Genome-Wide Identification and Functional Divergence of the Chloride Channel (CLC) Gene Family in Autotetraploid Alfalfa (Medicago sativa L.)
Abstract
1. Introduction
2. Results
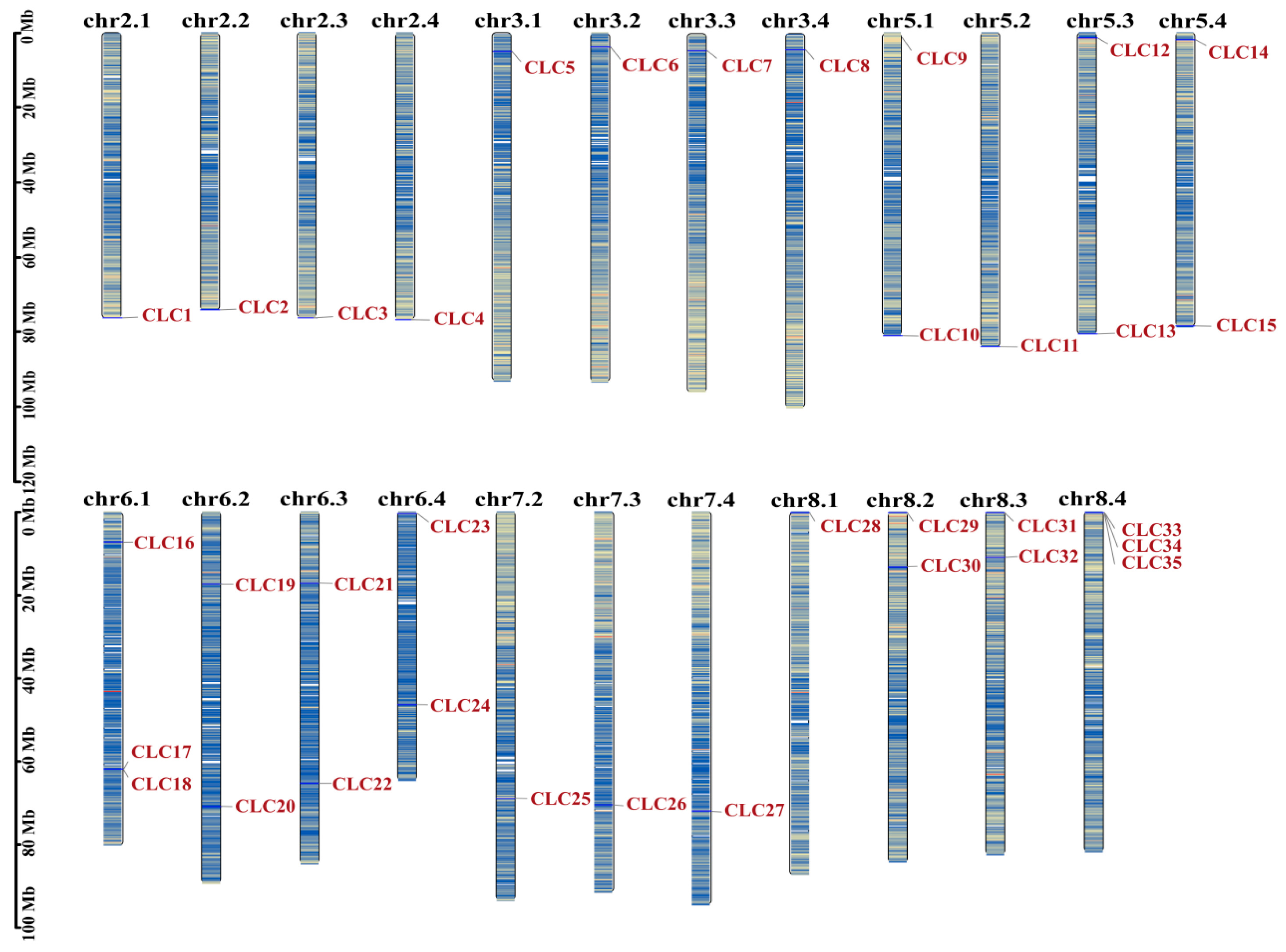
2.1. Identification and Classification of the MsCLCs Gene Family
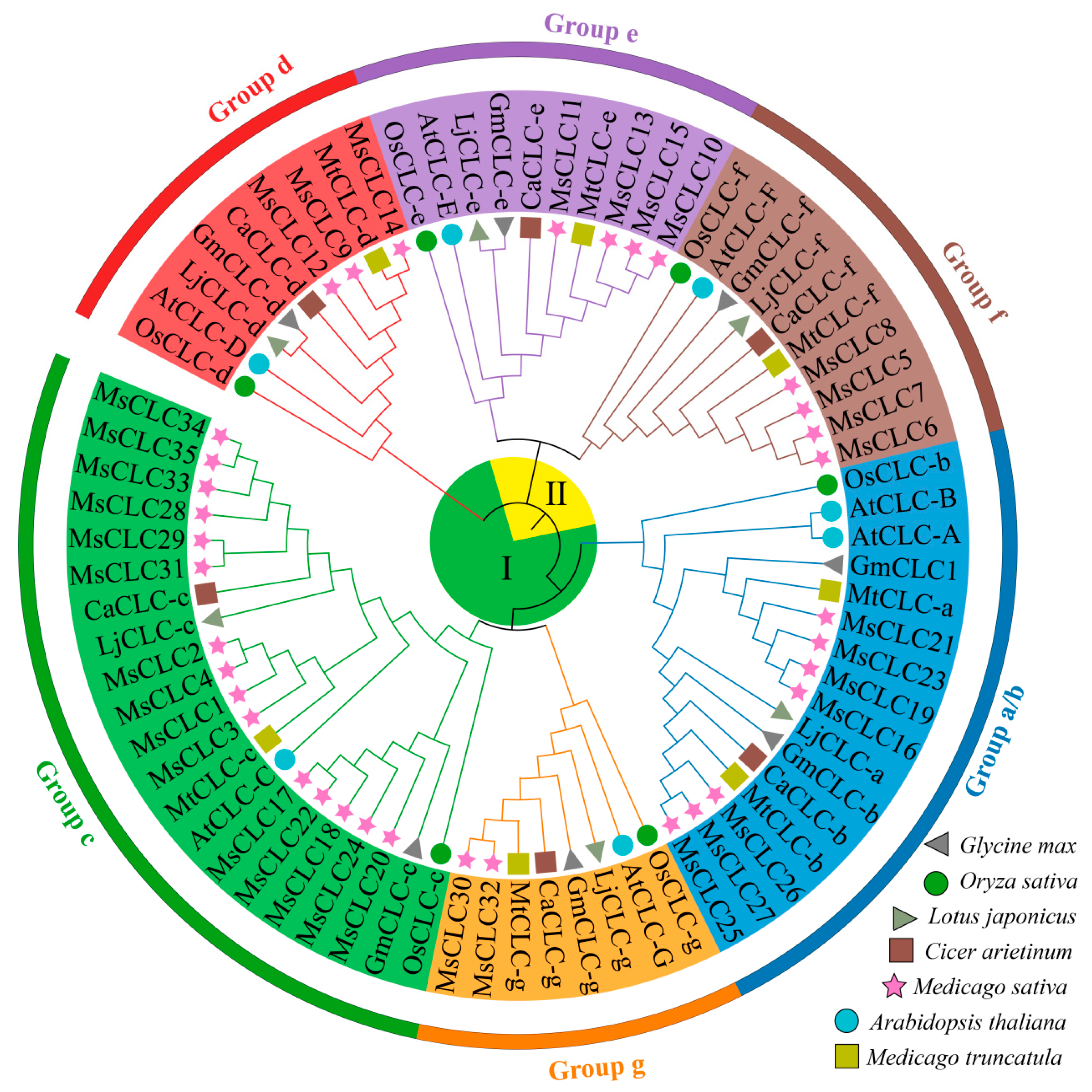
2.2. Phylogenetic Analysis and Multiple Sequence Alignment Analysis of MsCLC Proteins
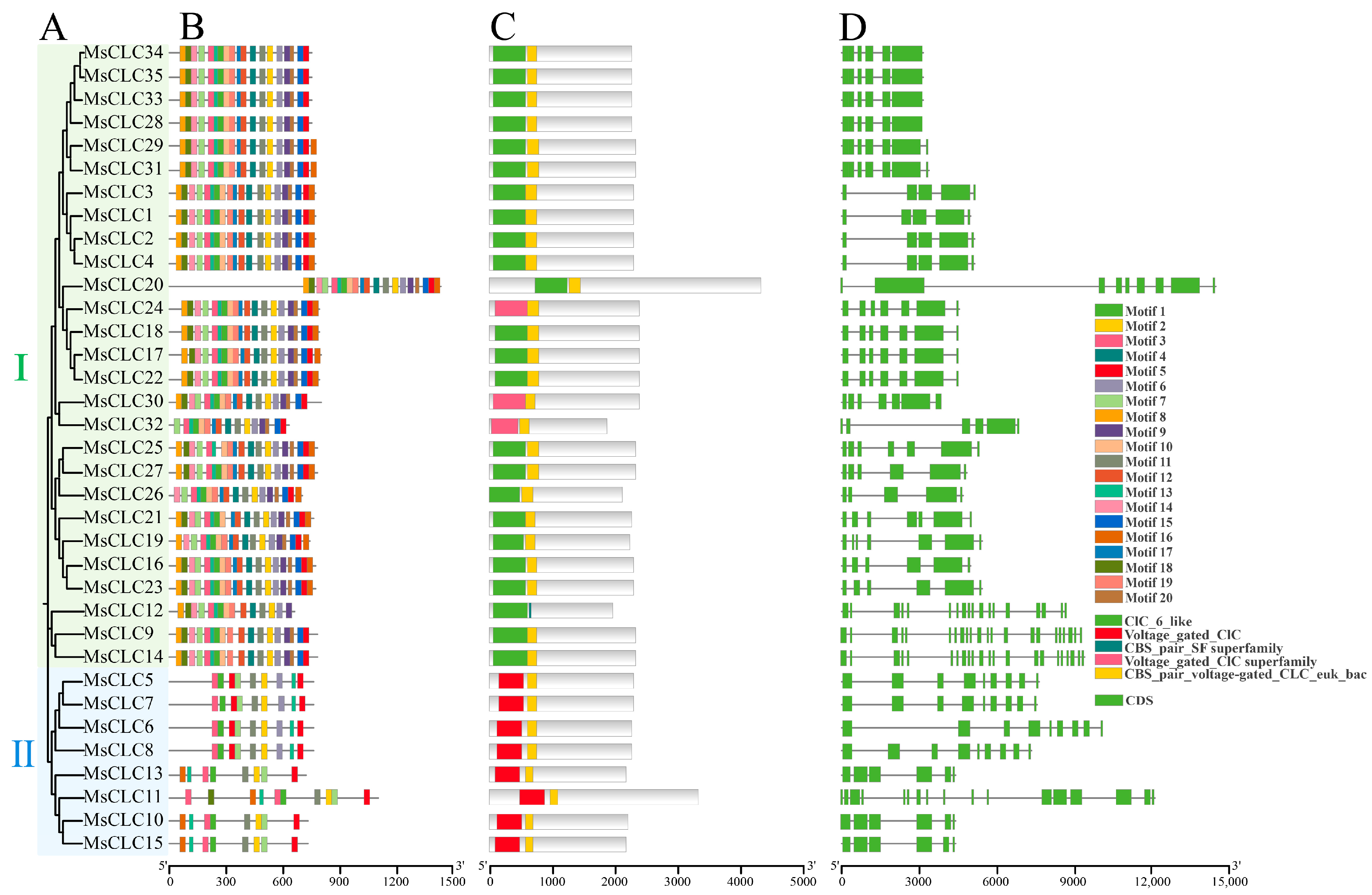
2.3. Conserved Motifs, Domains, and Gene Structures of MsCLC Genes Family
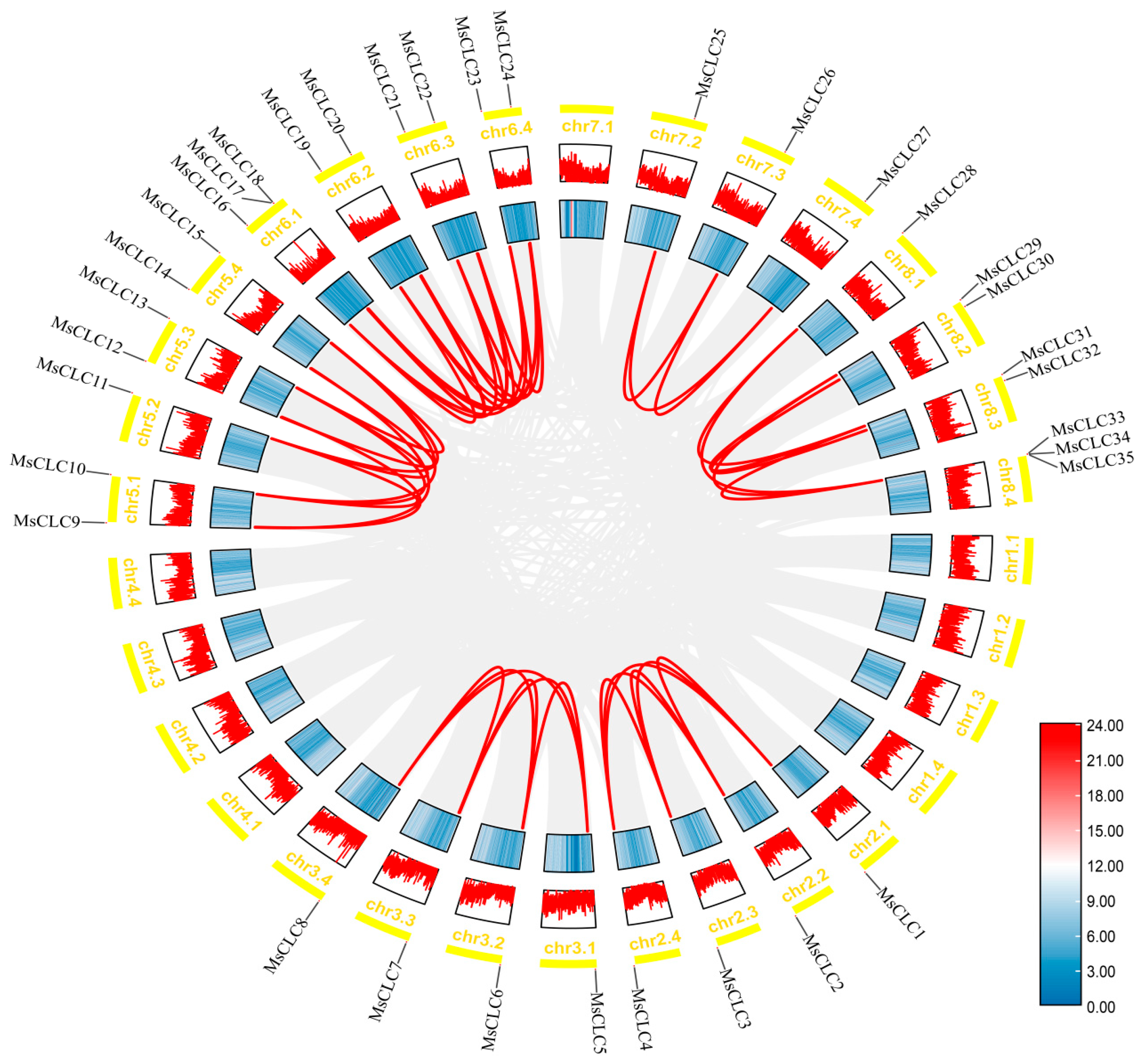
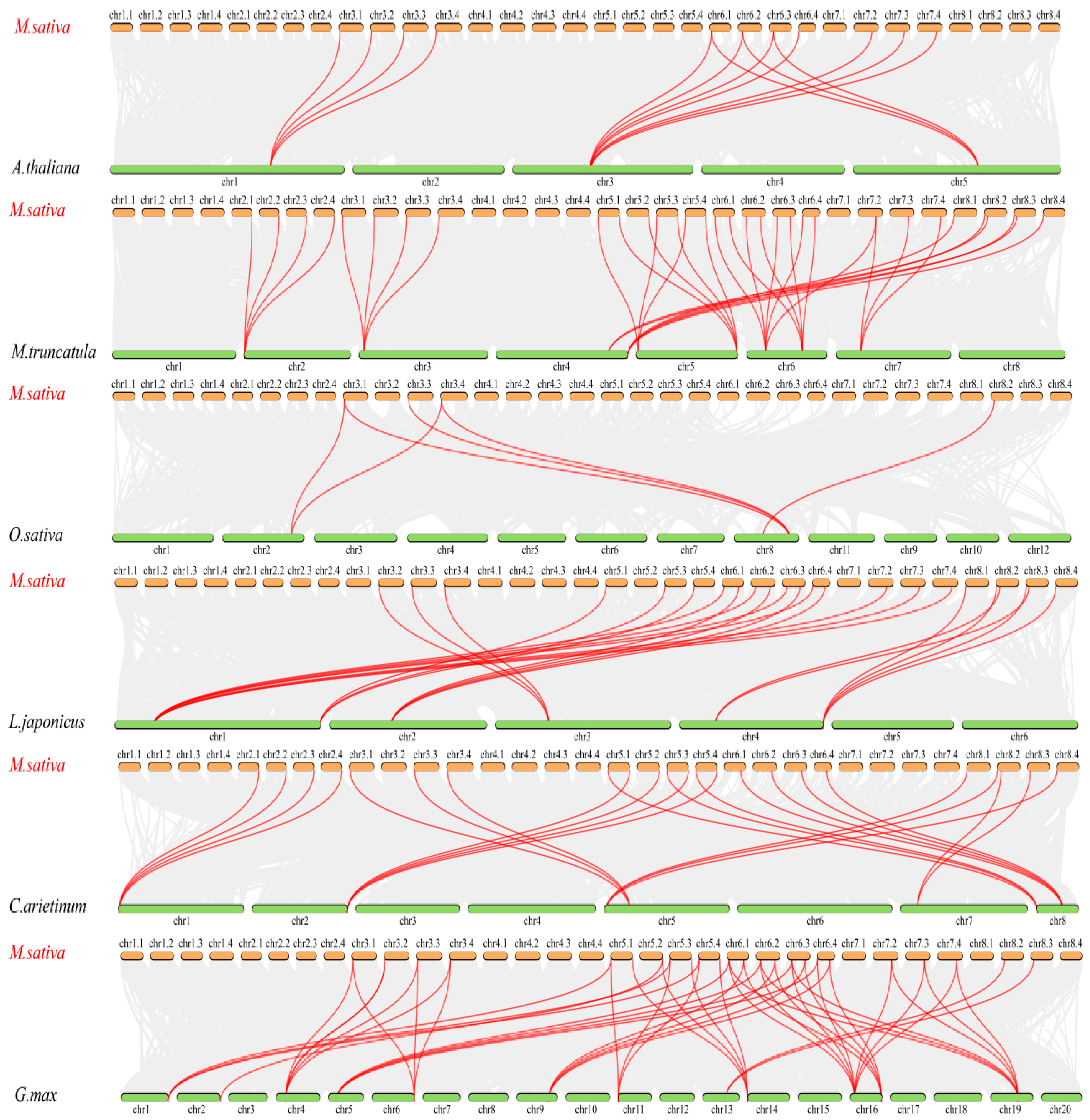
2.4. Gene Duplication Events and Colinearity Analysis of the MsCLCs Gene Family
2.5. Promoter Cis-Regulatory Elements Analysis of MsCLCs Gene Family
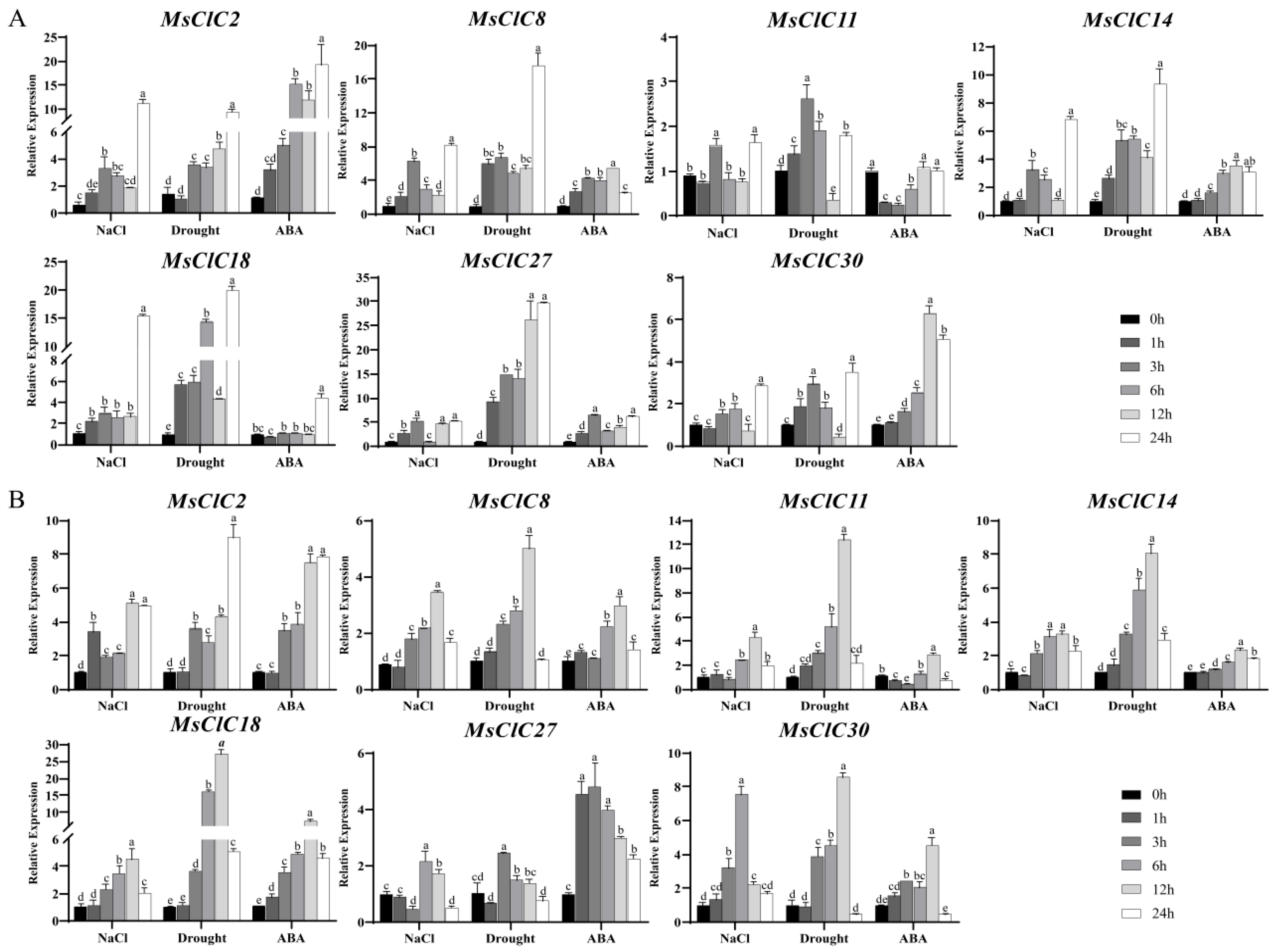
2.6. Analysis of the Expression Patterns of MsCLC Genes Under Different Abiotic Stress Treatments
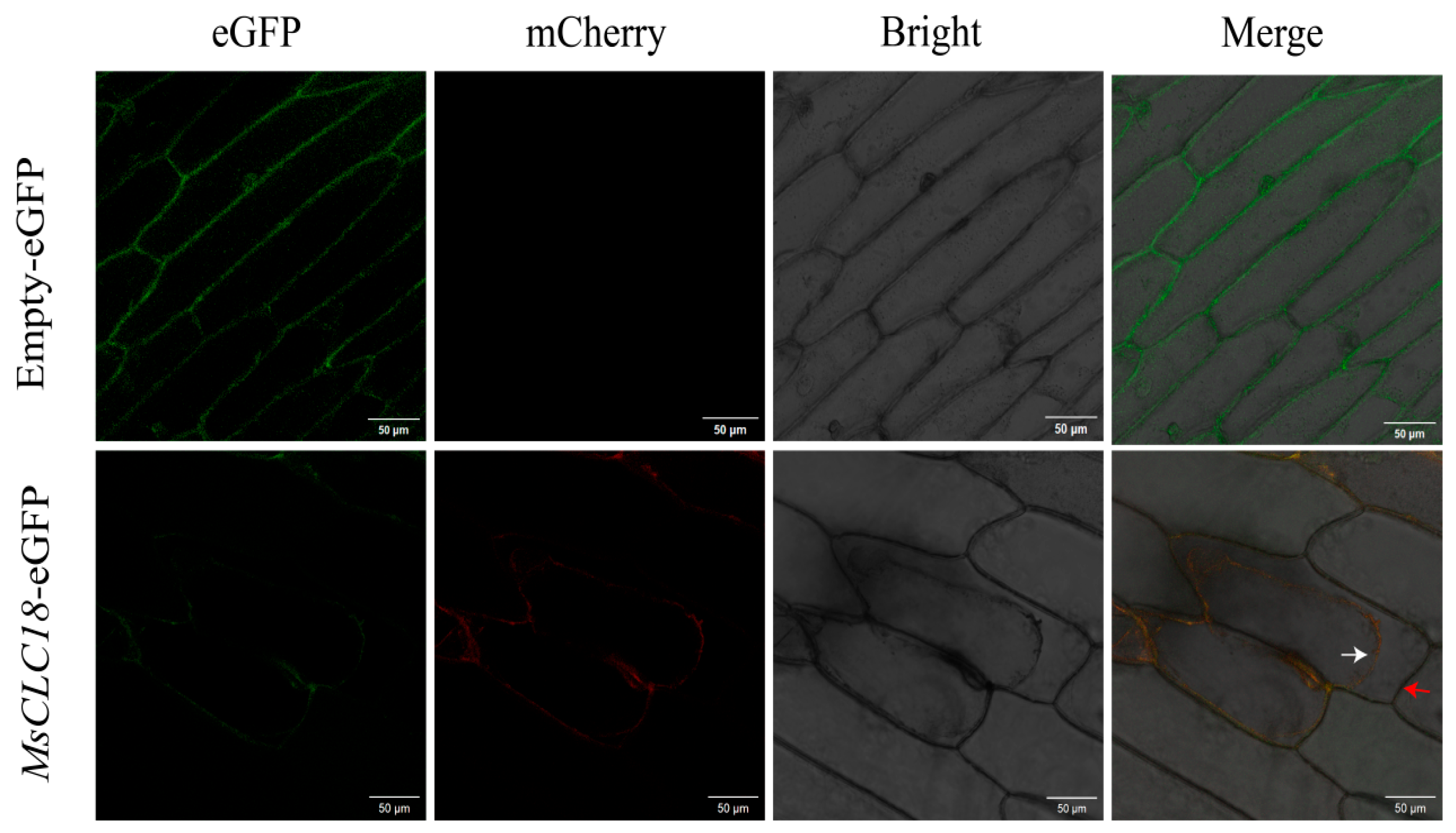
2.7. Subcellular Localization Analysis
3. Discussion
4. Materials and Methods
4.1. Identification and Classification of the MsCLCs Gene Family
4.2. Phylogenetic Analysis and Multiple Sequence Alignment Analysis of MsCLC Proteins
4.3. Conserved Motifs, Domains, and Gene Structures of MsCLCs Gene Family
4.4. Gene Duplication Events and Colinearity Analysis of the MsCLCs Gene Family
4.5. Promoter Cis-Regulatory Elements Analysis of MsCLCs Gene Family
4.6. Analysis of the Expression Patterns of MsCLC Genes Under Different Abiotic Stress Treatments
4.7. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, Y.; Tashpolat, N. Current Status and Development Trend of Soil Salinity Monitoring Research in China. Sustainability 2023, 15, 5874. [Google Scholar] [CrossRef]
- Almeida Machado, R.M.; Serralheiro, R.P. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Hossain, M. Present Scenario of Global Salt Affected Soils, its Management and Importance of Salinity Research. Int. Res. J. Biol. Sci. 2019, 1, 1–3. [Google Scholar]
- Otlewska, A.; Migliore, M.; Dybka-Stępień, K.; Manfredini, A.; Struszczyk-Świta, K.; Napoli, R.; Białkowska, A.; Canfora, L.; Pinzari, F. When Salt Meddles Between Plant, Soil, and Microorganisms. Front. Plant Sci. 2020, 11, 553087. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.H.; Li, Z.H. The Importance of Cl− Exclusion and Vacuolar Cl− Sequestration: Revisiting the Role of Cl− Transport in Plant Salt Tolerance Transport in Plant Salt Tolerance. Front. Plant Sci. 2019, 10, 482468. [Google Scholar] [CrossRef] [PubMed]
- Broyer, T.C.; Carlton, A.B.; Johnson, C.M.; Stout, P.R. Chlorine-A Micronutrient Element for Higher Plants. Plant Physiol. 1954, 29, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Chloride: Essential micronutrient and multifunctional beneficial ion. J. Exp. Bot. 2017, 38, 359–367. [Google Scholar] [CrossRef]
- Franco-Navarro, J.D.; Rosales, M.A.; Cubero-Font, P.; Calvo, P.; Álvarez, R.; Diaz-Espejo, A.; Colmenero-Flores, J.M. Chloride as a macronutrient increases water-use efficiency by anatomically driven reduced stomatal conductance and increased mesophyll diffusion to CO2. Plant J. 2019, 99, 815–831. [Google Scholar] [CrossRef]
- Kawakami, K.; Umena, Y.; Kamiya, N.; Shen, J.R. Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography. Proc. Natl. Acad. Sci. USA 2009, 106, 8567–8572. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Chloride in Soils and its Uptake and Movement within the Plant: A Review. Ann. Bot. 2001, 88, 967–988. [Google Scholar] [CrossRef]
- Geilfus, C.-M. Chloride: From Nutrient to Toxicant. Plant Cell Physiol. 2018, 59, 877–886. [Google Scholar] [CrossRef]
- Wahome, P.K. Mechanisms of salt (NaCl) stress tolerance in horticultural crops—A mini review. Acta Hortic. 2003, 609, 127–131. [Google Scholar] [CrossRef]
- Iqbal, M.S.; Kotula, L.; Malik, A.I.; Erskine, W. Salinity sensitivity in mungbean: Tissue ion accumulation in relation to growth and yield in contrasting genotypes. Plant Soil 2025, 512, 657–675. [Google Scholar] [CrossRef]
- Luo, Q.; Yu, B.; Liu, Y. Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J. Plant Physiol. 2005, 162, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Teakle, N.L.; Real, D.; Colmer, T.D. Growth and ion relations in response to combined salinity and waterlogging in the perennial forage legumes Lotus corniculatus and Lotus tenuis. Plant Soil 2006, 289, 369–383. [Google Scholar] [CrossRef]
- Farooq, M.; Gogoi, N.; Hussain, M.; Barthakur, S.; Paul, S.; Bharadwaj, N.; Migdadi, H.M.; Alghamdi, S.S.; Siddique, K.H.M. Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol. Biochem. 2017, 118, 199–217. [Google Scholar] [CrossRef]
- Rahman, M. Evaluation of mungbean genotypes for salt tolerance at seedling stage and alleviation of saline stress by gypsum. Fundam. Appl. Agric. 2016, 1, 39–43. [Google Scholar]
- Amador, B.; Yamada, S.; Yamaguchi, T.; Rueda-Puente, E.; Ávila Serrano, N.Y.; García-Hernández, J.; López-Aguilar, R.; Troyo-Dieguez, E.; Nieto-Garibay, A. Influence of Calcium Silicate on Growth, Physiological Parameters and Mineral Nutrition in Two Legume Species Under Salt Stress. J. Agron. Crop Sci. 2007, 193, 413–421. [Google Scholar] [CrossRef]
- Song, J.; Han, M.; Zhu, X.; Li, H.; Ning, Y.; Zhang, W.; Yang, H. MhCLC-c1, a Cl channel c homolog from Malus hupehensis, alleviates NaCl-induced cell death by inhibiting intracellular Cl− accumulation. BMC Plant Biol. 2023, 23, 306. [Google Scholar] [CrossRef]
- Henderson, S.W.; Baumann, U.; Blackmore, D.H.; Walker, A.R.; Walker, R.R.; Gilliham, M. Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots. BMC Plant Biol. 2014, 14, 273. [Google Scholar] [CrossRef]
- Visconti, F.; Intrigliolo, D.S.; Quiñones, A.; Tudela, L.; Bonet, L.; de Paz, J.M. Differences in specific chloride toxicity to Diospyros kaki cv. “Rojo Brillante” grafted on D. lotus and D. virginiana. Sci. Hortic. 2017, 214, 83–90. [Google Scholar] [CrossRef]
- Jentsch, T.J.; Günther, W.; Pusch, M.; Schwappach, B. Properties of voltage-gated chloride channels of the ClC gene family. J. Physiol. 1995, 482, 19s–25s. [Google Scholar] [CrossRef]
- Lv, Q.D.; Tang, R.J.; Liu, H.; Gao, X.S.; Li, Y.Z.; Zheng, H.Q.; Zhang, H.X. Cloning and molecular analyses of the chloride channel gene family. Plant Sci. 2009, 176, 650–661. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, J.; Jin, L.; Li, Z.; Xu, G.; Wang, R.; Zhang, J.; Zhai, N.; Chen, Q.; Liu, P.; et al. Identification and analysis of the chloride channel gene family members in tobacco (Nicotiana tabacum). Gene 2018, 676, 56–64. [Google Scholar] [CrossRef]
- Mao, P.; Run, Y.; Wang, H.; Han, C.; Zhang, L.; Zhan, K.; Xu, H.; Cheng, X. Genome-Wide Identification and Functional Characterization of the Chloride Channel TaCLC Gene Family in Wheat (Triticum aestivum L.). Front. Genet. 2022, 13, 846795. [Google Scholar] [CrossRef]
- Ma, J.; Li, S.; Zaman, S.; Anwar, A. CLC gene family in Solanum lycopersicum: Genome-wide identification, expression, and evolutionary analysis of tomato in response to salinity and Cd stress. Front. Plant Sci. 2025, 16, 1547723. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, T.J. CLC chloride channels and transporters: From genes to protein structure, pathology and physiology. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 3–36. [Google Scholar] [CrossRef] [PubMed]
- Accardi, A. Structure and gating of CLC channels and exchangers. J. Physiol. 2015, 593, 4129–4138. [Google Scholar] [CrossRef]
- De Angeli, A.; Monachello, D.; Ephritikhine, G.; Frachisse, J.M.; Thomine, S.; Gambale, F.; Barbier-Brygoo, H. CLC-mediated anion transport in plant cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Nedelyaeva, O.I.; Shuvalov, A.V.; Balnokin, Y.V. Chloride Channels and Transporters of the CLC Family in Plants. Russ. J. Plant Physiol. 2020, 67, 767–784. [Google Scholar] [CrossRef]
- Zifarelli, G.; Pusch, M. CLC transport proteins in plants. FEBS Lett. 2010, 584, 2122–2127. [Google Scholar] [CrossRef]
- Li, W.; Gao, S.; Zhao, Y.; Wu, Y.; Li, X.; Li, J.; Zhu, W.; Ma, Z.; Liu, W. GhCLCc-1, a Chloride Channel Gene from Upland Cotton, Positively Regulates Salt Tolerance by Modulating the Accumulation of Chloride Ions. Genes 2024, 15, 555. [Google Scholar] [CrossRef]
- Rajappa, S.; Krishnamurthy, P.; Huang, H.; Yu, D.; Friml, J.; Xu, J.; Kumar, P.P. The translocation of a chloride channel from the Golgi to the plasma membrane helps plants adapt to salt stress. Nat. Commun. 2024, 15, 3978. [Google Scholar] [CrossRef]
- Wei, P.; Wang, L.; Liu, A.; Yu, B.; Lam, H.M. GmCLC1 Confers Enhanced Salt Tolerance through Regulating Chloride Accumulation in Soybean. Front. Plant Sci. 2016, 7, 1082. [Google Scholar] [CrossRef]
- Nakamura, A.; Fukuda, A.; Sakai, S.; Tanaka, Y. Molecular cloning, functional expression and subcellular localization of two putative vacuolar voltage-gated chloride channels in rice (Oryza sativa L.). Plant Cell Physiol. 2006, 47, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Wang, C.; Ni, Y.; Liu, X. The Role of Chloride Channels in Plant Responses to NaCl. Int. J. Mol. Sci. 2023, 25, 19. [Google Scholar] [CrossRef] [PubMed]
- Franco-Navarro, J.D.; Brumós, J.; Rosales, M.A.; Cubero-Font, P.; Talón, M.; Colmenero-Flores, J.M. Chloride regulates leaf cell size and water relations in tobacco plants. J. Exp. Bot. 2016, 67, 873–891. [Google Scholar] [CrossRef] [PubMed]
- Franco-Navarro, J.D.; Díaz-Rueda, P.; Rivero-Núñez, C.M.; Brumós, J.; Rubio-Casal, A.E.; de Cires, A.; Colmenero-Flores, J.M.; Rosales, M.A. Chloride nutrition improves drought resistance by enhancing water deficit avoidance and tolerance mechanisms. J. Exp. Bot. 2021, 72, 5246–5261. [Google Scholar] [CrossRef]
- Wang, S.; Su, S.Z.; Wu, Y.; Li, S.P.; Shan, X.H.; Liu, H.K.; Wang, S.; Yuan, Y.P. Overexpression of maize chloride channel gene ZmCLC-d in Arabidopsis thaliana improved its stress resistance. Biol. Plant. 2015, 59, 55–64. [Google Scholar] [CrossRef]
- Russelle, M.P.; Birr, A.S. Large-scale assessment of symbiotic dinitrogen fixation by crops: Soybean and alfalfa in the Mississippi river basin. Agron. J. 2004, 96, 1754–1760. [Google Scholar] [CrossRef]
- Bora, K.S.; Sharma, A. Phytochemical and pharmacological potential of Medicago sativa: A review. Pharm. Biol. 2011, 49, 211–220. [Google Scholar] [CrossRef]
- Feng, Y.P.; Shi, Y.; Zhao, M.Y.; Shen, H.H.; Xu, L.C.; Luo, Y.K.; Liu, Y.Z.; Xing, A.J.; Kang, J.; Jing, H.C.; et al. Yield and quality properties of alfalfa (Medicago sativa L.) and their influencing factors in China. Eur. J. Agron. 2022, 141, 126637. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, G.; Lu, Q.; Xiong, D.; Li, G.; Du, S. Understanding the Limiting Climatic Factors on the Suitable Habitat of Chinese Alfalfa. Forests 2022, 13, 482. [Google Scholar] [CrossRef]
- Liu, J.; Yang, D.; Li, X.; Jin, Z.X.; Li, J.M. Deciphering the effect mechanism of chromosome doubling on the biomass increase in Elsholtzia splendens. Sci. Hortic. 2024, 326, 112751. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Li, H.; Li, S.; Song, J.; Guan, W. Comparison of the morphological and physiological characteristics of diploid and tetraploid Luculia pinceana Hook. BMC Plant Biol. 2025, 25, 536. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.P.; Hawkins, C.; Peel, M.D.; Yu, L.X. Genetic Loci Associated with Salt Tolerance in Advanced Breeding Populations of Tetraploid Alfalfa Using Genome-Wide Association Studies. Plant Genome 2019, 12, 180026. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef]
- Peng, W.; Cai, W.; Pan, J.; Su, X.; Dou, L. Molecular Mechanisms of Alfalfa Response to Abiotic Stresses. Plants 2025, 14, 487. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, X.; Zi, Y.; Zhao, G.; Zhu, L.; Hong, L.; Li, M.; Wang, S.; Long, R.; Kang, J.; et al. Characterization of the Heat Shock Transcription Factor Family in Medicago sativa L. and Its Potential Roles in Response to Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 12683. [Google Scholar] [CrossRef]
- Bakshi, A.; Moin, M.; Kumar, M.U.; Reddy, A.B.; Ren, M.; Datla, R.; Siddiq, E.A.; Kirti, P.B. Ectopic expression of Arabidopsis Target of Rapamycin (AtTOR) improves water-use efficiency and yield potential in rice. Sci. Rep. 2017, 7, 42835, Erratum in Sci. Rep. 2017, 7, 46124. [Google Scholar]
- Yang, J.; Yi, J.; Ma, S.; Wang, Y.; Song, J.; Li, S.; Feng, Y.; Sun, H.; Gao, C.; Yang, R.; et al. Integrated physiological, metabolomic, and transcriptomic analyses elucidate the regulation mechanisms of lignin synthesis under osmotic stress in alfalfa leaf (Medicago sativa L.). BMC Genom. 2024, 25, 174. [Google Scholar] [CrossRef] [PubMed]
- Zifarelli, G.; Pusch, M. CLC chloride channels and transporters: A biophysical and physiological perspective. Rev. Physiol. Biochem. Pharmacol. 2007, 158, 23–76. [Google Scholar] [PubMed]
- von der Fecht-Bartenbach, J.; Bogner, M.; Dynowski, M.; Ludewig, U. CLC-b-mediated NO-3/H+ exchange across the tonoplast of Arabidopsis vacuoles. Plant Cell Physiol. 2010, 51, 960–968. [Google Scholar] [CrossRef]
- De Angeli, A.; Monachello, D.; Ephritikhine, G.; Frachisse, J.M.; Thomine, S.; Gambale, F.; Barbier-Brygoo, H. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 2006, 442, 939–942. [Google Scholar] [CrossRef]
- Jossier, M.; Kroniewicz, L.; Dalmas, F.; Le Thiec, D.; Ephritikhine, G.; Thomine, S.; Barbier-Brygoo, H.; Vavasseur, A.; Filleur, S.; Leonhardt, N. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 2010, 64, 563–576. [Google Scholar] [CrossRef]
- von der Fecht-Bartenbach, J.; Bogner, M.; Krebs, M.; Stierhof, Y.D.; Schumacher, K.; Ludewig, U. Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 2007, 50, 466–474. [Google Scholar] [CrossRef]
- Hechenberger, M.; Schwappach, B.; Fischer, W.N.; Frommer, W.B.; Jentsch, T.J.; Steinmeyer, K. A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J. Biol. Chem. 1996, 271, 33632–33638. [Google Scholar] [CrossRef]
- Jentsch, T.J.; Pusch, M. CLC Chloride Channels and Transporters: Structure, Function, Physiology, and Disease. Physiol. Rev. 2018, 98, 1493–1590. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Mangena, P. Cell Mutagenic Autopolyploidy Enhances Salinity Stress Tolerance in Leguminous Crops. Cells 2023, 12, 2082. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Wei, Q.; Liu, Y.; Zhou, G.; Li, Q.; Yang, C.; Peng, S.-A. Overexpression of CsCLCc, a Chloride Channel Gene from Poncirus trifoliata, Enhances Salt Tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 1548–1557. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Dong, X.; Deng, H.; Ma, W.; Zhou, Q.; Liu, Z. Genome-wide identification of the MADS-box transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under abiotic stress. BMC Genom. 2021, 22, 603. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Thakur, T.; Gandass, N.; Mittal, K.; Jamwal, P.; Muthamilarasan, M.; Salvi, P. A rapid, efficient, and low-cost BiFC protocol and its application in studying in vivo interaction of seed-specific transcription factors, RISBZ and RPBF. Funct. Integr. Genom. 2021, 21, 593–603. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.; Jiang, G.; Du, P.; Wang, J.; He, H.; Li, H.; Li, H.; Wang, F.; Xie, Q. Genome-Wide Identification and Functional Divergence of the Chloride Channel (CLC) Gene Family in Autotetraploid Alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 2025, 26, 11442. https://doi.org/10.3390/ijms262311442
Fang Y, Jiang G, Du P, Wang J, He H, Li H, Li H, Wang F, Xie Q. Genome-Wide Identification and Functional Divergence of the Chloride Channel (CLC) Gene Family in Autotetraploid Alfalfa (Medicago sativa L.). International Journal of Molecular Sciences. 2025; 26(23):11442. https://doi.org/10.3390/ijms262311442
Chicago/Turabian StyleFang, Yanjun, Guangzhi Jiang, Pingping Du, Jiayin Wang, Huan He, Hongfei Li, Hongbin Li, Fei Wang, and Quanliang Xie. 2025. "Genome-Wide Identification and Functional Divergence of the Chloride Channel (CLC) Gene Family in Autotetraploid Alfalfa (Medicago sativa L.)" International Journal of Molecular Sciences 26, no. 23: 11442. https://doi.org/10.3390/ijms262311442
APA StyleFang, Y., Jiang, G., Du, P., Wang, J., He, H., Li, H., Li, H., Wang, F., & Xie, Q. (2025). Genome-Wide Identification and Functional Divergence of the Chloride Channel (CLC) Gene Family in Autotetraploid Alfalfa (Medicago sativa L.). International Journal of Molecular Sciences, 26(23), 11442. https://doi.org/10.3390/ijms262311442





