Int. 2019, 96, 291–301. [Google Scholar] [CrossRef]
- Qu, L.; Jiao, B. The interplay between immune and metabolic pathways in kidney disease. Cells 2023, 12, 1584. [Google Scholar] [CrossRef]
- Rosche, K.L.; Aljasham, A.T.; Kipfer, J.N.; Piatkowski, B.T.; Konjufca, V. Infection with Salmonella enterica serovar typhimurium leads to increased proportions of F4/80+ Red pulp macrophages and decreased proportions of B and T lymphocytes in the spleen. PLoS ONE 2015, 10, e0130092. [Google Scholar] [CrossRef] [PubMed]
- Swirski, F.K.; Nahrendorf, M.; Etzrodt, M.; Wildgruber, M.; Cortez-Retamozo, V.; Panizzi, P.; Figueiredo, J.L.; Kohler, R.H.; Chudnovskiy, A.; Waterman, P.; et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 2009, 325, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Vallelian, F.; Buzzi, R.M.; Pfefferlé, M.; Yalamanoglu, A.; Dubach, I.L.; Wassmer, A.; Wassmer, A.; Gentinetta, T.; Hansen, K.; Humar, R.; et al. Heme-stress activated NRF2 skews fate trajectories of bone marrow cells from dendritic cells towards red pulp-like macrophages in hemolytic anemia. Cell Death Differ. 2022, 29, 1450–1465. [Google Scholar] [CrossRef]
- Peeralil, S.; Joseph, T.C.; Murugadas, V.; Akhilnath, P.G.; Sreejith, V.N.; Lalitha, K.V. Vibrio harveyi virulence gene expression in vitro and in vivo during infection in black tiger shrimp Penaeus monodon. Dis. Aquat. Organ. 2020, 139, 153–160. [Google Scholar] [CrossRef]
- Chen, M.; Xu, M.; Chen, Z.; Wan, Q.; Guo, S. Protective effects of OmpA exposure via Bacillus subtilis spore bathing on Japanese eel (Anguilla japonica) against Edwardsiella anguillarum infection. Aquaculture 2024, 586, 740771. [Google Scholar] [CrossRef]
- Luo, J.; Lu, W.; Chen, Y.; Li, G.; Feng, J.; Huang, Y.; Yu, Y.; Cai, S.; Jian, J.; Yang, S.; et al. SQSTM1/p62 from Litopenaeus vannamei is involved in the immune response to Vibrio infection. Fish Shellfish Immunol. 2025, 158, 110161. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, Y.; Feng, J.; Hu, J.; Chen, H.; Guo, Z.; Gao, R.; Su, Y. ToxR modulates biofilm formation in fish pathogen Vibrio harveyi. Lett. Appl. Microbiol. 2022, 74, 288–299. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish, 5th ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Somboonwiwat, K.; Bachère, E.; Rimphanitchayakit, V.; Tassanakajon, A. Localization of anti-lipopolysaccharide factor (ALFPm3) in tissues of the black tiger shrimp, Penaeus monodon, and characterization of its binding properties. Dev. Comp. Immunol. 2008, 32, 1170–1176. [Google Scholar] [CrossRef]
- Cho, C.; Choi, S.; Kim, M.H.; Kim, B.S. Vibrio vulnificus PlpA facilitates necrotic host cell death induced by the pore forming MARTX toxin. J. Microbiol. 2022, 60, 224–233. [Google Scholar] [CrossRef]
- Ishida, K.; Shimohata, T.; Kanda, Y.; Nguyen, A.Q.; Masuda, R.; Yamazaki, K.; Uebanso, T.; Mawatari, K.; Kashimoto, T.; Takahashi, A. Characteristic metabolic changes in skeletal muscle due to Vibrio vulnificus infection in a wound infection model. mSystems 2023, 8, e0068222. [Google Scholar] [CrossRef]
- Lee, Y.; Roh, H.; Kim, A.; Park, J.; Lee, J.Y.; Kim, Y.J.; Kang, Y.R.; Kang, H.; Kim, S.; Kim, H.S.; et al. Molecular mechanisms underlying the vulnerability of Pacific abalone (Haliotis discus hannai) to Vibrio harveyi infection at higher water temperature. Fish Shellfish Immunol. 2023, 138, 108844. [Google Scholar] [CrossRef]
- Chen, Q.; Ma, B.; Xu, M.; Xu, H.; Yan, Z.; Wang, F.; Wang, Y.; Huang, Z.; Yin, S.; Zhao, Y.; et al. Comparative proteomics study of exosomes in Vibrio harveyi and Vibrio anguillarum. Microb. Pathog. 2023, 181, 106174. [Google Scholar] [CrossRef]
- Zhang, X.H.; He, X.; Austin, B. Vibrio harveyi: A serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wan, Q.; Xu, M.; Chen, M.; Chen, Z. Transcriptome analysis of host anti-Aeromonas hydrophila infection revealed the pathogenicity of A. hydrophila to American eels (Anguilla rostrata). Fish Shellfish Immunol. 2024, 148, 109504. [Google Scholar] [CrossRef]
- Lin, P.; Xu, M.; Yang, Q.; Chen, M.; Guo, S. Inoculation of Freund’s adjuvant in European eel (Anguilla anguilla) revealed key KEGG pathways and DEGs of host anti-Edwardsiella anguillarum infection. Fish Shellfish Immunol. 2023, 136, 108708. [Google Scholar] [CrossRef]
- He, W.; Wu, L.; Li, S.; Guo, S. Transcriptome RNA-seq revealed lncRNAs activated by Edwardsiella anguillarum post the immunization of OmpA protecting European eel (Anguilla anguilla) from being infected. Fish Shellfish Immunol. 2021, 118, 51–65. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, L.; He, L.; Tang, Y.; Guo, S.; Zhai, S. Transcriptomic analysis using dual RNA sequencing revealed a Pathogen-Host interaction after Edwardsiella anguillarum infection in European eel (Anguilla anguilla). Fish Shellfish Immunol. 2022, 120, 745–757. [Google Scholar] [CrossRef]
- Weber, R.A.; Yen, F.S.; Nicholson, S.P.V.; Alwaseem, H.; Bayraktar, E.C.; Alam, M.; Timson, R.C.; La, K.; Abu-Remaileh, M.; Molina, H.; et al. Maintaining iron homeostasis is the key role of lysosomal acidity for cell proliferation. Mol. Cell 2020, 77, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Ciaccio, C.; Coletta, A.; Coletta, M. Role of hemoglobin structural-functional relationships in oxygen transport. Mol. Aspects Med. 2022, 84, 101022. [Google Scholar] [CrossRef]
- Alibayov, B.; Scasny, A.; Khan, F.; Creel, A.; Smith, P.; Vidal, A.G.J.; Fitisemanu, F.M.; Padilla-Benavides, T.; Weiser, J.N.; Vidal, J.E. Oxidative reactions catalyzed by hydrogen peroxide produced by Streptococcus pneumoniae and other Streptococci cause the release and degradation of heme from hemoglobin. Infect. Immun. 2022, 90, e0047122. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yin, Z.; Zheng, Z.; Tang, Y.; Guo, S. Comprehensive relationship analysis of the Long Noncoding RNAs (lncRNAs) and the target mRNAs in response to the infection of Edwardsiella anguillarum in European eel (Anguilla anguilla) inoculated with Freund’s Adjuvant. Mar. Biotechnol. 2022, 24, 956–968. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Qiu, M.; Li, J.; Xiao, Y.; Lin, H.; Zheng, W.; Zhu, J.; Chen, N. Transcriptomic profiling reveals different innate immune responses in primary alveolar macrophages infected by two highly homologous porcine reproductive and respiratory syndrome viruses with distinct virulence. Microb. Pathog. 2021, 158, 105102. [Google Scholar] [CrossRef]
- Zhu, G.; Gao, C.; Wu, C.; Li, M.; Xu, J.R.; Liu, H.; Wang, Q. Comparative transcriptome analysis reveals distinct gene expression profiles in Brachypodium distachyon infected by two fungal pathogens. BMC Plant Biol. 2021, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar, J.; Sharma, S.; Hussain, M.A.; Shelly, A.; Das, B.; Yadav, A.K.; Mazumder, S. TLR22-mediated activation of TNF-α-caspase-1/IL-1β inflammatory axis leads to apoptosis of Aeromonas hydrophila-infected macrophages. Mol. Immunol. 2021, 137, 114–123. [Google Scholar] [CrossRef]
- Michaux, C.; Hansen, E.E.; Jenniches, L.; Gerovac, M.; Barquist, L.; Vogel, J. Single-Nucleotide RNA maps for the two major nosocomial pathogens Enterococcus faecalis and Enterococcus faecium. Front. Cell. Infect. Microbiol. 2020, 10, 600325. [Google Scholar] [CrossRef]
- Li, H.; Sun, Y.; Sun, L. A teleost CXCL10 Is both an immunoregulator and an antimicrobial. Front. Immunol. 2022, 13, 917697. [Google Scholar] [CrossRef]
- Feng, F.; Xu, W.; Lian, C.; Wang, L.; Wang, Z.; Chen, H.; Wang, X.; Wang, H.; Zhang, J. Tuberculosis to lung cancer: Application of tuberculosis signatures in identification of lung adenocarcinoma subtypes and marker screening. J. Cancer 2024, 15, 5329–5350. [Google Scholar] [CrossRef]
- Druszczyńska, M.; Godkowicz, M.; Kulesza, J.; Wawrocki, S.; Fol, M. Cytokine receptors-regulators of antimycobacterial immune response. Int. J. Mol. Sci. 2022, 23, 1112. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Chen, M.; Li, W.; Wan, Q.; Xu, M. Analysis of alternative splicing and long noncoding rnas after the Edwardsiella anguillarum infected the immunized European Eels (Anguilla anguilla) revealed the role of outer membrane protein A in OmpA subunit vaccine. Mar. Biotechnol. 2023, 25, 372–387. [Google Scholar] [CrossRef]
- Park, J.W.; Tokheim, C.; Shen, S.; Xing, Y. Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol. Biol. 2013, 1038, 171–179. [Google Scholar]
- Sieber, P.; Voigt, K.; Kämmer, P.; Brunke, S.; Schuster, S.; Linde, J. Comparative study on alternative splicing in human fungal pathogens suggests its involvement during host invasion. Front. Microbiol. 2018, 9, 2313. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, K.; Zhou, X.; Tang, X.; Yu, X.; Yao, W.; Wu, Z. Histopathology, antioxidant responses, transcriptome and gene expression analysis in triangle sail mussel Hyriopsis cumingii after bacterial infection. Dev. Comp. Immunol. 2021, 124, 104175. [Google Scholar] [CrossRef]
- He, S.; Valkov, E.; Cheloufi, S.; Murn, J. The nexus between RNA-binding proteins and their effectors. Nat. Rev. Genet. 2023, 24, 276–294. [Google Scholar] [CrossRef]
- Gray, L.G.; Mills, J.D.; Curry-Hyde, A.; Devore, S.; Friedman, D.; Thom, M.; Scott, C.; Thijs, R.D.; Aronica, E.; Devinsky, O.; et al. Identification of specific circular rna expression patterns and microRNA interaction networks in mesial temporal lobe epilepsy. Front. Genet. 2020, 11, 564301. [Google Scholar] [CrossRef]
- Jagadish, A.; Dubey, H.; Kamatchi, I.; Pradeep, A.R.; Subrahmanyam, G.; Mishra, R.K.; Ponnuvel, K.M. Transcriptome analysis of Nosema assamensis infecting muga silkworms (Antheraea assamensis) reveals insights into candidate pathogenicity related genes and molecular pathways required for pathogenesis. Ann. Parasitol. 2021, 67, 671–682. [Google Scholar]
- Wang, T.; Du, X.; Ji, L.; Han, Y.; Dang, J.; Wen, J.; Wang, Y.; Pu, Q.; Wu, M.; Liang, H. Pseudomonas aeruginosa T6SS-mediated molybdate transport contributes to bacterial competition during anaerobiosis. Cell Rep. 2021, 35, 108957. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, L.; Liu, Y.; Huang, T.; Liang, W.; Chen, M. Multiomics analyses reveal that NOD-like signaling pathway plays an important role against Streptococcus agalactiae in the spleen of tilapia. Fish Shellfish Immunol. 2019, 95, 336–348. [Google Scholar] [CrossRef]
- Black, C.S.; Whelan, T.A.; Garside, E.L.; MacMillan, A.M.; Fast, N.M.; Rader, S.D. Spliceosome assembly and regulation: Insights from analysis of highly reduced spliceosomes. RNA 2023, 29, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Björkman, K.; Sofou, K.; Darin, N.; Holme, E.; Kollberg, G.; Asin-Cayuela, J.; Holmberg Dahle, K.M.; Oldfors, A.; Moslemi, A.R.; Tulinius, M. Broad phenotypic variability in patients with complex I deficiency due to mutations in NDUFS1 and NDUFV1. Mitochondrion 2015, 21, 33–40. [Google Scholar] [CrossRef]
- He, L.; Wu, L.; Tang, Y.; Lin, P.; Zhai, S.; Xiao, Y.; Guo, S. Immunization of a novel outer membrane protein from Aeromonas hydrophila simultaneously resisting A. hydrophila and Edwardsiella anguillarum infection in European eels (Anguilla anguilla). Fish Shellfish Immunol. 2020, 97, 300–312. [Google Scholar] [CrossRef]
- Chen, X.; Petranovic, D. Role of frameshift ubiquitin B protein in Alzheimer’s disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 300–313. [Google Scholar] [CrossRef]
- Javanmard, D.; Najafi, M.; Babaei, M.R.; Karbalaie Niya, M.H.; Esghaei, M.; Panahi, M.; Safarnezhad Tameshkel, F.; Tavakoli, A.; Jazayeri, S.M.; Ghaffari, H.; et al. Investigation of CTNNB1 gene mutations and expression in hepatocellular carcinoma and cirrhosis in association with hepatitis B virus infection. Infect. Agent. Cancer 2020, 15, 37. [Google Scholar] [CrossRef]
- Meng, Y.; Kong, K.W.; Chang, Y.Q.; Deng, X.M.; Yang, T. Histone methyltransferase SETD2 inhibits M1 macrophage polarization and glycolysis by suppressing HIF-1α in sepsis-induced acute lung injury. Med. Microbiol. Immunol. 2023, 212, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]






| Samples | Raw Reads | Clean Reads | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| Con_0_1 | 50,874,554 | 50,146,920 | 0.0265 | 97.38 | 92.91 | 50.60 |
| Con_0_2 | 45,950,964 | 45,229,786 | 0.0267 | 97.31 | 92.74 | 50.90 |
| Con_0_3 | 47,524,136 | 46,814,670 | 0.0266 | 97.36 | 92.80 | 50.59 |
| Vv_36_1 | 71,776,048 | 69,105,472 | 0.0243 | 98.21 | 94.99 | 52.19 |
| Vv_36_2 | 62,302,708 | 60,657,518 | 0.0242 | 98.28 | 95.14 | 52.04 |
| Vv_36_3 | 62,143,894 | 60,086,848 | 0.0242 | 98.27 | 95.17 | 52.36 |
| Vh_36_1 | 46,999,710 | 46,238,298 | 0.0268 | 97.26 | 92.60 | 51.83 |
| Vh_36_2 | 48,078,512 | 47,406,682 | 0.0263 | 97.47 | 93.08 | 51.52 |
| Vh_36_3 | 44,869,076 | 44,249,230 | 0.0263 | 97.46 | 93.08 | 50.96 |
| Vv_60_1 | 66,197,906 | 63,080,504 | 0.0242 | 98.24 | 95.12 | 52.29 |
| Vv_60_2 | 63,084,782 | 60,768,442 | 0.0244 | 98.19 | 95.01 | 52.01 |
| Vv_60_3 | 63,182,062 | 61,359,548 | 0.0242 | 98.24 | 95.10 | 52.41 |
| Vh_60_1 | 43,751,960 | 43,172,338 | 0.0262 | 97.48 | 93.12 | 51.02 |
| Vh_60_2 | 46,005,832 | 45,378,744 | 0.0262 | 97.48 | 93.13 | 51.31 |
| Vh_60_3 | 44,664,684 | 44,013,716 | 0.0267 | 97.30 | 92.69 | 50.66 |
| Sample | Total Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|
| Con_0_1 | 50,146,920 | 41,792,062 (83.34%) | 1,493,400 (2.98%) | 40,298,662 (80.36%) |
| Con_0_2 | 45,229,786 | 37,877,970 (83.75%) | 1,262,581 (2.79%) | 36,615,389 (80.95%) |
| Con_0_3 | 46,814,670 | 38,945,320 (83.19%) | 1,355,266 (2.89%) | 37,590,054 (80.30%) |
| Vv_36_1 | 69,105,472 | 60,326,219 (87.30%) | 4,028,723 (5.83%) | 56,297,496 (81.47%) |
| Vv_36_2 | 60,657,518 | 52,353,692 (86.31%) | 2,953,420 (4.87%) | 49,400,272 (81.44%) |
| Vv_36_3 | 60,086,848 | 51,726,410 (86.09%) | 2,902,755 (4.83%) | 48,823,655 (81.26%) |
| Vh_36_1 | 46,238,298 | 39,785,883 (86.05%) | 1,453,818 (3.14%) | 38,332,065 (82.9%) |
| Vh_36_2 | 47,406,682 | 40,947,894 (86.38%) | 1,359,959 (2.87%) | 39,587,935 (83.51%) |
| Vh_36_3 | 44,249,230 | 37,733,480 (85.27%) | 1,393,405 (3.15%) | 36,340,075 (82.13%) |
| Vv_60_1 | 63,080,504 | 53,992,761 (85.59%) | 2,146,641 (3.4%) | 51,846,120 (82.19%) |
| Vv_60_2 | 60,768,442 | 51,576,117 (84.87%) | 1,862,310 (3.06%) | 49,713,807 (81.81%) |
| Vv_60_3 | 61,359,548 | 52,756,353 (85.98%) | 2,003,358 (3.26%) | 50,752,995 (82.71%) |
| Vh_60_1 | 43,172,338 | 37,080,541 (85.89%) | 1,204,051 (2.79%) | 35,876,490 (83.1%) |
| Vh_60_2 | 45,378,744 | 38,760,833 (85.42%) | 1,151,811 (2.54%) | 37,609,022 (82.88%) |
| Vh_60_3 | 44,013,716 | 36,931,235 (83.91%) | 1,275,855 (2.9%) | 35,655,380 (81.01%) |
| Gene Name | Gene Description | Pathways | Log2FC |
|---|---|---|---|
| loc118227146 | permeability factor 2-like | 7 | 2.99 |
| loc118229166 | proto-oncogene c-Fos-like | 7 | 1.78 |
| loc118223964 | C-X-C motif chemokine 10-like | 8 | 1.75 |
| loc118221018 | RAC-gamma serine/threonine-protein kinase-like | 9 | 1.38 |
| loc118207102 | interleukin-1 beta-like, transcript variant X2 | 10 | 3.31 |
| loc118234251 | interleukin-6-like | 10 | 3.42 |
| loc118228785 | NF-kappa-B inhibitor alpha-like | 10 | 1.04 |
| nfkbiab | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b | 10 | 1.36 |
| loc118237077 | transcription factor AP-1-like, transcript variant X1 | 10 | 1.79 |
| june | JunE proto-oncogene, AP-1 transcription factor subunit | 10 | 2.97 |
| tnfa | tumor necrosis factor a | 11 | 3.51 |
| tnfb | tumor necrosis factor b | 11 | 2.84 |
| Chromosome Number | Total AS Events | Total AS Genes | Average AS Events/Genes | Total DAS Events | Total DAS Genes | Average DAS Events/Genes |
|---|---|---|---|---|---|---|
| chrNC_049201 | 3506, 3370 | 1152, 1124 | 3.04, 3.00 | 229, 175 | 144, 127 | 1.59, 1.38 |
| chrNC_049202 | 3146, 3077 | 1005, 1027 | 3.13, 3.00 | 205, 144 | 155, 110 | 1.32, 1.31 |
| chrNC_049203 | 2798, 2575 | 876, 856 | 3.19, 3.01 | 198, 127 | 131, 82 | 1.51, 1.55 |
| chrNC_049204 | 2832, 2738 | 857, 849 | 3.30, 3.22 | 172, 126 | 117, 88 | 1.47, 1.43 |
| chrNC_049205 | 1999, 2015 | 681, 671 | 2.94, 3.00 | 135, 88 | 90, 59 | 1.50, 1.49 |
| chrNC_049206 | 2418, 2187 | 788, 745 | 3.07, 2.94 | 134, 77 | 96, 58 | 1.40, 1.33 |
| chrNC_049207 | 1917, 1890 | 646, 647 | 2.97, 2.92 | 107, 67 | 81, 54 | 1.32, 1.24 |
| chrNC_049208 | 2294, 2181 | 775, 756 | 2.96, 2.88 | 138, 103 | 110, 75 | 1.25, 1.37 |
| chrNC_049209 | 2346, 2242 | 745, 730 | 3.15, 3.07 | 136, 107 | 98, 77 | 1.39, 1.39 |
| chrNC_049210 | 1744, 1740 | 630, 639 | 2.77, 2.72 | 91, 73 | 74, 59 | 1.23, 1.24 |
| chrNC_049211 | 1957, 1862 | 618, 610 | 3.17, 3.05 | 114, 87 | 71, 67 | 1.61, 1.3 |
| chrNC_049212 | 1521, 1502 | 491, 491 | 3.10, 3.06 | 63, 53 | 44, 41 | 1.43, 1.29 |
| chrNC_049213 | 1827, 1855 | 578, 582 | 3.16, 3.19 | 115, 64 | 70, 44 | 1.64, 1.45 |
| chrNC_049214 | 1954, 1864 | 554, 531 | 3.53, 3.51 | 131, 104 | 78, 66 | 1.68, 1.58 |
| chrNC_049215 | 1312, 1372 | 427, 438 | 3.07, 3.13 | 73, 71 | 48, 44 | 1.52, 1.61 |
| chrNC_049216 | 1309, 1208 | 439, 420 | 2.98, 2.88 | 65, 48 | 48, 39 | 1.35, 1.23 |
| chrNC_049217 | 1777, 1779 | 545, 529 | 3.26, 3.36 | 105, 97 | 60, 60 | 1.75, 1.62 |
| chrNC_049218 | 1058, 1015 | 379, 369 | 2.79, 2.75 | 59, 44 | 42, 37 | 1.40, 1.19 |
| chrNC_049219 | 917, 974 | 296, 302 | 3.10, 3.23 | 47, 42 | 30, 30 | 1.57, 1.4 |
| total | 38,632, 37,446 | 12482, 12316 | 3.10, 3.04 | 2317, 1697 | 1579, 1214 | 1.46, 1.39 |
| Transcript_id | Gene Name | Gene Description | Primers (5′-3′) |
|---|---|---|---|
| rna-XM_035397752.1 | loc118216518 | β-actin(housekeeping) | F-AATCCACGAGACCACCTTCAA R-GTTGGCGTACAGGTCCTTACG |
| rna-XM_035421149.1 | ccl34a.3 | chemokine (C-C motif) ligand 34a, duplicate 3 | F: CTGCTGCAAAGAGGTCTCCA R: GCTGCATTTCTCCCCCTCTT |
| rna-XM_035406820.1 | ccr7 | chemokine (C-C motif) receptor 7 | F: ACCTGCTGGTGATGCTAACC R: AACACCCACTTGTCAAGGCA |
| rna-XM_035410631.1 | cd74a | CD74 molecule, major histocompatibility complex | F: TTCCTGGTGTGCATGCTCAT R: TTCACGAGCGGCTTCTTCTT |
| rna-XM_035435945.1 | ier3 | immediate early response 3 | F: TTTCGAGCAGGTTCCTGTCC R: ATCTGCAGGAACACCACGAG |
| rna-XM_035391350.1 | il7r | interleukin 7 receptor | F: CAAGTGCTGAAAACACCGGG R: CAGAAATGAGTCGGCAGGGT |
| rna-XM_035394107.1 | marco | macrophage receptor with collagenous structure | F: GAGGCACCGGAATAGTCGAG R: TGGTCCTGAGCCCATATGGA |
| rna-XM_035388377.1 | tktb | transketolase b | F: GCAGACCCAGACCTTTTCGA R: TGCGTCGATGTCAAAGGTCA |
| rna-XM_035387337.1 | loc118210870 | tumor necrosis factor receptor superfamily member 9-like | F: CACACCTGCCCCAAAACATG R: CCAAGAGCCGTACACCACTT |
| rna-XM_035389795.1 | loc118212156 | lysozyme C-like | F: GCATTTGGTGGCTTGTGGAC R: CTTCTCCAGCTTGCCCTCTC |
| rna-XM_035398056.1 | loc118216669 | ferritin, middle subunit-like | F: GGACTCTCACTTCTGCCACC R: CCACCGGCAGATCGTTGATA |
| rna-XM_035425000.1 | loc118231298 | H-2 class II histocompatibility antigen, A-U alpha chain-like | F: GGTCTGTTCACCTGCTTCGA R: TGTCTCCCAGGGGCTGAATA |
| MSTRG.16029.33 | loc118231311 | H-2 class II histocompatibility antigen, E-S beta chain-like | F: GGTCTGTTCACCTGCTTCGA R: TGTCTCCCAGGGGCTGAATA |
| rna-XM_035431152.1 | loc118234548 | biotinidase-like, transcript variant X1 | F: TTCGCCTCCTACTACCAGCT R: TAGGCTTTCCCTTGAGCTGC |
| AS ID | Gene Name | Gene Description | Chromosome | Primers (5′-3′) |
|---|---|---|---|---|
| SE_4646 | loc118208085 | serine-aspartate repeat-containing protein F-like, transcript variant X7 | NC_049211.1 | Fi-ggcttctctggaggtggttt Ri-ctgaaaatgcatcagttaataagtc 136 bp Fs-caagcagcagcacaggaaaatgg Rs = Ri 62 bp |
| SE_10321 | tnfsf13 | TNF superfamily member 13, transcript variant X3 | NC_049209.1 | Fi-cccctctcctcacactcatacg Ri-agtaggaccctccagtcgac 126 bp Fs-cagctcggtaaacgatgaggaggac Rs = Ri 120 bp |
| SE_5690 | znf341 | zinc finger protein 341, transcript variant X4 | NC_049211.1 | Fi-tctactggaacaacagccgc Ri-cttcaaatattgcctgcgcca 104 bp Fs-actatacaacctatggggctaaatctacagag Rs = Ri 105 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Sun, G.; Hong, S.; Lin, Q.; Yang, J.; Guo, S. Comparative Transcriptomic Analysis Underlies the Differential Virulence of Vibrio harveyi and Vibrio vulnificus in American Eels (Anguilla rostrata). Int. J. Mol. Sci. 2025, 26, 11763. https://doi.org/10.3390/ijms262411763
Yang Q, Sun G, Hong S, Lin Q, Yang J, Guo S. Comparative Transcriptomic Analysis Underlies the Differential Virulence of Vibrio harveyi and Vibrio vulnificus in American Eels (Anguilla rostrata). International Journal of Molecular Sciences. 2025; 26(24):11763. https://doi.org/10.3390/ijms262411763
Chicago/Turabian StyleYang, Qiuhua, Guanghua Sun, Sijia Hong, Qi Lin, Jinjin Yang, and Songlin Guo. 2025. "Comparative Transcriptomic Analysis Underlies the Differential Virulence of Vibrio harveyi and Vibrio vulnificus in American Eels (Anguilla rostrata)" International Journal of Molecular Sciences 26, no. 24: 11763. https://doi.org/10.3390/ijms262411763
APA StyleYang, Q., Sun, G., Hong, S., Lin, Q., Yang, J., & Guo, S. (2025). Comparative Transcriptomic Analysis Underlies the Differential Virulence of Vibrio harveyi and Vibrio vulnificus in American Eels (Anguilla rostrata). International Journal of Molecular Sciences, 26(24), 11763. https://doi.org/10.3390/ijms262411763






