Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR in Tuberculomas and Peripheral Blood Mononuclear Cells for Pulmonary Tuberculosis Patients
Abstract
1. Introduction
2. Results
2.1. Primer Validation
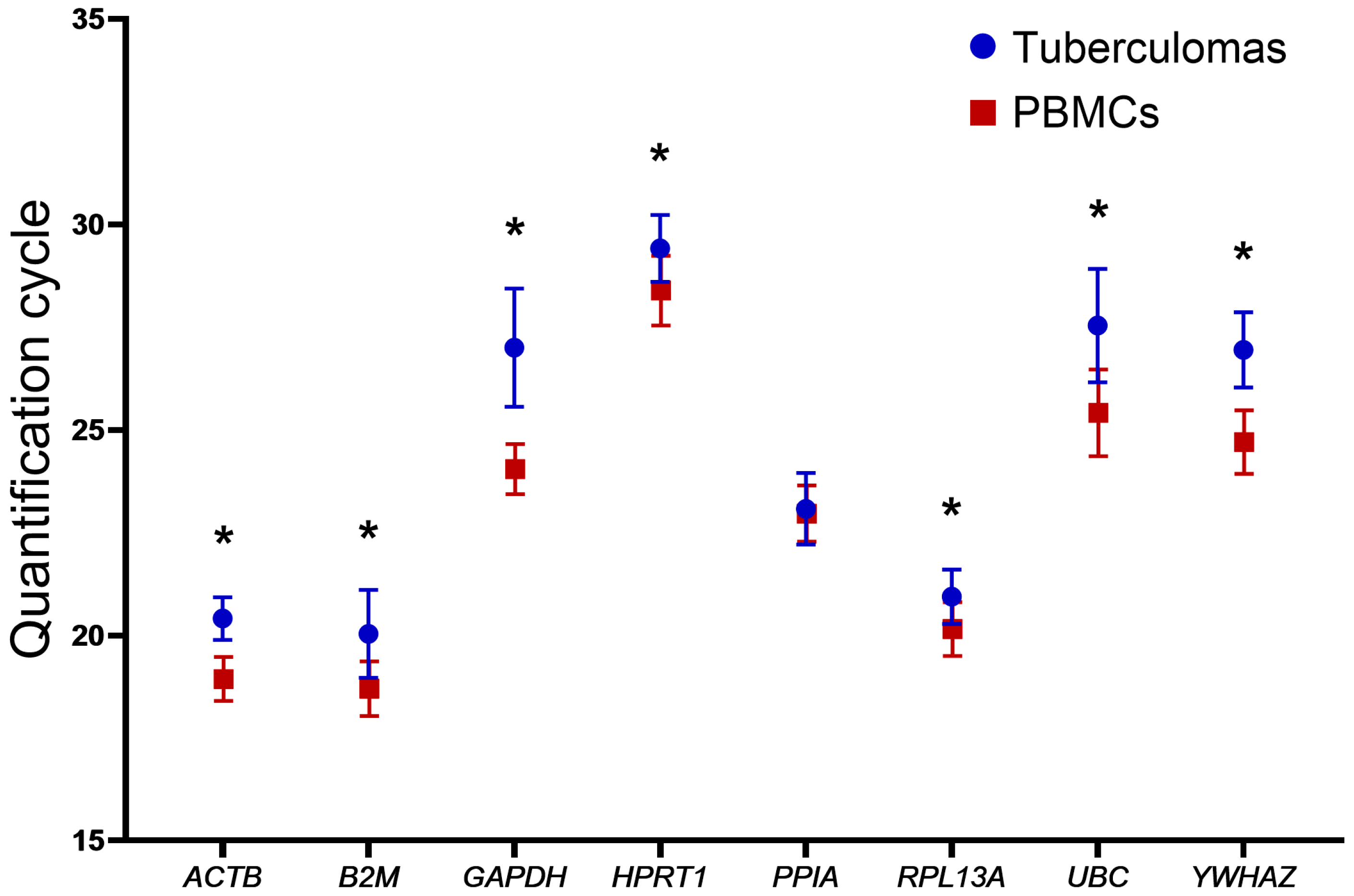
2.2. Quantification Cycle (Cq) Stability Analysis of Genes in Groups “Tuberculomas” and “PBMCs”
2.3. Ranking Gene-Expression Stability in Tuberculoma Tissue and PBMCs Using the geNorm, NormFinder, BestKeeper and Delta CT Algorithms
2.3.1. Ranking Selected Housekeeping Genes Using the geNorm Analysis
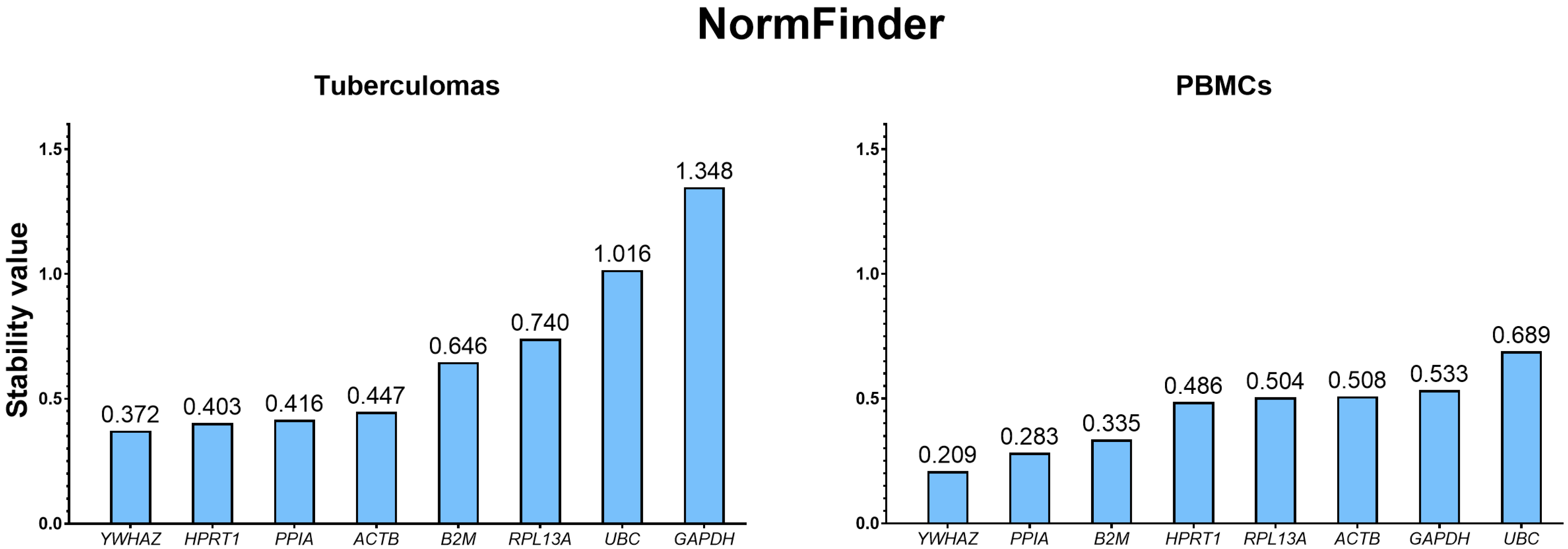
2.3.2. Ranking of Selected Housekeeping Genes Using the NormFinder Analysis
2.3.3. Ranking of Selected Housekeeping Genes Using the Delta CT Analysis
2.3.4. Ranking of Selected Housekeeping Genes Using the BestKeeper Analysis
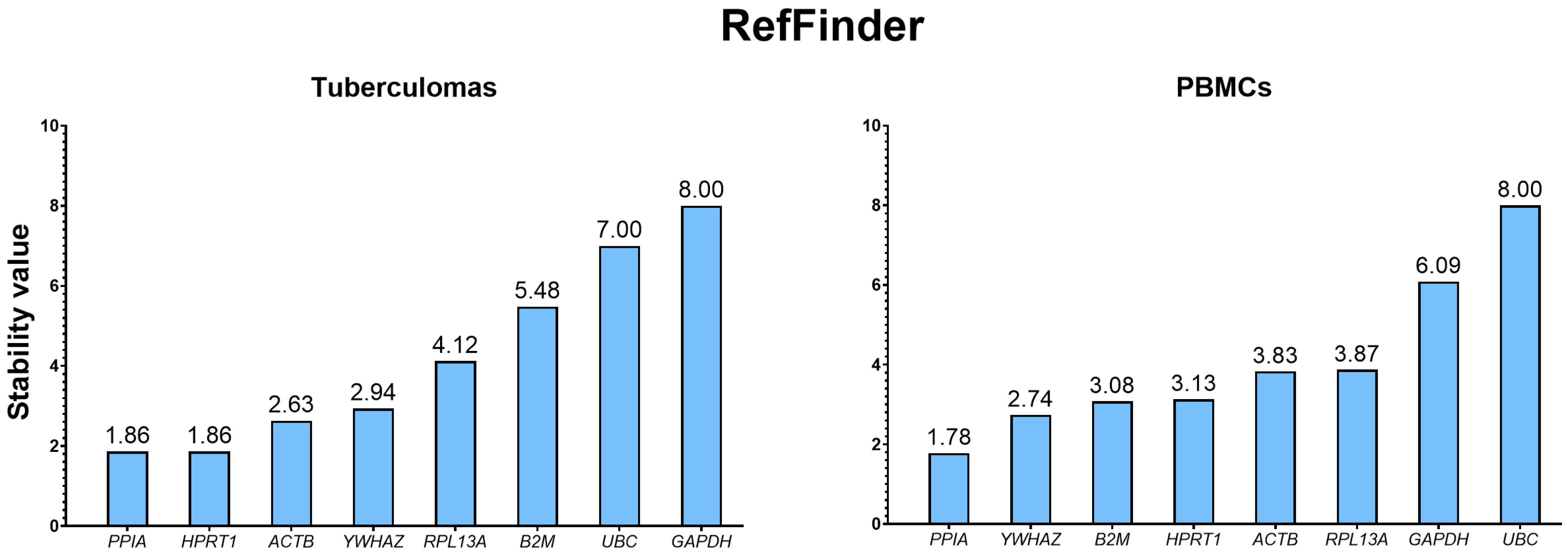
2.3.5. Integrated Ranking Based on RefFinder
2.4. Correlation Analysis of Selected Housekeeping Genes in Groups “Tuberculomas” and “PBMCs”
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Lung Tissue Sampling
4.3. Isolation of Peripheral Blood Mononuclear Cells
4.4. Total RNA Extraction, RNA Quantity, Purity and Integrity Analysis and cDNA Synthesis
4.5. Quantitative Real-Time PCR
4.6. Primer Characteristics
4.7. Assessment of Reference-Gene Stability
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PBMCs | peripheral blood mononuclear cells |
| ACTB | β-actin |
| B2M | β2-microglobulin |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| HPRT1 | hypoxanthine phosphoribosyl-transferase 1 |
| PPIA | peptidylprolyl isomerase A |
| RPL13A | ribosomal protein L13a |
| UBC | ubiquitin C |
| YWHAZ | tyrosine 3-monooxygenase tryptophan 5-monooxygenase activation protein |
References
- Abbasnia, S.; Hajimiri, S.; Jafari Rad, M.; Ariaee, N.; Mosavat, A.; Hashem Asnaashari, A.M.; Derakhshan, M.; Amel Jamehdar, S.; Ghazvini, K.; Mohammadi, F.S.; et al. Gene Expression Study of Host and Mycobacterium Tuberculosis Interactions in the Manifestation of Acute Tuberculosis. Appl. Biochem. Biotechnol. 2023, 195, 3641–3652. [Google Scholar] [CrossRef]
- Pavlova, E.N.; Lepekha, L.N.; Rybalkina, E.Y.; Tarasov, R.V.; Sychevskaya, K.A.; Voronezhskaya, E.E.; Masyutin, A.G.; Ergeshov, A.E.; Erokhina, M.V. High and Low Levels of ABCB1 Expression Are Associated with Two Distinct Gene Signatures in Lung Tissue of Pulmonary TB Patients with High Inflammation Activity. Int. J. Mol. Sci. 2023, 24, 14839. [Google Scholar] [CrossRef]
- Hai, H.T.; Thanh Hoang Nhat, L.; Tram, T.T.B.; Vinh, D.D.; Nath, A.P.; Donovan, J.; Thu, N.T.A.; Van Thanh, D.; Bang, N.D.; Ha, D.T.M.; et al. Whole Blood Transcriptional Profiles and the Pathogenesis of Tuberculous Meningitis. eLife 2024, 13, RP92344. [Google Scholar] [CrossRef]
- Wang, L.-H.; Xu, M.-L. Non-Invasive Diagnosis of Pulmonary Tuberculosis and Predictive Potential for Treatment Outcomes via miR-146a and miR-155 Levels. Diagn. Microbiol. Infect. Dis. 2025, 112, 116795. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Angulo, J.M.; Nogueira, B.M.F.; Arriaga, M.B.; Barreto-Duarte, B.; Araújo-Pereira, M.; Fernandes, C.D.; Vinhaes, C.L.; Villalva-Serra, K.; Nunes, V.M.; Miguez-Pinto, J.P.; et al. Host-Directed Therapies in Pulmonary Tuberculosis: Updates on Anti-Inflammatory Drugs. Front. Med. 2022, 9, 970408. [Google Scholar] [CrossRef] [PubMed]
- Wallis, R.S.; Ginindza, S.; Beattie, T.; Arjun, N.; Likoti, M.; Sebe, M.; Edward, V.A.; Rassool, M.; Ahmed, K.; Fielding, K.; et al. Lung and Blood Early Biomarkers for Host-Directed Tuberculosis Therapies: Secondary Outcome Measures from a Randomized Controlled Trial. PLoS ONE 2022, 17, e0252097. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xin, T.; Liu, Z.; Wang, Q.; Ma, L. Construction of ceRNA Regulatory Networks for Active Pulmonary Tuberculosis. Sci. Rep. 2024, 14, 10595. [Google Scholar] [CrossRef]
- Jhilmeet, N.; Lowe, D.M.; Riou, C.; Scriba, T.J.; Coussens, A.; Goliath, R.; Wilkinson, R.J.; Wilkinson, K.A. The Effect of Antiretroviral Treatment on Selected Genes in Whole Blood from HIV-Infected Adults Sensitised by Mycobacterium Tuberculosis. PLoS ONE 2018, 13, e0209516. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.; Yang, X.; Kuang, H.; Feng, T.; Deng, C.; Li, X.; Ye, M.; Tan, X.; Gong, L.; et al. Differential Gene Expression in PBMCs: Insights into the Mechanism How Pulmonary Tuberculosis Increases Lung Cancer Risk. Gene 2025, 940, 149199. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Potashnikova, D.; Gladkikh, A.; Vorobjev, I.A. Selection of Superior Reference Genes’ Combination for Quantitative Real-Time PCR in B-Cell Lymphomas. Ann. Clin. Lab. Sci. 2015, 45, 64–72. [Google Scholar]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Banfi, G.; Lombardi, G. Free Circulating miRNAs Measurement in Clinical Settings. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2018; Volume 87, pp. 113–139. ISBN 978-0-12-815203-4. [Google Scholar]
- Da Conceição Braga, L.; Gonçalves, B.Ô.P.; Coelho, P.L.; Da Silva Filho, A.L.; Silva, L.M. Identification of Best Housekeeping Genes for the Normalization of RT-qPCR in Human Cell Lines. Acta Histochem. 2022, 124, 151821. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR. BioTechniques 2004, 37, 112–119. [Google Scholar] [CrossRef]
- Gu, W.; Tu, X.; Lu, W.; Yin, Y.; Meng, Q.; Wang, X.; Zhang, F.; Fu, Y. Identification of RNU44 as an Endogenous Reference Gene for Normalizing Cell-Free RNA in Tuberculosis. Open Forum Infect. Dis. 2022, 9, ofac540. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A Web-Based Tool for Comprehensively Analyzing and Identifying Reference Genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Toorani, T.; Mackie, P.M.; Mastromonaco, G.F. Validation of Reference Genes for Use in Untreated Bovine Fibroblasts. Sci. Rep. 2021, 11, 10253. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Furukawa, T.; Plotkin, S.A. Human Cytomegalovirus Stimulates Host Cell RNA Synthesis. J. Virol. 1975, 15, 297–304. [Google Scholar] [CrossRef]
- Piechaczyk, M.; Blanchard, J.M.; Marty, L.; Dani, C.; Panabieres, F.; El Sabouty, S.; Fort, P.; Jeanteur, P. Post-Transcrptional Regulation of Glycenadehyde-3-Phosphate-Dehydrogenase Gene Expression in Rat Tissues. Nucleic Acids Res. 1984, 12, 6951. [Google Scholar] [CrossRef]
- Stout, J.T.; Chen, H.Y.; Brennand, J.; Caskey, C.T.; Brinster, R.L. Expression of Human HPRT in the Central Nervous System of Transgenic Mice. Nature 1985, 317, 250–252. [Google Scholar] [CrossRef]
- Bémeur, C.; Ste-Marie, L.; Desjardins, P.; Hazell, A.S.; Vachon, L.; Butterworth, R.; Montgomery, J. Decreased β-Actin mRNA Expression in Hyperglycemic Focal Cerebral Ischemia in the Rat. Neurosci. Lett. 2004, 357, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Selvey, S.; Thompson, E.W.; Matthaei, K.; Lea, R.A.; Irving, M.G.; Griffiths, L.R. β-Actin—An Unsuitable Internal Control for RT-PCR. Mol. Cell. Probes 2001, 15, 307–311. [Google Scholar] [CrossRef]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a Housekeeping Gene: Analysis of GAPDH mRNA Expression in a Panel of 72 Human Tissues. Physiol. Genom. 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Franko, N.; Vrščaj, L.A.; Zore, T.; Ostanek, B.; Marc, J.; Lojk, J. TBP, PPIA, YWHAZ and EF1A1 Are the Most Stably Expressed Genes during Osteogenic Differentiation. Int. J. Mol. Sci. 2022, 23, 4257. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yao, B.; Zhang, H.; Guo, H.; Hu, D.; Wang, Q.; Zhao, Y. Identification of Novel Reference Genes Using Sika Deer Antler Transcriptome Expression Data and Their Validation for Quantitative Gene Expression Analysis. Genes Genom. 2014, 36, 573–582. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Bustin, S.A.; Ruijter, J.M.; van den Hoff, M.J.B.; Kubista, M.; Pfaffl, M.W.; Shipley, G.L.; Tran, N.; Rodiger, S.; Untergasser, A.; Mueller, R.; et al. MIQE 2.0: Revision of the Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines. Clin. Chem. 2025, 71, 634–651. [Google Scholar] [CrossRef]
- Montero-Melendez, T.; Perretti, M. Gapdh Gene Expression Is Modulated by Inflammatory Arthritis and Is Not Suitable for qPCR Normalization. Inflammation 2014, 37, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, B.; Zhou, Y.; Wang, X.; Wu, W.; Wang, Z.; Dai, Z.; Cheng, Q.; Yang, K. B2M Overexpression Correlates with Malignancy and Immune Signatures in Human Gliomas. Sci. Rep. 2021, 11, 5045. [Google Scholar] [CrossRef]
- Chiou, S.-J.; Ko, H.-J.; Hwang, C.-C.; Hong, Y.-R. The Double-Edged Sword of Beta2-Microglobulin in Antibacterial Properties and Amyloid Fibril-Mediated Cytotoxicity. Int. J. Mol. Sci. 2021, 22, 6330. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; Marchese, M.; Hellemans, J.; Betsou, F.; Skov Frisk, N.L.; Dalgaard, L.T.; Lakkisto, P.; Foy, C.; Scherer, A.; Garcia Bermejo, M.L.; et al. Consensus Guidelines for the Validation of qRT-PCR Assays in Clinical Research by the CardioRNA Consortium. Mol. Ther.—Methods Clin. Dev. 2022, 24, 171–180. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference Genes in Real-Time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Geigges, M.; Gubser, P.M.; Unterstab, G.; Lecoultre, Y.; Paro, R.; Hess, C. Reference Genes for Expression Studies in Human CD8+ Naïve and Effector Memory T Cells under Resting and Activating Conditions. Sci. Rep. 2020, 10, 9411. [Google Scholar] [CrossRef]
- Jeon, R.-H.; Lee, W.-J.; Son, Y.-B.; Bharti, D.; Shivakumar, S.B.; Lee, S.-L.; Rho, G.-J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of mRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. BioMed Res. Int. 2019, 2019, 3093545. [Google Scholar] [CrossRef]
- Guaita-Cespedes, M.; Grillo-Risco, R.; Hidalgo, M.R.; Fernández-Veledo, S.; Burks, D.J.; De La Iglesia-Vayá, M.; Galán, A.; Garcia-Garcia, F. Deciphering the Sex Bias in Housekeeping Gene Expression in Adipose Tissue: A Comprehensive Meta-Analysis of Transcriptomic Studies. Biol. Sex Differ. 2023, 14, 20. [Google Scholar] [CrossRef]
- Koshechkin, V.A. Phthisiatry: Textbook; GEOTAR-Media: Moscow, Russia, 2019; ISBN 978-5-9704-7329-0. [Google Scholar]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A Flexible Statistical Power Analysis Program for the Social, Behavioral, and Biomedical Sciences. Behav. Res. Methods. 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]




| Gene | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|
| PPIA † | GTTTATGTGTCAGGGTGGTG | CGTATGCTTTAGGATGAAGTTCTC | 103 |
| B2M † | GGGTTTCATCCATCCGACATTG | ACACGGCAGGCATACTCATCTTTT | 161 |
| ACTB †† | CTGGAACGGTGAAGGTGACA | AAGGGACTTCCTGTAACAATGCA | 140 |
| GAPDH †† | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG | 87 |
| HPRT1 †† | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT | 94 |
| RPL13A †† | CCTGGAGGAGAAGAGGAAAGAGA | TTGAGGACCTCTGTGTATTTGTCAA | 126 |
| UBC †† | ATTTGGGTCGCAGTTCTTG | TGCCTTGACATTCTCGATGGT | 133 |
| YWHAZ †† | ACTTTTGGTACATTGTGGCTTCAA | CCGCCAGGACAAACCAGTAT | 94 |
| Gene Symbol | Gene Name | Slope | Efficiency, % | R2 |
|---|---|---|---|---|
| ACTB | β-actin | −3.3863 | 97.4 | 0.988 |
| B2M | β2-microglobulin | −3.5406 | 91.6 | 0.999 |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase | −3.3093 | 100.5 | 0.996 |
| HPRT1 | hypoxanthine phosphoribosyl-transferase 1 | −3.4958 | 93.2 | 0.998 |
| PPIA | peptidylprolyl isomerase A | −3.3265 | 99.8 | 0.998 |
| RPL13A | ribosomal protein L13a | −3.3978 | 96.9 | 0.996 |
| UBC | ubiquitin C | −3.3656 | 98.2 | 0.999 |
| YWHAZ | tyrosine 3-monooxygenase tryptophan 5-monooxygenase activation protein | −3.4538 | 94.8 | 0.996 |
| Genes | ACTB | B2M | GAPDH | HPRT1 | PPIA | RPL13A | UBC | YWHAZ |
|---|---|---|---|---|---|---|---|---|
| CqMean in tuberculomas | 20.41 | 20.04 | 27.01 | 29.42 | 23.08 | 20.94 | 27.54 | 26.95 |
| SD in tuberculomas | 0.518 | 1.071 | 1.437 | 0.811 | 0.872 | 0.661 | 1.379 | 0.913 |
| CqMean in PBMCs | 18.94 | 18.70 | 24.05 | 28.40 | 22.97 | 20.16 | 25.42 | 24.70 |
| SD in PBMCs | 0.534 | 0.662 | 0.612 | 0.849 | 0.682 | 0.656 | 1.055 | 0.773 |
| Difference in CqMean † | 1.47 | 1.34 | 2.96 | 1.02 | 0.11 | 0.78 | 2.12 | 2.25 |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0005 | 0.6580 | 0.0008 | <0.0001 | <0.0001 |
| Genes | CV | Standard Deviation | Correlation Coefficient | p-Value |
|---|---|---|---|---|
| ACTB | 1.78 | 0.36 | 0.777 | 0.001 |
| RPL13A | 2.39 | 0.5 | 0.542 | 0.011 |
| HPRT1 | 2.11 | 0.62 | 0.863 | 0.001 |
| PPIA | 3.04 | 0.7 | 0.884 | 0.001 |
| YWHAZ | 2.65 | 0.71 | 0.893 | 0.001 |
| B2M | 4.28 | 0.86 | 0.855 | 0.001 |
| UBC | 4.1 | 1.13 | 0.759 | 0.001 |
| GAPDH | 4.37 | 1.18 | 0.513 | 0.017 |
| Genes | CV | Standard Deviation | Correlation Coefficient | p-Value |
|---|---|---|---|---|
| ACTB | 2.3 | 0.44 | 0.683 | 0.003 |
| B2M | 2.43 | 0.45 | 0.873 | 0.001 |
| RPL13A | 2.36 | 0.48 | 0.723 | 0.001 |
| GAPDH | 2.05 | 0.49 | 0.678 | 0.003 |
| PPIA | 2.26 | 0.52 | 0.895 | 0.001 |
| HPRT1 | 2.25 | 0.64 | 0.853 | 0.001 |
| YWHAZ | 2.62 | 0.65 | 0.961 | 0.001 |
| UBC | 3.34 | 0.85 | 0.857 | 0.001 |
| Sample Group | Rank | geNorm | NormFinder | Delta CT | |||
|---|---|---|---|---|---|---|---|
| Gene | M Value | Gene | Stability Value | Gene | Stability Value | ||
| Tuberculomas | 1 | HPRT1/PPIA | 0.358 | YWHAZ | 0.372 | PPIA | 0.77 |
| 2 | HPRT1 | 0.403 | HPRT1 | 0.78 | |||
| 3 | ACTB | 0.499 | PPIA | 0.416 | YWHAZ | 0.82 | |
| 4 | RPL13A | 0.533 | ACTB | 0.447 | ACTB | 0.84 | |
| 5 | YWHAZ | 0.574 | B2M | 0.646 | B2M | 0.93 | |
| 6 | B2M | 0.622 | RPL13A | 0.740 | RPL13A | 0.95 | |
| 7 | UBC | 0.808 | UBC | 1.016 | UBC | 1.22 | |
| 8 | GAPDH | 0.971 | GAPDH | 1.348 | GAPDH | 1.46 | |
| PBMCs | 1 | HPRT1/PPIA | 0.251 | YWHAZ | 0.209 | PPIA | 0.52 |
| 2 | PPIA | 0.283 | YWHAZ | 0.53 | |||
| 3 | RPL13A | 0.302 | B2M | 0.335 | B2M | 0.58 | |
| 4 | YWHAZ | 0.383 | HPRT1 | 0.486 | HPRT1 | 0.62 | |
| 5 | B2M | 0.459 | RPL13A | 0.504 | RPL13A | 0.64 | |
| 6 | ACTB | 0.533 | ACTB | 0.508 | ACTB | 0.67 | |
| 7 | GAPDH | 0.576 | GAPDH | 0.533 | GAPDH | 0.68 | |
| 8 | UBC | 0.630 | UBC | 0.689 | UBC | 0.79 | |
| Sample Group | Rank | BestKeeper | RefFinder | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SD | Gene | CV | Gene | Correlation Coefficient | Gene | Geomean | ||
| Tuberculomas | 1 | ACTB | 0.36 | ACTB | 1.78 | YWHAZ | 0.893 | PPIA | 1.86 |
| 2 | RPL13A | 0.5 | HPRT1 | 2.11 | PPIA | 0.884 | HPRT1 | 1.86 | |
| 3 | HPRT1 | 0.62 | RPL13A | 2.39 | HPRT1 | 0.863 | ACTB | 2.63 | |
| 4 | PPIA | 0.7 | YWHAZ | 2.65 | B2M | 0.855 | YWHAZ | 2.94 | |
| 5 | YWHAZ | 0.71 | PPIA | 3.04 | ACTB | 0.777 | RPL13A | 4.12 | |
| 6 | B2M | 0.86 | UBC | 4.1 | UBC | 0.759 | B2M | 5.48 | |
| 7 | UBC | 1.13 | B2M | 4.28 | RPL13A | 0.542 | UBC | 7.00 | |
| 8 | GAPDH | 1.18 | GAPDH | 4.37 | GAPDH | 0.513 | GAPDH | 8.00 | |
| PBMCs | 1 | ACTB | 0.44 | GAPDH | 2.05 | YWHAZ | 0.961 | PPIA | 1.78 |
| 2 | B2M | 0.45 | HPRT1 | 2.25 | PPIA | 0.895 | YWHAZ | 2.74 | |
| 3 | RPL13A | 0.48 | PPIA | 2.26 | B2M | 0.873 | B2M | 3.08 | |
| 4 | GAPDH | 0.49 | ACTB | 2.3 | UBC | 0.857 | HPRT1 | 3.13 | |
| 5 | PPIA | 0.52 | RPL13A | 2.36 | HPRT1 | 0.853 | ACTB | 3.83 | |
| 6 | HPRT1 | 0.64 | B2M | 2.43 | RPL13A | 0.723 | RPL13A | 3.87 | |
| 7 | YWHAZ | 0.65 | YWHAZ | 2.62 | ACTB | 0.683 | GAPDH | 6.09 | |
| 8 | UBC | 0.85 | UBC | 3.34 | GAPDH | 0.678 | UBC | 8.00 | |
| Tuberculomas | Categories | n | Percent, % |
| Gender | M | 10 | 48 |
| F | 11 | 52 | |
| Therapy duration, months | 2–6 | 6 | 29 |
| 7–12 | 4 | 19 | |
| >12 | 11 | 52 | |
| PBMCs | Categories | n | Percent, % |
| Gender | M | 9 | 53 |
| F | 8 | 47 | |
| Therapy duration, months | 2–6 | 8 | 47 |
| 7–12 | 7 | 41 | |
| >12 | 2 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarasova, E.K.; Pavlova, E.N.; Rybalkina, E.Y.; Scherbakova, E.A.; Tarasov, R.V.; Erokhina, M.V. Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR in Tuberculomas and Peripheral Blood Mononuclear Cells for Pulmonary Tuberculosis Patients. Int. J. Mol. Sci. 2025, 26, 11219. https://doi.org/10.3390/ijms262211219
Tarasova EK, Pavlova EN, Rybalkina EY, Scherbakova EA, Tarasov RV, Erokhina MV. Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR in Tuberculomas and Peripheral Blood Mononuclear Cells for Pulmonary Tuberculosis Patients. International Journal of Molecular Sciences. 2025; 26(22):11219. https://doi.org/10.3390/ijms262211219
Chicago/Turabian StyleTarasova, Ekaterina K., Ekaterina N. Pavlova, Ekaterina Yu. Rybalkina, Ekaterina A. Scherbakova, Ruslan V. Tarasov, and Maria V. Erokhina. 2025. "Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR in Tuberculomas and Peripheral Blood Mononuclear Cells for Pulmonary Tuberculosis Patients" International Journal of Molecular Sciences 26, no. 22: 11219. https://doi.org/10.3390/ijms262211219
APA StyleTarasova, E. K., Pavlova, E. N., Rybalkina, E. Y., Scherbakova, E. A., Tarasov, R. V., & Erokhina, M. V. (2025). Validation of Housekeeping Genes for Normalizing RNA Expression in Real-Time PCR in Tuberculomas and Peripheral Blood Mononuclear Cells for Pulmonary Tuberculosis Patients. International Journal of Molecular Sciences, 26(22), 11219. https://doi.org/10.3390/ijms262211219





