Glutathione S-Transferase (GST) Activities and Gene Expression Patterns of Different GST Classes in Musca domestica L. Depending on Sex and Stage of Development
Abstract
1. Introduction
2. Results
2.1. No-Choice Feeding Bioassay
2.2. Glutathione-S-Transferase Activities
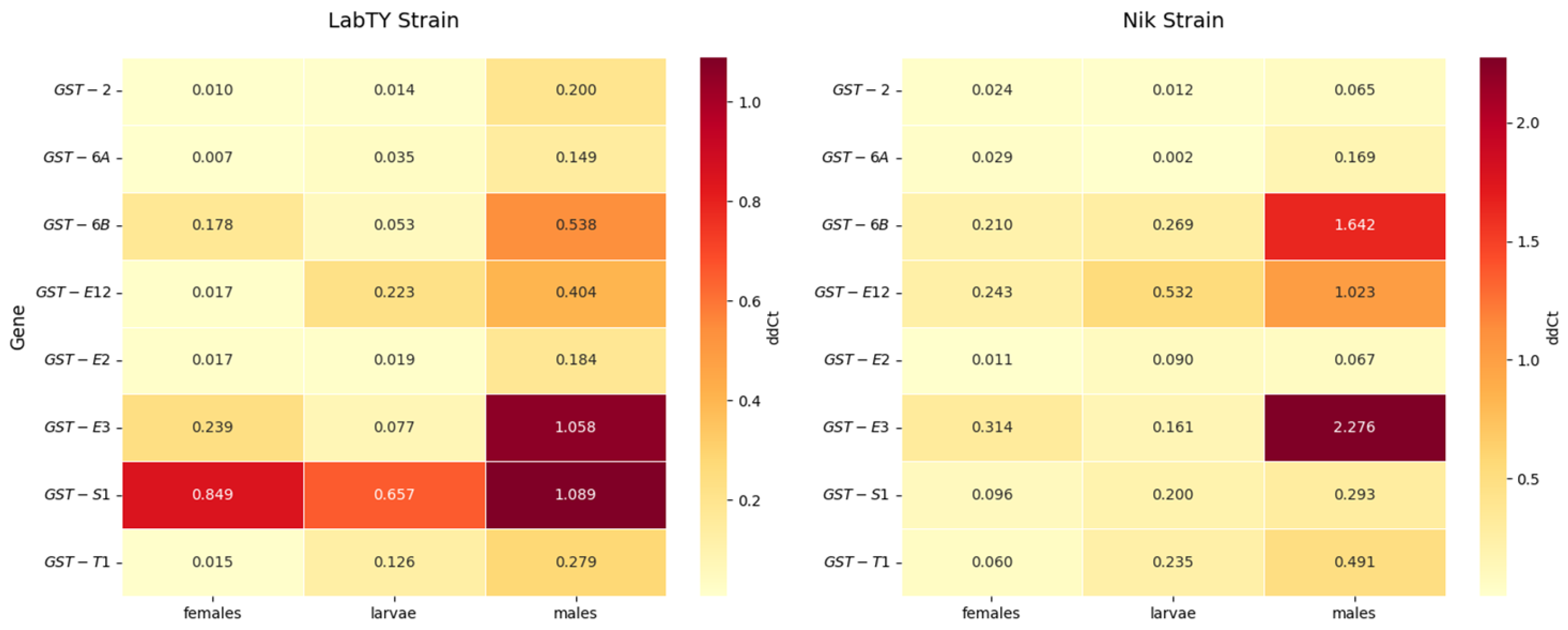
2.3. Gene Expression Levels of GST Isoforms
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. No-Choice Feeding Bioassay
4.3. Glutathione-S-Transferase Activity
4.4. Gene Expression Levels of GST Isoforms
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GSTs | Glutathione-S-transferases |
| LC50 | Lethal concentrations for 50% mortality |
| LC95 | Lethal concentrations for 95% mortality |
| RR | Resistance ratio |
| CI | Confidence intervals |
| WHO | World Health Organization |
| ddCt | Relative quantity of gene expression |
References
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef]
- Koirala, B.K.S.; Moural, T.; Zhu, F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int. J. Biol. Sci. 2022, 18, 5713–5723. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.H.; Clark, A.G.; Syvanen, M. Identification and cloning of a key insecticide-metabolizing glutathione S-transferase (MdGST-6A) from a hyper insecticide-resistant strain of the housefly Musca domestica. Insect Biochem. Mol. Biol. 2001, 31, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, J.A.; Fan, R.; Naz, H.; Bamisile, B.S.; Hafeez, M.; Ghani, M.I.; Wei, Y.; Xu, Y.; Chen, X. Insights into insecticide-resistance mechanisms in invasive species: Challenges and control strategies. Front. Physiol. 2023, 13, 1112278. [Google Scholar] [CrossRef]
- Nakamura, C.; Yajima, S.; Miyamoto, T.; Sue, M. Structural analysis of an epsilon-class glutathione transferase from housefly, Musca domestica. Biochem. Biophys. Res. Commun. 2013, 430, 1206–1211. [Google Scholar] [CrossRef] [PubMed]
- Nkya, T.E.; Akhouayri, I.; Kisinza, W.; David, J.P. Impact of environment on mosquito response to pyrethroid insecticides: Facts, evidences and prospects. Insect Biochem. Mol. Biol. 2013, 43, 407–416. [Google Scholar] [CrossRef]
- Scian, M.; Le Trong, I.; Mazari, A.M.; Mannervik, B.; Atkins, W.M.; Stenkamp, R.E. Comparison of epsilon- and delta-class glutathione S-transferases: The crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2089–2098. [Google Scholar] [CrossRef]
- Aloke, C.; Onisuru, O.O.; Achilonu, I. Glutathione S-transferase: A versatile and dynamic enzyme. Biochem. Biophys. Res. Commun. 2024, 734, 150774. [Google Scholar] [CrossRef]
- Venu, V.; Alias, Z. Substrate specificities and kinetic parameters of recombinant Drosophila melanogaster glutathione S-transferases E6 and E7. In Drosophila melanogaster—Model for Recent Advances in Genetics and Therapeutics; Perveen, F., Ed.; IntechOpen: London, UK, 2018; pp. 227–239. [Google Scholar] [CrossRef]
- Fang, S. Insect glutathione S-transferase: A review of comparative genomic studies and response to xenobiotics. Bull. Insectol. 2012, 65, 265–271. [Google Scholar]
- Friedman, R. Genomic organization of the glutathione S-transferase family in insects. Mol. Phylogenet. Evol. 2011, 61, 924–932. [Google Scholar] [CrossRef]
- Liu, S.; Rao, X.J.; Li, M.Y.; Feng, M.F.; He, M.Z.; Li, S.G. Glutathione S-transferase genes in the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae): Identi-fication and expression profiles. Arch. Insect Biochem. Physiol. 2015, 90, 1–13. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Z.; Lin, X.; Feng, Q.; Zheng, S. Structure and expression of glutathione S-transferase genes from the midgut of the Common cutworm, Spodoptera litura (Noctuidae) and their response to xenobiotic compounds and bacteria. J. Insect Physiol. 2011, 57, 1033–1044. [Google Scholar] [CrossRef]
- Yamamoto, K.; Aso, Y.; Yamada, N. Catalytic function of an ε-class glutathione S-transferase of the silkworm. Insect Mol. Biol. 2013, 22, 523–531. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.X.; Wang, W.L.; Zhang, B.X.; Li, S.G. Identification and characterisation of seventeen glutathione S-transferase genes from the cabbage white butterfly Pieris rapae. Pestic. Biochem. Physiol. 2017, 143, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Lumjuan, N.; Rajatileka, S.; Changsom, D.; Wicheer, J.; Leelapat, P.; Prapanthadara, L.A.; Somboon, P.; Lycett, G.; Ranson, H. The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem. Mol. Biol. 2011, 41, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hall, P.; Zhou, X.; Ranson, H.; Hemingway, J.; Meehan, E. Structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Liu, J.; Wang, W.; Mota-Sanchez, D.; He, S.; Shi, Y.; Yang, X. Glutathione S-transferase genes are involved in lambda-cyhalothrin resistance in Cydia pomonella via sequestration. J. Agric. Food Chem. 2022, 70, 2265–2279. [Google Scholar] [CrossRef]
- Jing, T.-X.; Wu, Y.-X.; Li, T.; Wei, D.-D.; Smagghe, G.; Wang, J.-J. Identification and expression profiles of fifteen delta-class glutathione S-transferase genes from a stored-product pest, Liposcelis entomophila (Enderlein) (Psocoptera: Liposcelididae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 206, 35–41. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Gao, H.; Guo, X.; Xu, B. Identification, genomic organization, and oxidative stress response of a sigma class glutathione S-transferase gene (AccGSTS1) in the honey bee, Apis cerana cerana. Cell Stress Chaperones. 2013, 18, 415–426. [Google Scholar] [CrossRef]
- Geden, C.J.; Nayduch, D.; Scott, J.G.; Burgess, E.R., IV; Gerry, A.C.; Kaufman, P.E.; Thomson, J.; Pickens, V.; Machtinger, E.T. House fly (Diptera: Muscidae): Biology, pest status, current management prospects, and research needs. J. Integr. Pest Manag. 2021, 12, 39. [Google Scholar] [CrossRef]
- You, C.; Li, Z.; Yin, Y.; Na, N.; Gao, X. Time of day-specific changes in metabolic detoxification and insecticide tolerance in the house fly, Musca domestica L. Front. Physiol. 2022, 12, 803682. [Google Scholar] [CrossRef]
- Nayduch, D.; Neupane, S.; Pickens, V.; Purvis, T.; Olds, C. House flies are underappreciated yet important reservoirs and vectors of microbial threats to animal and human health. Microorganisms 2023, 11, 583. [Google Scholar] [CrossRef]
- Scott, J.G.; Warren, W.C.; Beukeboom, L.W.; Bopp, D.; Clark, A.G.; Giers, S.D.; Hediger, M.; Jones, A.K.; Kasai, S.; Leichter, C.A.; et al. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014, 15, 466. [Google Scholar] [CrossRef] [PubMed]
- El Sherif, D.F.; Soliman, N.H.; Alshallash, K.S.; Ahmed, N.; Ibrahim, M.A.R.; Al-Shammery, K.A.; Al-Khalaf, A.A. The binary mixtures of lambda-cyhalothrin, chlorfenapyr, and abamectin, against the house fly larvae, Musca domestica L. Molecules 2022, 27, 3084. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G. Evolution of resistance to pyrethroid insecticides in Musca domestica. Pest Manag. Sci. 2017, 73, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Sue, M.; Mikawa, T.; Ueda, T.; Nomoto, Y.; Miyamoto, T. A novel function of housefly glutathione S-transferase 6B—Its effect on the retention and increase of insecticidal activity of the insecticide prothiofos. J. Pestic. Sci. 2006, 31, 139–145. [Google Scholar] [CrossRef]
- Sue, M.; Yajima, S. Crystal structure of the delta-class glutathione transferase in Musca domestica. Biochem. Biophys. Res. Commun. 2018, 502, 345–350. [Google Scholar] [CrossRef]
- Fournier, D.; Bride, J.M.; Poirie, M.; Bergé, J.B.; Plapp, F.W., Jr. Insect glutathione S-transferases. Biochemical characteristics of the major forms from houseflies susceptible and resistant to insecticides. J. Biol. Chem. 1992, 267, 1840–1845. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Ma, Z.; Shan, C.; Gao, X. Multiple mutations and overexpression of the MdaE7 carboxylesterase gene associated with male-linked malathion resistance in housefly, Musca domestica (Diptera: Muscidae). Sci. Rep. 2018, 8, 224. [Google Scholar] [CrossRef]
- Denlinger, D.S.; Hudson, S.B.; Keweshan, N.S.; Gompert, Z.; Bernhardt, S.A. Standing genetic variation in laboratory populations of insecticide-susceptible Phlebotomus papatasi and Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae) for the evolution of resistance. Evol. Appl. 2021, 14, 1248–1262. [Google Scholar] [CrossRef]
- St Pierre, S.E.; Ponting, L.; Stefancsik, R.; McQuilton, P.; FlyBase Consortium. FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014, 42, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, X.; Liang, P. Beta-cypermethrin resistance associated with high carboxylesterase activities in a strain of house fly, Musca domestica (Diptera: Muscidae). Pestic. Biochem. Physiol. 2007, 89, 65–72. [Google Scholar] [CrossRef]
- Aponte, A.; Penilla, R.P.; Rodríguez, A.D.; Ocampo, C.B. Mechanisms of pyrethroid resistance in Aedes (Stegomyia) aegypti from Colombia. Acta Trop. 2019, 191, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, M. Glutathione S-transferase and insecticide resistance in laboratory strains and field populations of Musca domestica. J. Econ. Entomol. 2005, 98, 1341–1348. [Google Scholar] [CrossRef]
- Ahmadi, E.; Khajehali, J.; Rameshgar, F. Evaluation of resistance to permethrin, cypermethrin and deltamethrin in different populations of Musca domestica (L.), collected from the Iranian dairy cattle farms. J. Asia Pac. Entomol. 2020, 23, 277–284. [Google Scholar] [CrossRef]
- Wang, K.Y.; Zhang, Y.; Wang, H.Y.; Xia, X.M. Role of glutathione-S-transferases in the resistance forming to insecticides of three different classes in housefly (Musca domestica). Resist. Pest Manag. Newsl. 2012, 21, 28. [Google Scholar]
- Sokolyanskaya, M.P. Development of pyrethroid resistance in larvae of housefly Musca domestica. Agrohimija 2014, 3, 54–59. [Google Scholar]
- Ramadan, M.M.; Selem, G.; Khater, K.S.; Elsobki, A. Monitoring of development of resistance to pyrethroids in Musca domestica L. population, using toxicological and biochemical features. Sci. J. Agric. Sci. 2021, 3, 219–229. [Google Scholar] [CrossRef]
- Kouamo, M.F.M.; Ibrahim, S.S.; Hearn, J.; Riveron, J.M.; Kusimo, M.; Tchouakui, M.; Ebai, T.; Tchapga, W.; Wondji, M.J.; Irving, H.; et al. Genome-wide transcriptional analysis and functional validation linked a cluster of epsilon glutathione S-transferases with insecticide resistance in the major malaria vector Anopheles funestus across Africa. Genes 2021, 12, 561. [Google Scholar] [CrossRef]
- Tao, F.; Si, F.L.; Hong, R.; He, X.; Li, X.Y.; Qiao, L.; He, Z.B.; Yan, Z.T.; He, S.L.; Chen, B. Glutathione S-transferase (GST) genes and their function associated with pyrethroid resistance in the malaria vector Anopheles sinensis. Pest Manag. Sci. 2022, 78, 4127–4139. [Google Scholar] [CrossRef] [PubMed]
- Waheibi, N.S. GST Detoxifying Enzymes and Pyrethroid Insecticide Resistance Evaluation in the Red Palm Weevil in the UAE. Master of Science Thesis, United Arab Emirates University, Al Ain, United Arab Emirates, 2019. [Google Scholar]
- Lumjuan, N.; McCarroll, L.; Prapanthadara, L.A.; Hemingway, J.; Ranson, H. Elevated activity of an Epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 861–871. [Google Scholar] [CrossRef]
- Chen, X.D.; Sandoval-Mojica, A.F.; Bonilla, S.I.; Ebert, T.A.; Gossett, H.; Pelz-Stelinski, K.S.; Stelinski, L.L. Fenpropathrin resistance in Asian citrus psyllid, Diaphorina citri Kuwayama: Risk assessment and changes in expression of CYP and GST genes associated with resistance. Int. J. Pest Manag. 2023, 69, 54–63. [Google Scholar] [CrossRef]
- Thornton, B.J. Sex-Dependent Changes in Activity of Detoxification Enzymes, Insecticide Susceptibility, and Alterations in Protein Expression Induced by Atrazine in Drosophila melanogaster. Ph.D. Dissertation, The University of Nebraska-Lincoln, Lincoln, NE, USA, 2009. [Google Scholar]
- Markussen, M.D.; Kristensen, M. Spinosad resistance in female Musca domestica L. from a field-derived population. Pest Manag. Sci. 2012, 68, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Ma, Z.; Zhai, D.; Gao, X.; Shi, X. Cytochrome P450 monooxygenases-mediated sex differential spinosad resistance in house flies Musca domestica (Diptera: Muscidae). Pestic. Biochem. Physiol. 2019, 157, 178–185. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, M.; Ma, Z.; You, C.; Gao, X.; Shi, X. Esterase mediated spinosad resistance in house flies Musca domestica (Diptera: Muscidae). Ecotoxicology 2020, 29, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Valle, D.; Montella, I.R.; Ribeiro, R.A.; Viana-Medeiros, P.F.; Martin, A.J., Jr.; Lima, J.B.P. Quantification Methodology for Enzyme Activity Related to Insecticide Resistance in Aedes aegypti; Ministry of Health of Brazil; Fundação Oswaldo Cruz: Brasília, Brazil, 2006; p. 128.
- Krestonoshina, K.; Melnichuk, A.; Kinareikina, A.; Maslakova, K.; Yangirova, L.; Silivanova, E. The P450-monooxygenase activity and CYP6D1 expression in the chlorfenapyr-resistant strain of Musca domestica L. Insects 2024, 15, 461. [Google Scholar] [CrossRef]
- Codd, V.; Dolezel, D.; Stehlik, J.; Piccin, A.; Garner, K.J.; Racey, S.N.; Straatman, K.R.; Louis, E.J.; Costa, R.; Sauman, I.; et al. Circadian rhythm gene regulation in the housefly Musca domestica. Genetics 2007, 177, 1539–1551. [Google Scholar] [CrossRef]
- Mekapogu, A.R. Finney’s Probit Analysis Spreadsheet Calculator (Version 2021). Available online: https://probitanalysis.wordpress.com/ (accessed on 29 September 2025).


| Insecticide | Strain | N | LC50 (95% CI) a | LC50 (95% CI) a | Slope (±SE) | χ2 | RR (95% CI) |
|---|---|---|---|---|---|---|---|
| Chlorpyrifos | LabTY ♀ | 195 | 12.227 (8.055–18.561) | 84.627 (55.748–128.465) | 2.19 (±0.09) | 0.21 | - |
| LabTY ♂ | 195 | 7.592 (5.479–10.520) | 25.043 (18.073–34.702 | 3.21 (±0.07) | 0.32 | - | |
| Nik ♀ | 195 | 14.962 (11.497–19.472) | 32.653 (25.091–42.494) | 4.89 (±0.06) | 0.52 | 1.22 (1.05–1.43) | |
| Nik ♂ | 195 | 6.363 (4.794–8.445) # | 15.659 (11.798–20.782) | 4.27 (±0.06) | 0.85 | 0.84 (0.80–0.88) | |
| Deltamethrin | LabTY ♀ | 240 | 2.078 (1.295–3.334) | 26.880 (16.755–43.123) | 1.65 (±0.11) | 0.17 | - |
| LabTY ♂ | 233 | 1.004 (0.688–1.465) | 5.240 (3.591–7.647) | 2.40 (±0.08) | 0.61 | - | |
| Nik ♀ | 195 | 5.707 (3.811–8.545) * | 24.012 (16.037–35.953) | 2.64 (±0.09) | 0.88 | 2.75 (2.56–2.95) | |
| Nik ♂ | 195 | 6.730 (4.036–11.224) * | 56.806 (34.065–94.728) | 1.78 (±0.11) | 0.98 | 6.73 (5.85–7.68) | |
| Chlorfenapyr | LabTY ♀ | 384 | 23.625 (17.385–32.103) | 96.306 (70.872–130.868) | 2.72 (±0.06) | 0.05 | - |
| LabTY ♂ | 401 | 9.745 (7.320–12.975) # | 58.781 (44.149–78.262) | 1.64 (±0.10) | 0.72 | - | |
| Nik ♀ | 230 | 27.609 (19.906–38.293) | 139.574 (100.632–193.584) | 2.44 (±0.07) | 0.55 | 1.17 (1.15–1.19) | |
| Nik ♂ | 230 | 14.332 (10.526–19.513) # | 55.686 (40.900–75.817) | 2.84 (±0.07) | 0.87 | 1.47 (1.44–1.50) |
| GST Class | D. melanogaster | M. domestica | |||
|---|---|---|---|---|---|
| Protein Accession | Gene ID | mRNA Accession | Gene Name | ||
| Epsilon | DMGSTE4-8 | XP005185166 | LOC101895036 | XM_005185109.4 | GST-2 |
| XP005185169 | LOC101895555 | XM_005185112.4 | GST-6B | ||
| XP005190027 | LOC101887250 | XM_005189970.4 | GST-1 | ||
| NP_001295979.2 | - | NM_001309050.2 | GST-6A | ||
| DMGSTE13A-B | XP005179508 | LOC101900016 | XM_005179451.4 | GST-1 | |
| XP005179509 | LOC109611606 | XM_005179452.4 | GST-1 | ||
| DMGSTE11 | XP005184662 | LOC101895316 | XM_005184605.4 (X1) | GST-E11 | |
| XP005184663 | XM_005184606.4 (X2) | ||||
| XP005184664 | LOC101895607 | XM_005184607.4 (X1) | GST-1 | ||
| DMGSTE14 | XP005180753 | LOC101888181 | XM_005180696.4 | GST1-like | |
| XP005180754 | LOC101888349 | XM_005180697.4 | GST1-like | ||
| Delta | DMGSTD11A-B | XP005180103 | LOC101897797 | XM_005180046.4 (X1) | GST-D11 |
| DMGSTD10, 1A-B | XP005180099 | LOC101897094 | XM_005180042.4 (X1) | GST4-like | |
| NP_001295926.1 | LOC101897277 | NM_001308997.1 | GST-2 | ||
| Theta | DMGSTT4 | XP005191555 | LOC101897781 | XM_005191498.4 | GST-T1 |
| DMGSTT3A-B | XP005177600 | LOC101900949 | XM_005177543.4 | GST-T3 | |
| Sigma | DMGSTS1 | NP_001273827.1 | - | NM_001286898.1 | GST |
| Gene Name | GeneID | Nucleotide Sequence (5′→3′) of Primers (Forward/Reverse) | Ta, ° | Length of PCR Product (bp) | GenBank ID |
|---|---|---|---|---|---|
| RP49 * | LOC101894827 | GTTATGCCAAATTGTCGCACA GGCGGGTACGTTTGTTGG | 59.5 | 123 | XM_020038490.2 |
| EF-1 | LOC101899175 | TAAGGAAGGTAACGCTGAAGG CAAGGGCAAACGCAAAGG | 59.5 | 91 | XM_005181459.4 |
| GST-6A | 101887423 | ATTCGACGACAAAATGGG CCTTAGCCATCAAATTAACC | 61.5 | 131 | NM_001309050.2 |
| GST-6B | LOC131804118 | ACCTGTTCGTGCTTGTTTGC CGAGTGTGGGCACTGTATGT | 63.5 | 137 | XM_059126509.1 |
| GST-S1 (GST) | 101890455 | TGGAAGTTAACGGCAAGCGT CCGGCAAAGTAGACATCGGC | 61.5 | 313 | NM_001286898.1 |
| GST-T1 | LOC101897781 | GTGTAGCCATATTCCGCCACT GCATCGTACCATTTTGAGAGCT | 59.5 | 391 | XM_005191498.4 |
| GST-E2 (GST-2) | LOC101895036 | AAGCGATCACAACAGCCAAC CATCGTCTTCCAGTGTAGGCA | 61.5 | 240 | XM_005185109.4 |
| GST-E3 (GST-6B) | LOC101895555 | ATCCCCAACATACAGTGCCC TCCCTGAAATAAGACACCAGCT | 61.5 | 182 | XM_005185112.4 |
| GST-2 | LOC101897277 | AGAACGGACAACAAGTAGCTCC AATGGCACGAGATTCCCACA | 59.5 | 247 | NM_001308997.1 |
| GST-E12 | LOC101900672 | GGTCACTTGTTTGCCCGTCT GCACCCTCCTCGTTTGTATCC | 61.5 | 302 | XM_059131016.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garbaly, V.; Krestonoshina, K.; Kinareikina, A.; Bobreshova, S.; Avdeeva, A.; Ismagilova, J.; Zaitsev, I.; Silivanova, E. Glutathione S-Transferase (GST) Activities and Gene Expression Patterns of Different GST Classes in Musca domestica L. Depending on Sex and Stage of Development. Int. J. Mol. Sci. 2025, 26, 11366. https://doi.org/10.3390/ijms262311366
Garbaly V, Krestonoshina K, Kinareikina A, Bobreshova S, Avdeeva A, Ismagilova J, Zaitsev I, Silivanova E. Glutathione S-Transferase (GST) Activities and Gene Expression Patterns of Different GST Classes in Musca domestica L. Depending on Sex and Stage of Development. International Journal of Molecular Sciences. 2025; 26(23):11366. https://doi.org/10.3390/ijms262311366
Chicago/Turabian StyleGarbaly, Vladislava, Kseniya Krestonoshina, Anna Kinareikina, Svetlana Bobreshova, Anastasiya Avdeeva, Juliya Ismagilova, Ivan Zaitsev, and Elena Silivanova. 2025. "Glutathione S-Transferase (GST) Activities and Gene Expression Patterns of Different GST Classes in Musca domestica L. Depending on Sex and Stage of Development" International Journal of Molecular Sciences 26, no. 23: 11366. https://doi.org/10.3390/ijms262311366
APA StyleGarbaly, V., Krestonoshina, K., Kinareikina, A., Bobreshova, S., Avdeeva, A., Ismagilova, J., Zaitsev, I., & Silivanova, E. (2025). Glutathione S-Transferase (GST) Activities and Gene Expression Patterns of Different GST Classes in Musca domestica L. Depending on Sex and Stage of Development. International Journal of Molecular Sciences, 26(23), 11366. https://doi.org/10.3390/ijms262311366







