De Novo Detection of Clonal Structure and Evolution in Single-Cell and Spatial Transcriptomes
Abstract
1. Introduction
2. Results
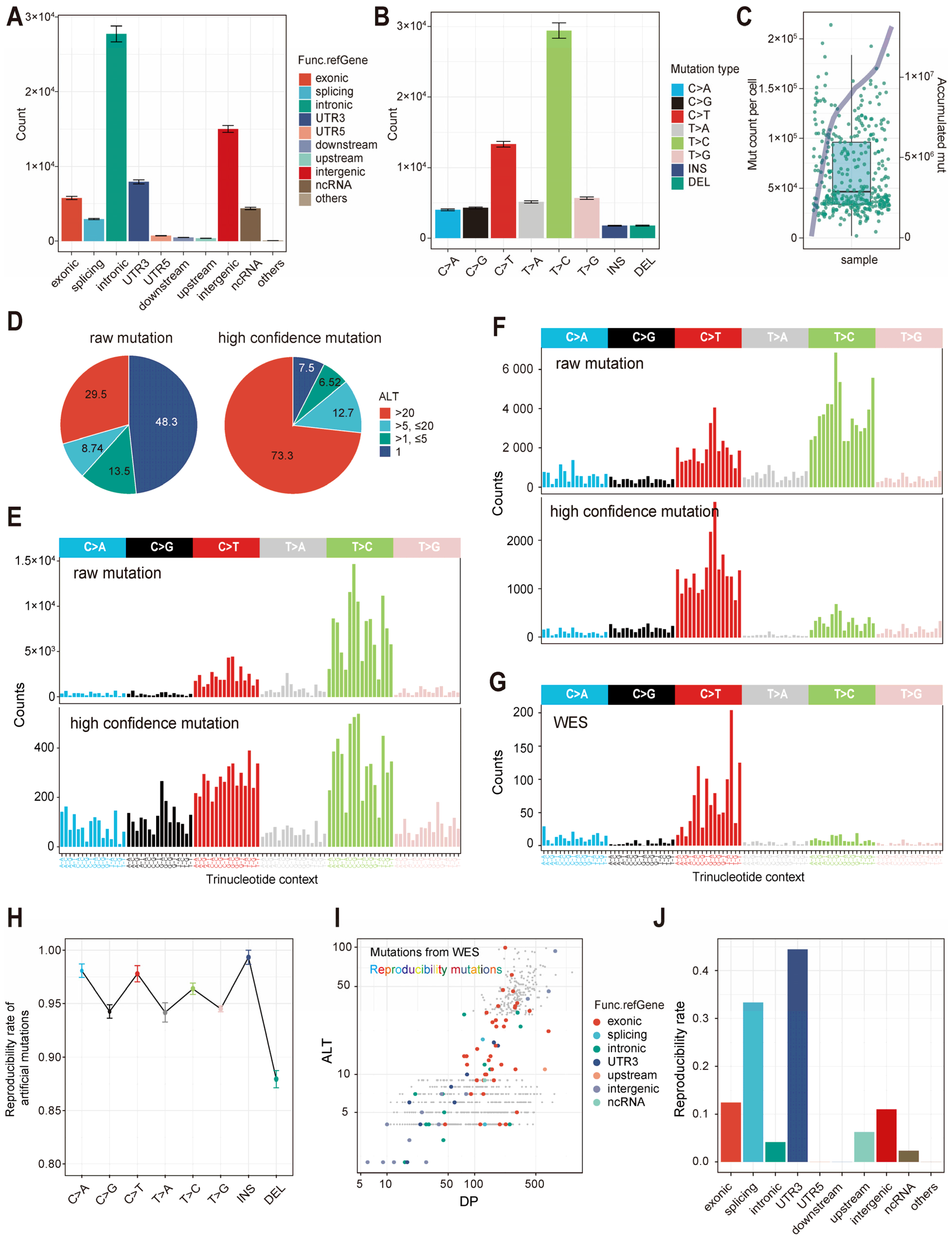
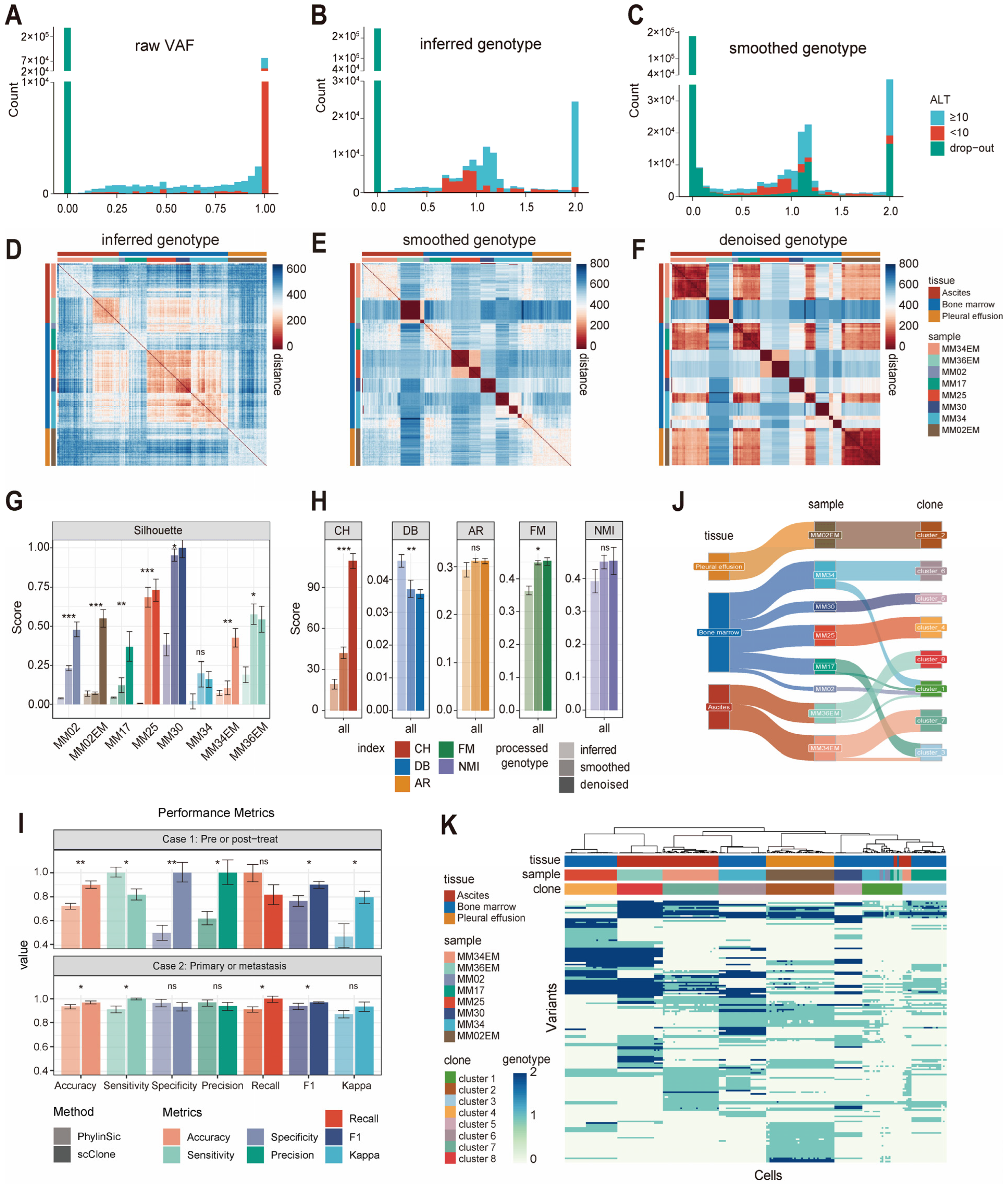
2.1. Benchmark: Assessing the Effectiveness and Usability of scClone
2.2. Identification of Subclones Among Single Cells by scClone
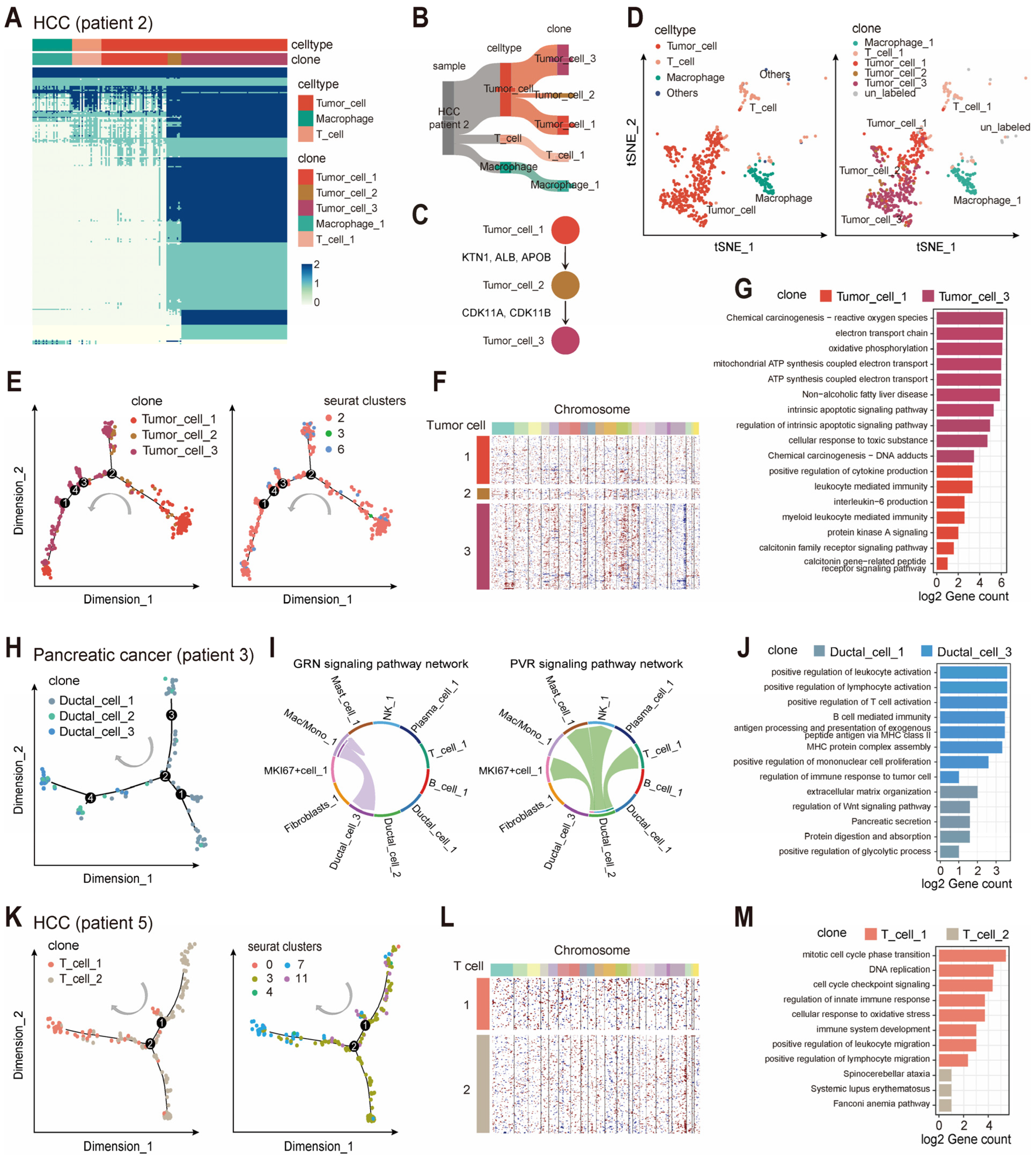
2.3. scClone Describes the Evolutionary Trajectories of Immune Cells
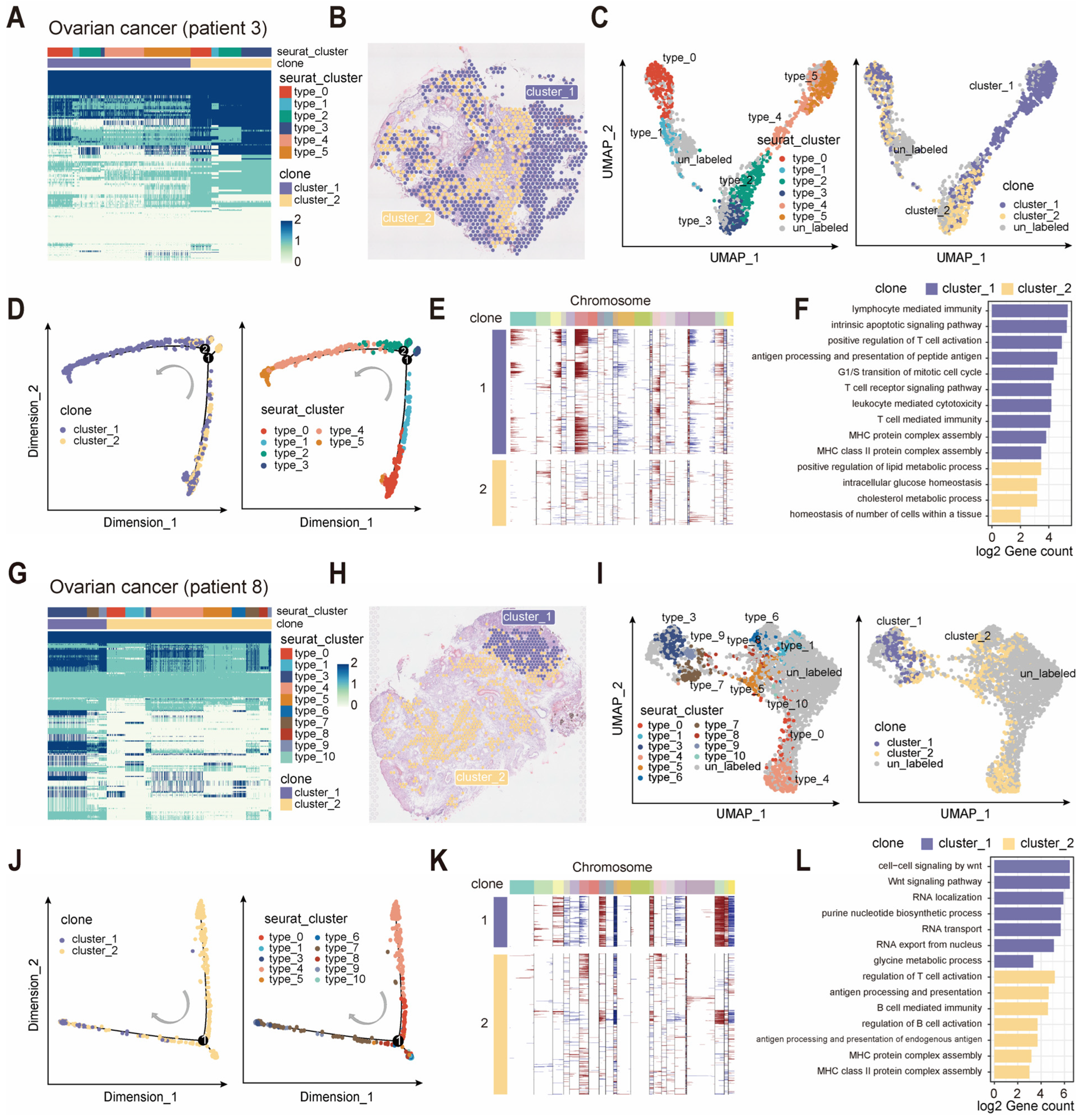
2.4. scClone Enables Clonal Cluster Identification in Spatial Transcriptomics
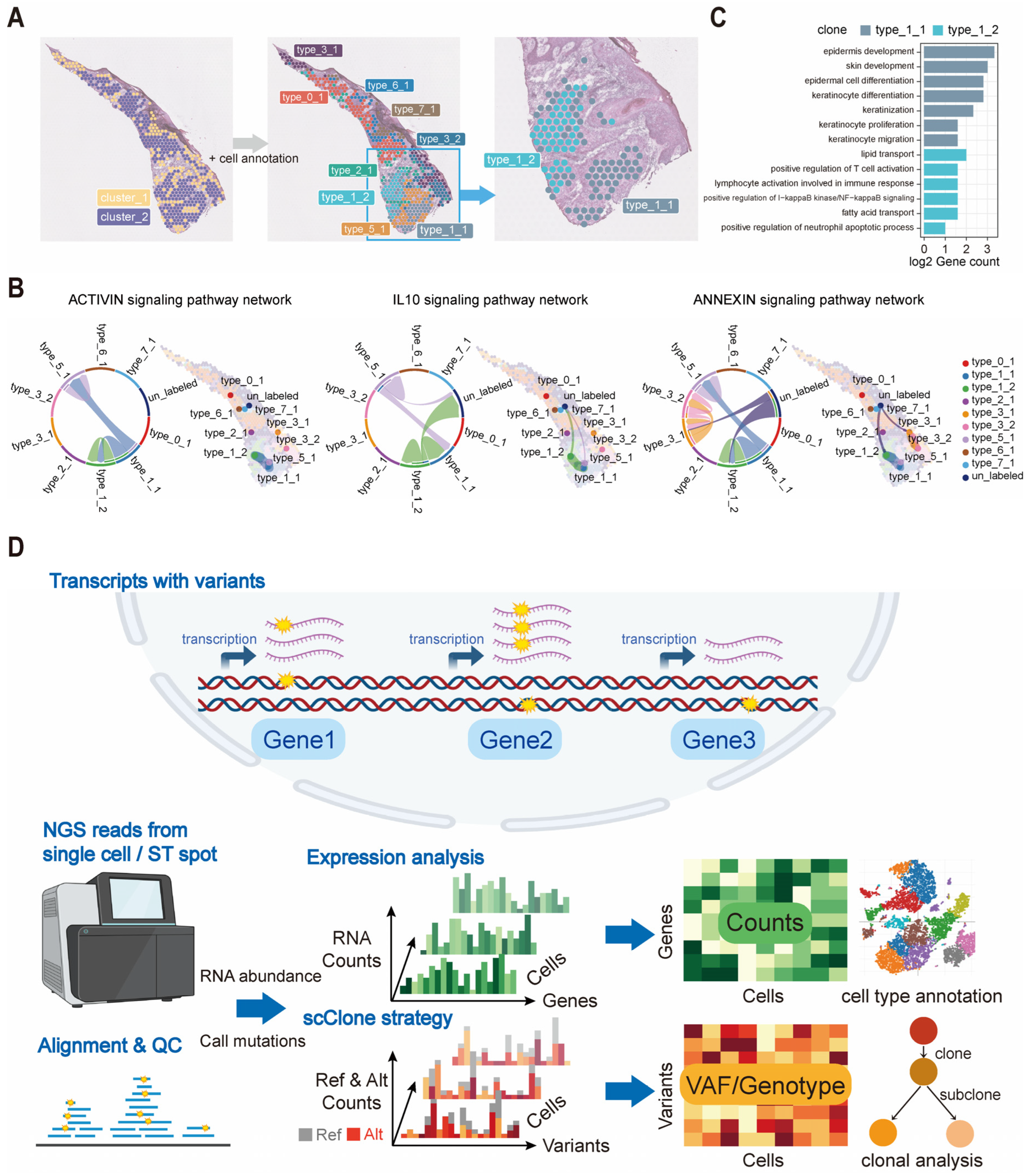
2.5. Integration of scClone and Transcriptomic Information Reveals High-Resolution Clonal Structures
3. Discussion
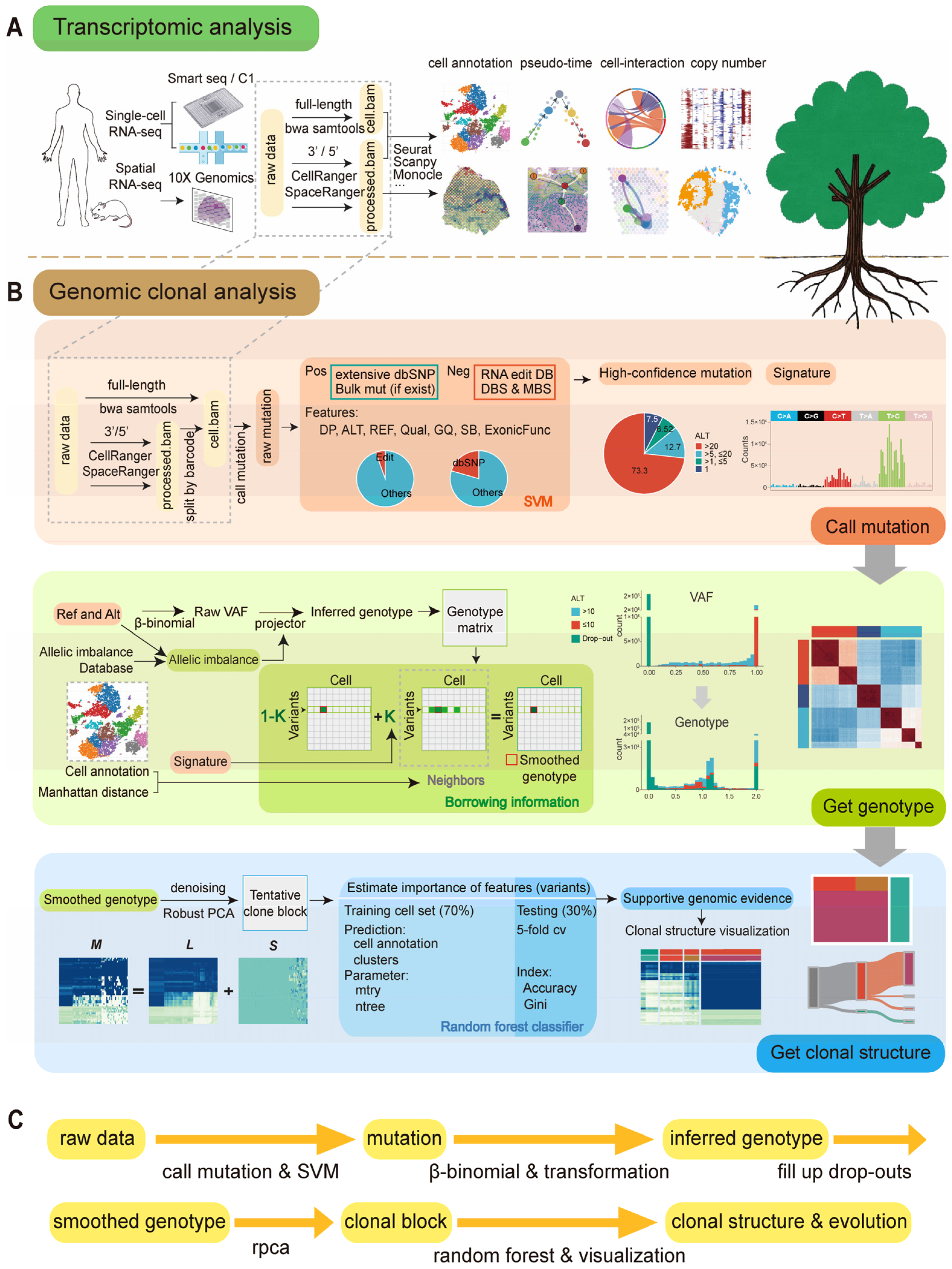
4. Materials and Methods
4.1. Data Acquisition
4.2. scClone Workflow
5. Conclusions
- This work introduces scClone, a full-process clonal evolution inference toolkit based on mutation detection that relies solely on single-cell transcriptome sequencing. It includes a reliable mutation detection pipeline, a series of genotype inference algorithms, and clonal structure visualization.
- scClone achieves promising results across various cell types from different platforms and is compared with mainstream transcriptome analysis methods.
- scClone can be applied to spatial transcriptomics and identifies subclonal structures on histological sections that traditional methods fail to detect.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aparicio, S.; Caldas, C. The implications of clonal genome evolution for cancer medicine. N. Engl. J. Med. 2013, 368, 842–851. [Google Scholar] [CrossRef]
- Venkatesan, S.; Swanton, C. Tumor Evolutionary Principles: How Intratumor Heterogeneity Influences Cancer Treatment and Outcome. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e141–e149. [Google Scholar] [CrossRef]
- Frieda, K.L.; Linton, J.M.; Hormoz, S.; Choi, J.; Chow, K.K.; Singer, Z.S.; Budde, M.W.; Elowitz, M.B.; Cai, L. Synthetic recording and in situ readout of lineage information in single cells. Nature 2017, 541, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Bai, S.; Xie, G.; Shi, Y.; Zhao, L.; Yang, G.; Tian, F.; He, K.Y.; Wang, L.; Li, X.; et al. Accurate tumor clonal structures require single-cell analysis. Ann. N. Y. Acad. Sci. 2022, 1517, 213–224. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef]
- Moore, L.; Cagan, A.; Coorens, T.H.H.; Neville, M.D.C.; Sanghvi, R.; Sanders, M.A.; Oliver, T.R.W.; Leongamornlert, D.; Ellis, P.; Noorani, A.; et al. The mutational landscape of human somatic and germline cells. Nature 2021, 597, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Lee-Six, H.; Obro, N.F.; Shepherd, M.S.; Grossmann, S.; Dawson, K.; Belmonte, M.; Osborne, R.J.; Huntly, B.J.P.; Martincorena, I.; Anderson, E.; et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018, 561, 473–478. [Google Scholar] [CrossRef]
- Su, X.; Zhao, L.; Shi, Y.; Zhang, R.; Long, Q.; Bai, S.; Luo, Q.; Lin, Y.; Zou, X.; Ghazanfar, S.; et al. Clonal evolution in liver cancer at single-cell and single-variant resolution. J. Hematol. Oncol. 2021, 14, 22. [Google Scholar] [CrossRef]
- Tang, J.; Tu, K.; Lu, K.; Zhang, J.; Luo, K.; Jin, H.; Wang, L.; Yang, L.; Xiao, W.; Zhang, Q.; et al. Single-cell exome sequencing reveals multiple subclones in metastatic colorectal carcinoma. Genome Med. 2021, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Lahnemann, D.; Koster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Haerty, W.; Kumar, P.; Li, Y.I.; Hu, T.X.; Teng, M.J.; Goolam, M.; Saurat, N.; Coupland, P.; Shirley, L.M.; et al. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 2015, 12, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.S.; Kester, L.; Spanjaard, B.; Bienko, M.; van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015, 33, 285–289. [Google Scholar] [CrossRef]
- Nam, A.S.; Kim, K.T.; Chaligne, R.; Izzo, F.; Ang, C.; Taylor, J.; Myers, R.M.; Abu-Zeinah, G.; Brand, R.; Omans, N.D.; et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 2019, 571, 355–360. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.V.; Pai, R.K.; Snyder, M.P. Simul-seq: Combined DNA and RNA sequencing for whole-genome and transcriptome profiling. Nat. Methods 2016, 13, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ferdinand, J.R.; Loudon, K.W.; Bowyer, G.S.; Laidlaw, S.; Muyas, F.; Mamanova, L.; Neves, J.B.; Bolt, L.; Fasouli, E.S.; et al. Mapping single-cell transcriptomes in the intra-tumoral and associated territories of kidney cancer. Cancer Cell 2022, 40, 1583–1599.e10. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.R.; Steif, A.; Laks, E.; Zahn, H.; Lai, D.; McPherson, A.; Farahani, H.; Kabeer, F.; O’Flanagan, C.; Biele, J.; et al. clonealign: Statistical integration of independent single-cell RNA and DNA sequencing data from human cancers. Genome Biol. 2019, 20, 54. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Gao, R.; Bai, S.; Henderson, Y.C.; Lin, Y.; Schalck, A.; Yan, Y.; Kumar, T.; Hu, M.; Sei, E.; Davis, A.; et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 2021, 39, 599–608. [Google Scholar] [CrossRef]
- Fan, J.; Lee, H.O.; Lee, S.; Ryu, D.E.; Lee, S.; Xue, C.; Kim, S.J.; Kim, K.; Barkas, N.; Park, P.J.; et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell RNA-seq data. Genome Res. 2018, 28, 1217–1227. [Google Scholar] [CrossRef]
- Liu, X.; Griffiths, J.I.; Bishara, I.; Liu, J.; Bild, A.H.; Chang, J.T. Phylogenetic inference from single-cell RNA-seq data. Sci. Rep. 2023, 13, 12854. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Silberstein, L.; Scadden, D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 2014, 11, 740–742. [Google Scholar] [CrossRef]
- Behm, M.; Ohman, M. RNA Editing: A Contributor to Neuronal Dynamics in the Mammalian Brain. Trends Genet. 2016, 32, 165–175. [Google Scholar] [CrossRef]
- Gommans, W.M.; Mullen, S.P.; Maas, S. RNA editing: A driving force for adaptive evolution? Bioessays 2009, 31, 1137–1145. [Google Scholar] [CrossRef]
- Ryu, D.; Kim, S.J.; Hong, Y.; Jo, A.; Kim, N.; Kim, H.J.; Lee, H.O.; Kim, K.; Park, W.Y. Alterations in the Transcriptional Programs of Myeloma Cells and the Microenvironment during Extramedullary Progression Affect Proliferation and Immune Evasion. Clin. Cancer Res. 2020, 26, 935–944, Erratum in Clin. Cancer Res. 2020, 26, 5049. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstrahle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514.e22. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.M.; O’Mahony, J.J.; Sanjeev, K.; Baranov, P.V. Darned in 2013: Inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res 2013, 41, D258–D261. [Google Scholar] [CrossRef]
- Gagnidze, K.; Rayon-Estrada, V.; Harroch, S.; Bulloch, K.; Papavasiliou, F.N. A New Chapter in Genetic Medicine: RNA Editing and its Role in Disease Pathogenesis. Trends Mol. Med. 2018, 24, 294–303. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Ohishi, W.; Cologne, J.B.; Fujiwara, S.; Suzuki, G.; Hayashi, T.; Niwa, Y.; Akahoshi, M.; Ueda, K.; Tsuge, M.; Chayama, K. Serum interleukin-6 associated with hepatocellular carcinoma risk: A nested case-control study. Int. J. Cancer 2014, 134, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.W.; Oh, B.S.; Kwon, J.H.; You, C.R.; Chung, K.W.; Kay, C.S.; Jung, H.S. Serum interleukin-6 and C-reactive protein as a prognostic indicator in hepatocellular carcinoma. Cytokine 2012, 60, 686–693. [Google Scholar] [CrossRef]
- Yang, J.; Zeng, L.; Chen, R.; Zheng, S.; Zhou, Y.; Chen, R. Characterization of heterogeneous metabolism in hepatocellular carcinoma identifies new therapeutic target and treatment strategy. Front. Immunol. 2023, 14, 1076587. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, Y.; Han, W.; Yuan, J.; Xie, W.; Liu, K.; Qiu, Y.; Wang, X.; Li, X. Potentiality of alpha-fetoprotein (AFP) and soluble intercellular adhesion molecule-1 (sICAM-1) in prognosis prediction and immunotherapy response for patients with hepatocellular carcinoma. Bioengineered 2021, 12, 9435–9451. [Google Scholar] [CrossRef]
- Zhang, S.; Fang, W.; Zhou, S.; Zhu, D.; Chen, R.; Gao, X.; Li, Z.; Fu, Y.; Zhang, Y.; Yang, F.; et al. Single cell transcriptomic analyses implicate an immunosuppressive tumor microenvironment in pancreatic cancer liver metastasis. Nat. Commun. 2023, 14, 5123. [Google Scholar] [CrossRef]
- Liu, F.; Huang, J.; He, F.; Ma, X.; Fan, F.; Meng, M.; Zhuo, Y.; Zhang, L. CD96, a new immune checkpoint, correlates with immune profile and clinical outcome of glioma. Sci. Rep. 2020, 10, 10768. [Google Scholar] [CrossRef]
- Ye, W.; Luo, C.; Liu, F.; Liu, Z.; Chen, F. CD96 Correlates With Immune Infiltration and Impacts Patient Prognosis: A Pan-Cancer Analysis. Front. Oncol. 2021, 11, 634617. [Google Scholar] [CrossRef]
- Denisenko, E.; de Kock, L.; Tan, A.; Beasley, A.B.; Beilin, M.; Jones, M.E.; Hou, R.; Muiri, D.O.; Bilic, S.; Mohan, G.; et al. Spatial transcriptomics reveals discrete tumour microenvironments and autocrine loops within ovarian cancer subclones. Nat. Commun. 2024, 15, 2860. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.M.; Markowetz, F. OncoNEM: Inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Jahn, K.; Kuipers, J.; Beerenwinkel, N. Tree inference for single-cell data. Genome Biol. 2016, 17, 86. [Google Scholar] [CrossRef]
- Zafar, H.; Tzen, A.; Navin, N.; Chen, K.; Nakhleh, L. SiFit: Inferring tumor trees from single-cell sequencing data under finite-sites models. Genome Biol. 2017, 18, 178. [Google Scholar] [CrossRef]
- Zafar, H.; Navin, N.; Chen, K.; Nakhleh, L. SiCloneFit: Bayesian inference of population structure, genotype, and phylogeny of tumor clones from single-cell genome sequencing data. Genome Res. 2019, 29, 1847–1859. [Google Scholar] [CrossRef]
- Vu, T.N.; Nguyen, H.N.; Calza, S.; Kalari, K.R.; Wang, L.; Pawitan, Y. Cell-level somatic mutation detection from single-cell RNA sequencing. Bioinformatics 2019, 35, 4679–4687. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Rostom, R.; Huang, Y.; Kunz, D.J.; Danecek, P.; Bonder, M.J.; Hagai, T.; Lyu, R.; HipSci, C.; Wang, W.; et al. Cardelino: Computational integration of somatic clonal substructure and single-cell transcriptomes. Nat. Methods 2020, 17, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.Y.; Li, P.; Rodin, R.E.; Kim, S.N.; Dou, Y.; Kenny, C.J.; Akula, S.K.; Hodge, R.D.; Bakken, T.E.; Miller, J.A.; et al. Parallel RNA and DNA analysis after deep sequencing (PRDD-seq) reveals cell type-specific lineage patterns in human brain. Proc. Natl. Acad. Sci. USA 2020, 117, 13886–13895. [Google Scholar] [CrossRef]
- Salehi, S.; Steif, A.; Roth, A.; Aparicio, S.; Bouchard-Cote, A.; Shah, S.P. ddClone: Joint statistical inference of clonal populations from single cell and bulk tumour sequencing data. Genome Biol. 2017, 18, 44. [Google Scholar] [CrossRef]
- Jun, S.H.; Toosi, H.; Mold, J.; Engblom, C.; Chen, X.; O’Flanagan, C.; Hagemann-Jensen, M.; Sandberg, R.; Aparicio, S.; Hartman, J.; et al. Reconstructing clonal tree for phylo-phenotypic characterization of cancer using single-cell transcriptomics. Nat. Commun. 2023, 14, 982. [Google Scholar] [CrossRef]
- Dou, J.; Tan, Y.; Kock, K.H.; Wang, J.; Cheng, X.; Tan, L.M.; Han, K.Y.; Hon, C.C.; Park, W.Y.; Shin, J.W.; et al. Single-nucleotide variant calling in single-cell sequencing data with Monopogen. Nat. Biotechnol. 2023, 42, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Muyas, F.; Sauer, C.M.; Valle-Inclan, J.E.; Li, R.; Rahbari, R.; Mitchell, T.J.; Hormoz, S.; Cortes-Ciriano, I. De novo detection of somatic mutations in high-throughput single-cell profiling data sets. Nat. Biotechnol. 2023, 42, 758–767. [Google Scholar] [CrossRef]
- Picardi, E.; D’Erchia, A.M.; Lo Giudice, C.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef] [PubMed]
- Kravitz, S.N.; Ferris, E.; Love, M.I.; Thomas, A.; Quinlan, A.R.; Gregg, C. Random allelic expression in the adult human body. Cell Rep. 2023, 42, 111945. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578, Erratum in Nat. Protoc. 2014, 9, 2513. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Ewing, A.D.; Houlahan, K.E.; Hu, Y.; Ellrott, K.; Caloian, C.; Yamaguchi, T.N.; Bare, J.C.; P’ng, C.; Waggott, D.; Sabelnykova, V.Y.; et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods 2015, 12, 623–630. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Wang, S.; Wu, C.; Wu, T.; Zhao, X.; Ning, W.; Wang, G.; Wang, J.; Chen, J.; Diao, K.; et al. The repertoire of copy number alteration signatures in human cancer. Brief. Bioinform. 2023, 24, bbad053. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Kallberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Lu, T.; Park, S.; Zhu, J.; Wang, Y.; Zhan, X.; Wang, X.; Wang, L.; Zhu, H.; Wang, T. Overcoming Expressional Drop-outs in Lineage Reconstruction from Single-Cell RNA-Sequencing Data. Cell Rep. 2021, 34, 108589. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, S.; Su, X.; Chen, Z.; Han, Z.-G. De Novo Detection of Clonal Structure and Evolution in Single-Cell and Spatial Transcriptomes. Int. J. Mol. Sci. 2025, 26, 11428. https://doi.org/10.3390/ijms262311428
Bai S, Su X, Chen Z, Han Z-G. De Novo Detection of Clonal Structure and Evolution in Single-Cell and Spatial Transcriptomes. International Journal of Molecular Sciences. 2025; 26(23):11428. https://doi.org/10.3390/ijms262311428
Chicago/Turabian StyleBai, Shihao, Xianbin Su, Ziyao Chen, and Ze-Guang Han. 2025. "De Novo Detection of Clonal Structure and Evolution in Single-Cell and Spatial Transcriptomes" International Journal of Molecular Sciences 26, no. 23: 11428. https://doi.org/10.3390/ijms262311428
APA StyleBai, S., Su, X., Chen, Z., & Han, Z.-G. (2025). De Novo Detection of Clonal Structure and Evolution in Single-Cell and Spatial Transcriptomes. International Journal of Molecular Sciences, 26(23), 11428. https://doi.org/10.3390/ijms262311428





