Dual-Receptor Recognition, Lysis Inhibition, Endolysin Release, and Reaction–Diffusion as Alternative Explanations. Comment on Rojero et al. Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914
Abstract
1. Introduction
2. “The Bypassing of the Evolution of Host Resistance”
3. “The Clearing of Semi-Turbid Plaques”
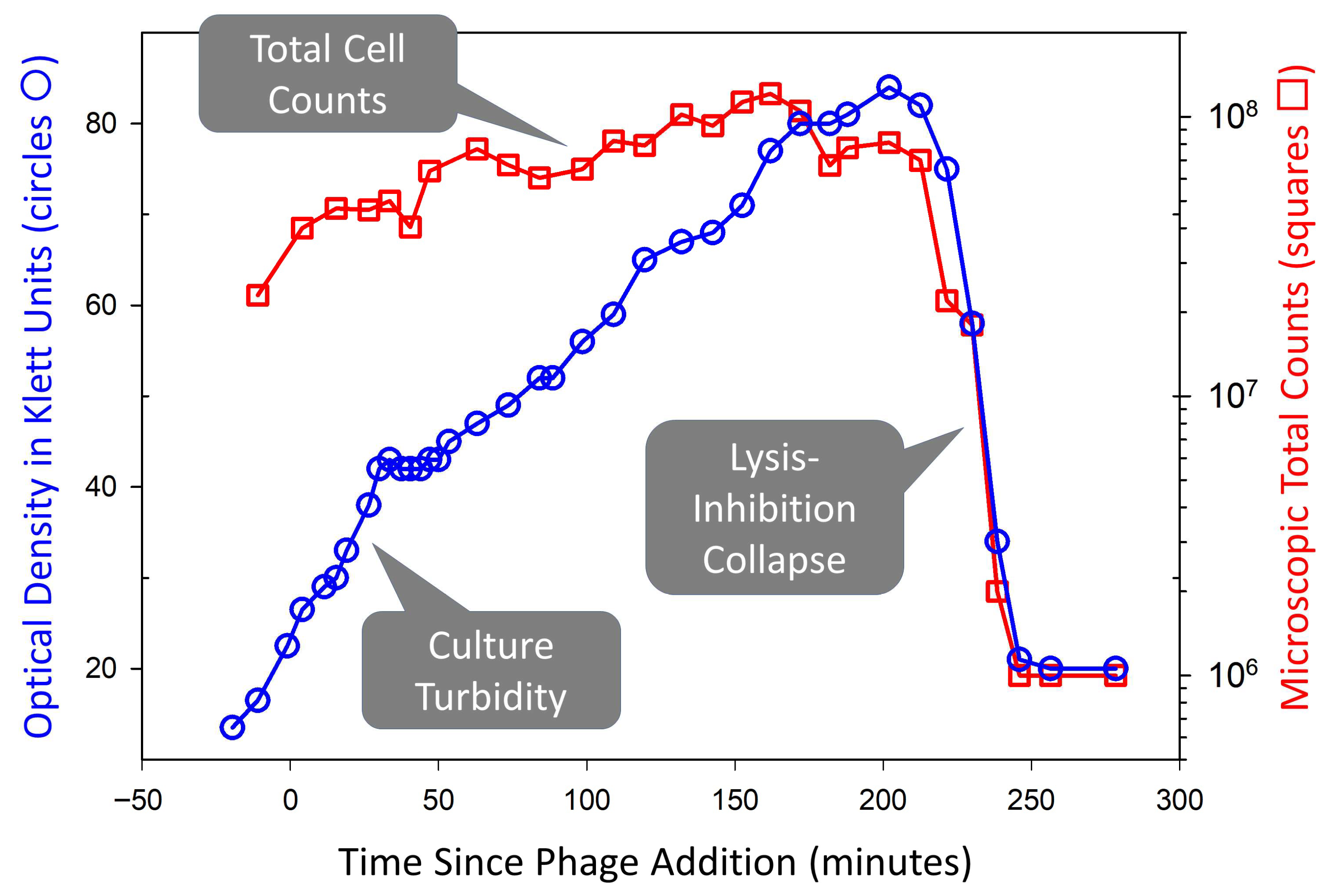
4. “The Formation of Satellite Plaques”
5. “Multi-Day Plaque Enlargement”
5.1. Excised Plaque Assay
5.2. Impact of Lysis-Released Endolysin?
5.3. The Spot-on-Lawn Assay
5.4. An Important Role of Phage 0524phi7-1 Endolysin?
6. Alternative to Swimming Phages
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Rojero, M.; Weaver-Rosen, M.; Serwer, P. Bypassing evolution of bacterial resistance to phages: The example of hyper-aggressive phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Lee, J.J.; Gerbino, K.R.; Meyer, J.R. Comparison of bacterial suppression by phage cocktails, dual-receptor generalists, and coevolutionarily trained phages. Evol. Appl. 2023, 16, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics 1984, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Morona, R.; Henning, U. Host range mutants of bacteriophage Ox2 can use two different outer membrane proteins of Escherichia coli K-12 as receptors. J. Bacteriol. 1984, 159, 724–730. [Google Scholar] [CrossRef]
- Schwarzer, D.; Buettner, F.F.; Browning, C.; Nazarov, S.; Rabsch, W.; Bethe, A.; Oberbeck, A.; Bowman, V.D.; Stummeyer, K.; Muhlenhoff, M.; et al. A multivalent adsorption apparatus explains the broad host range of phage phi92: A comprehensive genomic and structural analysis. J. Virol. 2012, 86, 10384–10398. [Google Scholar] [CrossRef]
- Ramanculov, E.; Young, R. An ancient player unmasked: T4 rI encodes a t-specific antiholin. Mol. Microbiol. 2001, 41, 575–583. [Google Scholar] [CrossRef]
- Tran, T.A.; Struck, D.K.; Young, R. The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. J. Bacteriol. 2007, 189, 7618–7625. [Google Scholar] [CrossRef]
- Burch, L.H.; Zhang, L.; Chao, F.G.; Xu, H.; Drake, J.W. The bacteriophage T4 rapid-lysis genes and their mutational proclivities. J. Bacteriol. 2011, 193, 3537–3545. [Google Scholar] [CrossRef]
- Chen, Y.; Young, R. The last r locus unveiled: T4 RIII is a cytoplasmic antiholin. J. Bacteriol. 2016, 198, 2448–2457. [Google Scholar] [CrossRef]
- Abedon, S.T. Look who’s talking: T-even phage lysis inhibition, the granddaddy of virus-virus intercellular communication research. Viruses 2019, 11, 951. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis of lysis inhibited bacteriophage T4-infected cells. J. Bacteriol. 1992, 174, 8073–8080. [Google Scholar] [CrossRef]
- Asami, K.; Xing, X.H.; Tanji, Y.; Unno, H. Synchronized disruption of Escherichia coli cells by T4 phage infection. J. Ferment. Bioeng. 1997, 83, 511–516. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage T4 resistance to lysis-inhibition collapse. Genet. Res. 1999, 74, 1–11. [Google Scholar] [CrossRef]
- Golec, P.; Wiczk, A.; Majchrzyk, A.; Łos, J.M.; Węgrzyn, G.; Łos, M. A role for accessory genes rI.-1 and rI.1 in the regulation of lysis inhibition by bacteriophage T4. Virus Genes 2010, 41, 459–468. [Google Scholar] [CrossRef]
- Freedman, M.L.; Krisch, R.E. Enlargement of Escherichia coli after bacteriophage infection I. description of phenomenon. J. Virol. 1971, 8, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Freedman, M.L.; Krisch, R.E. Enlargement of Escherichia coli after bacteriophage infection II. proposed mechanism. J. Virol. 1971, 8, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Yin, J. Impact of spatial structure on phage population growth. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 94–113. [Google Scholar]
- Symonds, N. The properties of a star mutant of phage T2. J. Gen. Microbiol. 1958, 18, 330–345. [Google Scholar] [CrossRef] [PubMed]
- Krylov, V.N. Star mutants of the bacteriophage T4B. Genetika 1971, 7, 112–119. [Google Scholar]
- Krylov, V.N.; Plotnikova, T.C. Genetic and physiological study of amber mutants in gene stII of T4B phage. Genetika 1972, 8, 85–95. [Google Scholar]
- Krylov, V.N.; Yankovsky, N.K. Mutations in the new gene stIlI of bacteriophage T4B suppressing the lysis defect of gene stIl and gene e mutants. J. Virol. 1975, 15, 22–26. [Google Scholar] [CrossRef]
- Delbrück, M. Bacterial viruses or bacteriophages. Biol. Rev. 1946, 21, 30–40. [Google Scholar] [CrossRef]
- Yin, J. A quantifiable phenotype of viral propagation. Biochem. Biophys. Res. Com. 1991, 174, 1009–1014. [Google Scholar] [CrossRef]
- Doermann, A.H. Lysis and lysis inhibition with Escherichia coli bacteriophage. J. Bacteriol. 1948, 55, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Hays, S.G.; Seed, K.D. Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite. eLife 2020, 9, e53200. [Google Scholar] [CrossRef] [PubMed]
- Sultan-Alolama, M.I.; Amin, A.; Vijayan, R.; El-Tarabily, K.A. Isolation, characterization, and comparative genomic analysis of bacteriophage Ec_MI-02 from pigeon feces infecting Escherichia coli O157:H7. Int. J. Mol. Sci. 2023, 24, 9506. [Google Scholar] [CrossRef] [PubMed]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating bacterial infections with bacteriophage-based enzybiotics: In vitro, in vivo and clinical application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef]
- Jacob, F.; Fuerst, C.R. The mechanism of lysis by phage studied with defective lysogenic bacteria. J. Gen. Microbiol. 1958, 18, 518–526. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Alreja, A.B.; Appel, A.E.; Zhu, J.C.; Riley, S.P.; Gonzalez-Juarbe, N.; Nelson, D.C. SP-CHAP, an endolysin with enhanced activity against biofilm pneumococci and nasopharyngeal colonization. mBio 2024, 15, e0006924. [Google Scholar] [CrossRef]
- Liu, H.; Wei, X.; Wang, Z.; Huang, X.; Li, M.; Hu, Z.; Zhang, K.; Hu, Q.; Peng, H.; Shang, W.; et al. LysSYL: A broad-spectrum phage endolysin targeting Staphylococcus species and eradicating S. aureus biofilms. Microb. Cell Fact. 2024, 23, 89. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Xu, H.; Bao, X.; Hong, W.; Wang, A.; Wang, K.; Dong, H.; Hou, J.; Govinden, R.; Deng, B.; Chenia, H.Y. Biological characterization and evolution of bacteriophage T7-Δholin during the serial passage process. Front. Microbiol. 2021, 12, 705310. [Google Scholar]
- Son, B.; Kong, M.; Lee, Y.; Ryu, S. Development of a novel chimeric endolysin, Lys109 with enhanced lytic activity against Staphylococcus aureus. Front. Microbiol. 2020, 11, 615887. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Nakonieczna, A.; Topolska-Wos, A.; Lobocka, M. New bacteriophage-derived lysins, LysJ and LysF, with the potential to control Bacillus anthracis. Appl. Microbiol. Biotechnol. 2024, 108, 76. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Chang, W.L.; Gutiérrez, D.; Lavigne, R.; Martínez, B.; Rodríguez, A.; Govers, S.K.; Aertsen, A.; Hirl, C.; Biebl, M.; et al. ‘Artilysation’ of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Sci. Rep. 2016, 6, 35382. [Google Scholar] [CrossRef]
- Koch, A.L. The growth of viral plaques during the enlargement phase. J. Theor. Biol. 1964, 6, 413–431. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage exploitation of bacterial biofilms: Phage preference for less mature targets? FEMS Microbiol. Lett. 2016, 363, fnv246. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophages and Biofilms: Ecology, Phage Therapy, Plaques; Nova Science Publishers: Hauppauge, NY, USA, 2011. [Google Scholar]
- Yin, J.; McCaskill, J.S. Replication of viruses in a growing plaque: A reaction-diffusion model. Biophys. J. 1992, 61, 1540–1549. [Google Scholar] [CrossRef]
- You, L.; Yin, J. Amplification and spread of viruses in a growing plaque. J. Theor. Biol. 1999, 200, 365–373. [Google Scholar] [CrossRef]
- Bryan, D.; El-Shibiny, A.; Hobbs, Z.; Porter, J.; Kutter, E.M. Bacteriophage T4 infection of stationary phase E. coli: Life after log from a phage perspective. Front. Microbiol. 2016, 7, 1391. [Google Scholar] [CrossRef]
- Hershey, A.D.; Rotman, R. Genetic recombination between host range and plaque-type mutants of bacteriophage in a single bacterial culture. Genetics 1949, 34, 44–71. [Google Scholar] [CrossRef] [PubMed]
- Rajnovic, D.; Munoz-Berbel, X.; Mas, J. Fast phage detection and quantification: An optical density-based approach. PLoS ONE 2019, 14, e0216292. [Google Scholar] [CrossRef] [PubMed]
- Schito, G.C. Development of coliphage N4: Ultrastructural studies. J. Virol. 1974, 13, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.; Ryu, S. Elucidation of molecular function of phage protein responsible for optimization of host cell lysis. BMC Microbiol. 2024, 24, 532. [Google Scholar] [CrossRef]
- Hershey, A.D. Mutation of bacteriophage with respect to type of plaque. Genetics 1946, 31, 620–640. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedon, S.T. Dual-Receptor Recognition, Lysis Inhibition, Endolysin Release, and Reaction–Diffusion as Alternative Explanations. Comment on Rojero et al. Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914. Int. J. Mol. Sci. 2025, 26, 11368. https://doi.org/10.3390/ijms262311368
Abedon ST. Dual-Receptor Recognition, Lysis Inhibition, Endolysin Release, and Reaction–Diffusion as Alternative Explanations. Comment on Rojero et al. Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914. International Journal of Molecular Sciences. 2025; 26(23):11368. https://doi.org/10.3390/ijms262311368
Chicago/Turabian StyleAbedon, Stephen T. 2025. "Dual-Receptor Recognition, Lysis Inhibition, Endolysin Release, and Reaction–Diffusion as Alternative Explanations. Comment on Rojero et al. Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914" International Journal of Molecular Sciences 26, no. 23: 11368. https://doi.org/10.3390/ijms262311368
APA StyleAbedon, S. T. (2025). Dual-Receptor Recognition, Lysis Inhibition, Endolysin Release, and Reaction–Diffusion as Alternative Explanations. Comment on Rojero et al. Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914. International Journal of Molecular Sciences, 26(23), 11368. https://doi.org/10.3390/ijms262311368






