Characteristics and Phylogenetic Analysis of the Complete Chloroplast Genome of Hibiscus sabdariffa L.
Abstract
1. Introduction
2. Results
2.1. Chloroplast Genome Structure of Hibiscus sabdariffa L.
2.2. Functional Annotation of Chloroplast Genes
2.3. Codon Preference Analysis
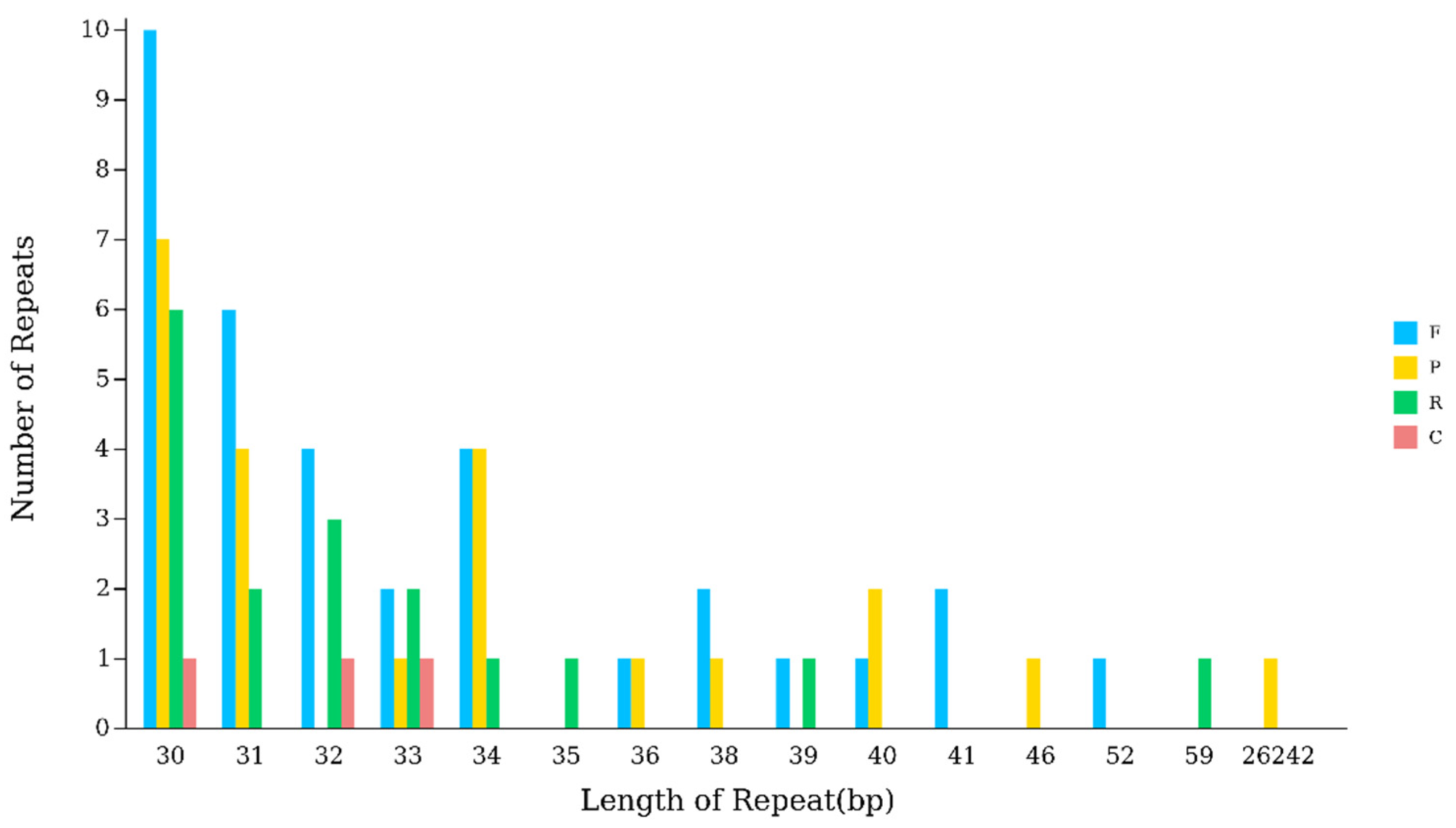
2.4. Repeat Sequence Analysis
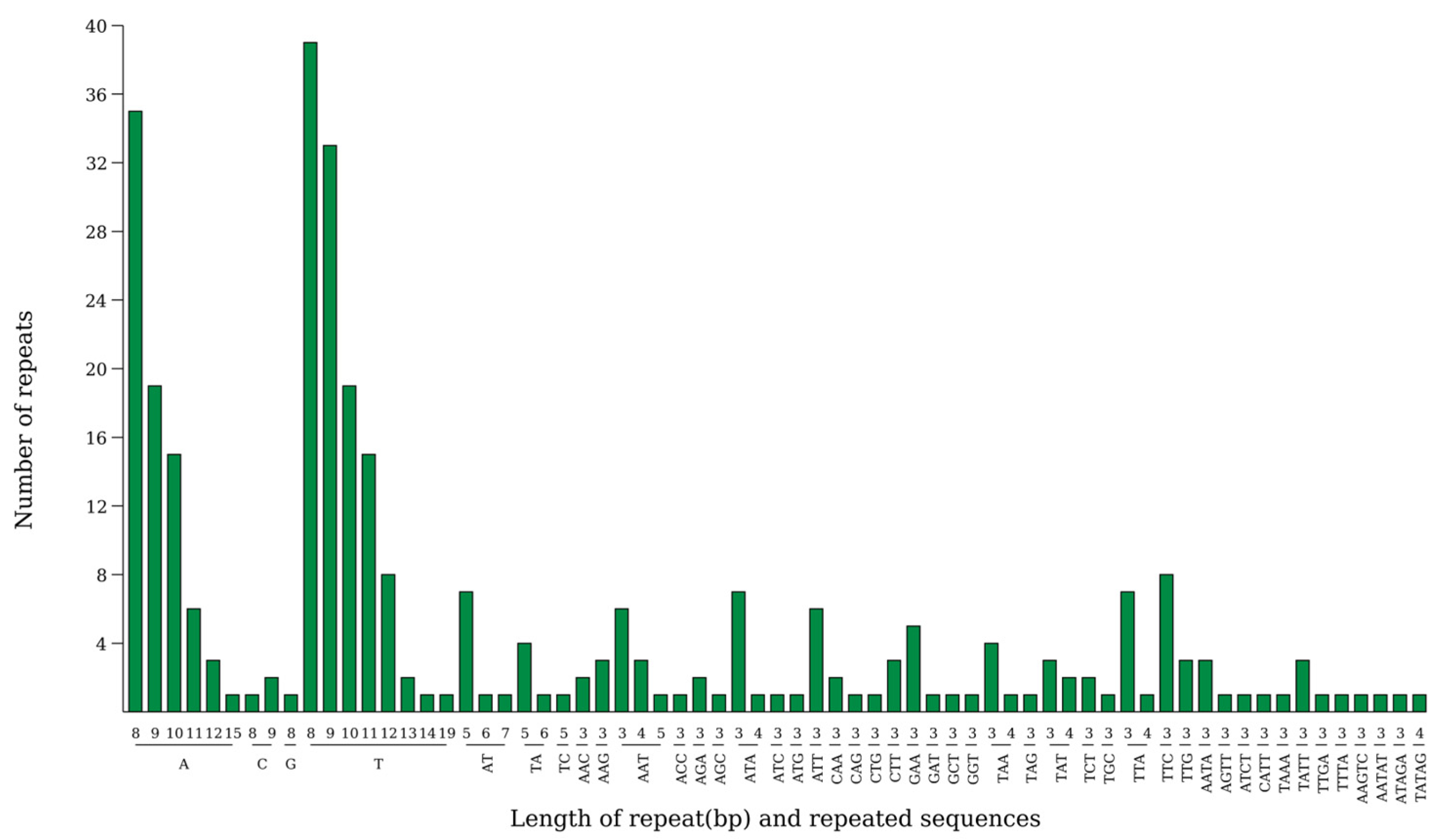
2.5. Simple Sequence Repeat (SSR) Analysis
2.6. Ka/Ks Analysis
2.7. Nucleic Acid Diversity Pi Analysis
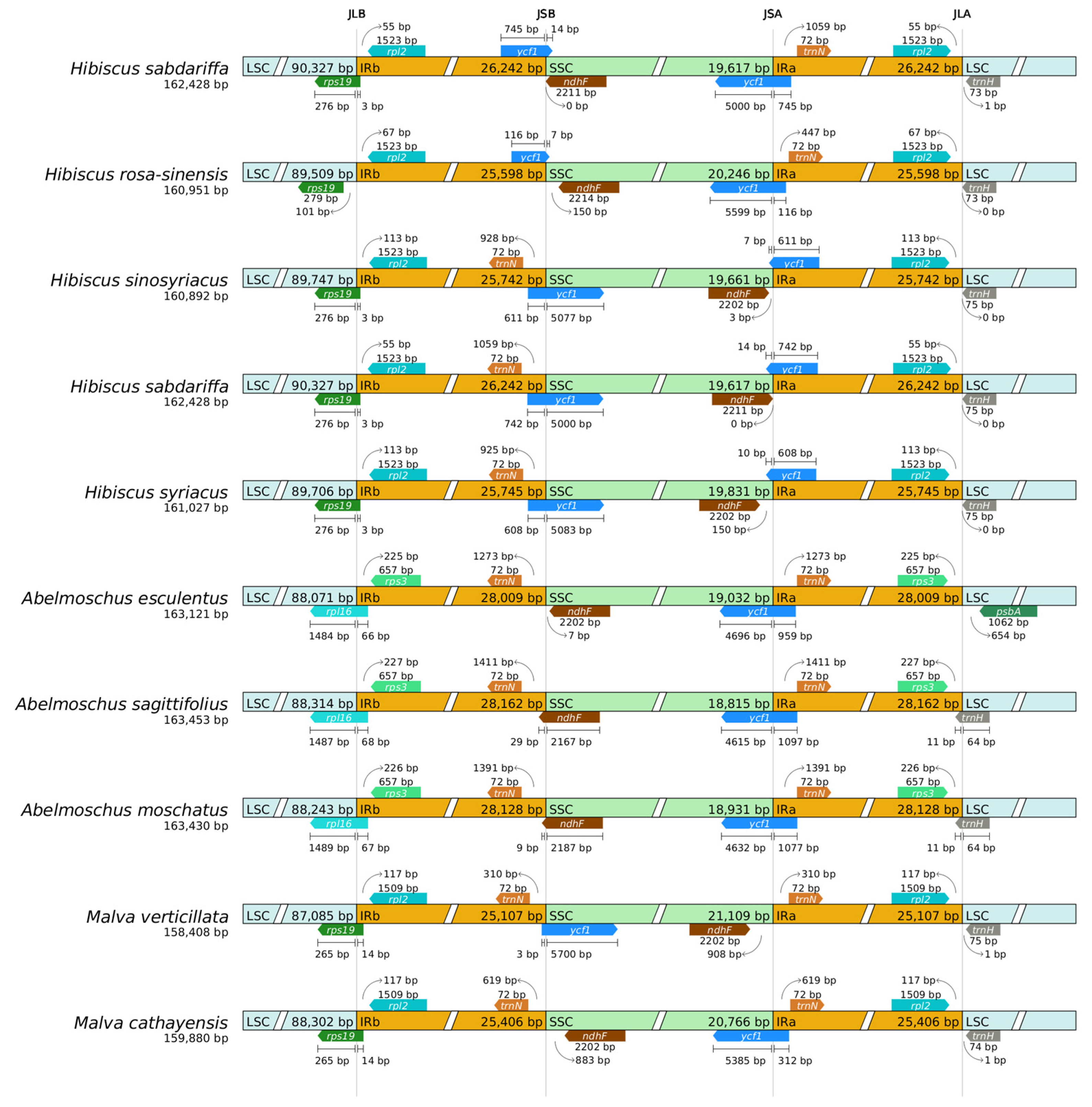
2.8. Boundary Analysis
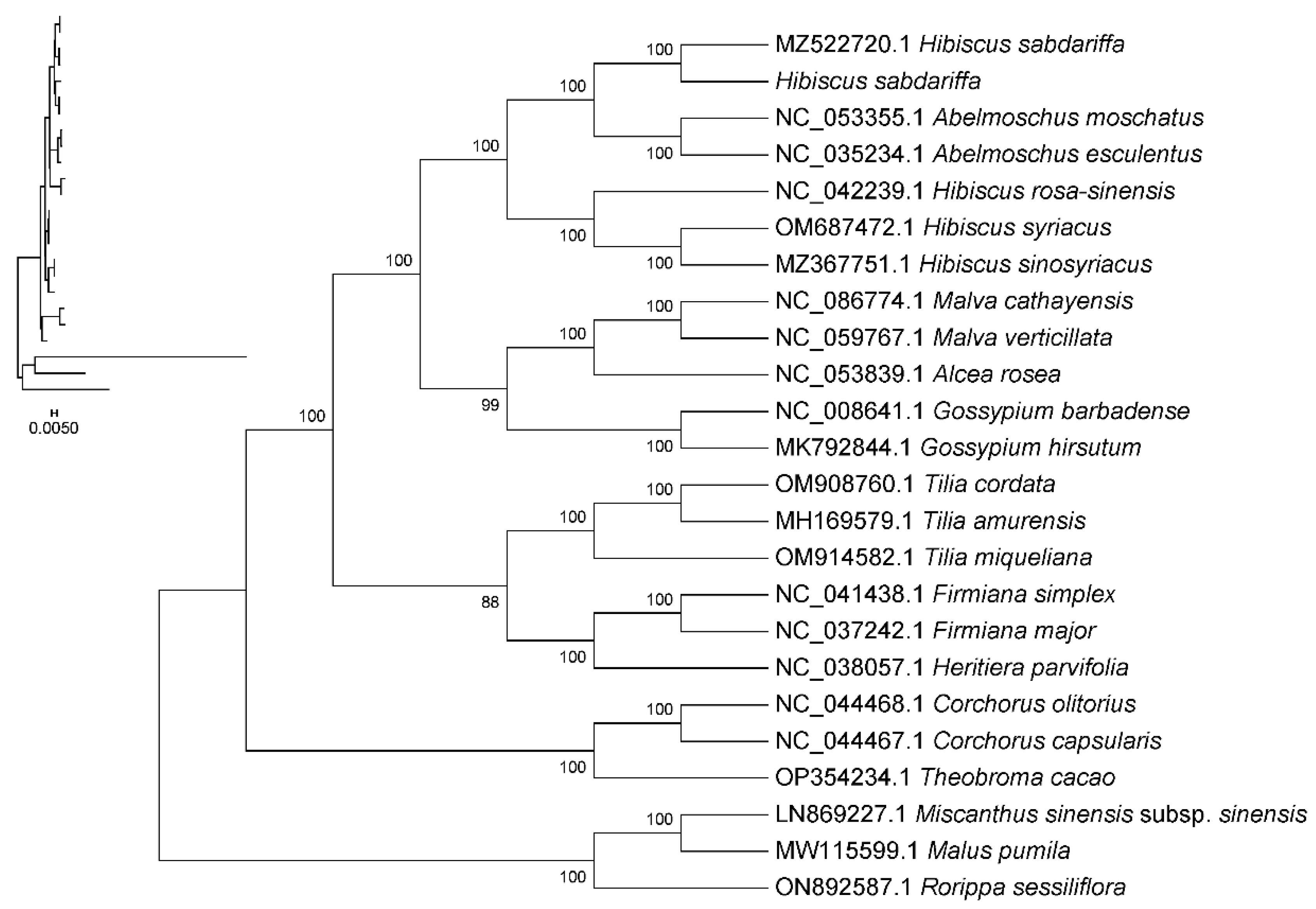
2.9. Phylogenetic Analysis
3. Discussion
3.1. Comparison of Chloroplast Genome Structure with Existing Study
3.2. Conservation and Difference Analysis of Chloroplast Gene Functional Annotation in Hibiscus sabdariffa
3.3. Evolutionary Significance of Repetitive Sequences and SSRs in the Hibiscus sabdariffa Chloroplast Genome
3.4. Ka/Ks Ratio and Nucleotide Diversity: Insights into Selection Pressure and Variation Hotspots
3.5. Boundary Dynamics of Chloroplast Genomes and Their Taxonomic Implications
3.6. Evolutionary and Functional Implications of the ycf1 Gene
3.7. Phylogenetic Position of Hibiscus sabdariffa and Evolutionary Relationships Within Malvaceae
4. Materials and Methods
4.1. Materials and Sequences
4.2. Genome Assembly and Result Control
4.3. Comprehensive Data Analysis
4.3.1. Codon Preference Analysis
4.3.2. Repetitive Sequence Analysis
4.3.3. Simple Sequence Repeat (SSR) Analysis
4.3.4. Ka/Ks Ratio Analysis
4.3.5. Nucleotide Diversity (π) Analysis
4.4. Chloroplast Genome Annotation and Genetic Map Construction
4.4.1. Raw Data Filtering
4.4.2. Chloroplast Genome Annotation
4.4.3. Chloroplast Genome Map Construction
4.4.4. Phylogenetic Tree Construction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maganha, E.G.; Halmenschlager, R.D.C.; Rosa, R.M.; Henriques, J.A.P.; Ramos, A.L.L.D.P.; Saffi, J. Pharmacological Evidences for the Extracts and Secondary Metabolites from Plants of the Genus. Hibiscus. Food Chem. 2010, 118, 1–10. [Google Scholar] [CrossRef]
- Sharma, H.K.; Sarkar, M.; Choudhary, S.B.; Kumar, A.A.; Maruthi, R.T.; Mitra, J.; Karmakar, P.G. Diversity Analysis Based on Agro-Morphological Traits and Microsatellite Based Markers in Global Germplasm Collections of Roselle (Hibiscus sabdariffa L.). Ind. Crops Prod. 2016, 89, 303–315. [Google Scholar] [CrossRef]
- Tran-Thi, N.Y.; Kasim, N.S.; Yuliana, M.; Huynh, L.H.; Chang, W.-L.; Ju, Y.-H. Polysaccharides-Induced Precipitation of Protein from Defatted Roselle Seed and Its Characterization: Antinutritional Factors and Functional Properties. J. Taiwan Inst. Chem. Eng. 2013, 44, 152–155. [Google Scholar] [CrossRef]
- Subbiah, G.; Acharya, S.K.; Sudhakara Reddy, M.; Sharma, S.; Singh, B.P.; Suchithra, R.; Kumar, K.P. Roselle Plant Stem Fibers: A Sustainable Biomaterial for Composite Reinforcement and Antimicrobial Applications. Results Eng. 2025, 26, 104852. [Google Scholar] [CrossRef]
- Morris, J.B.; Dierig, D.; Heinitz, C.; Hellier, B.; Bradley, V.; Marek, L. Vulnerability of U.S. New and Industrial Crop Genetic Resources. Ind. Crops Prod. 2023, 206, 117364. [Google Scholar] [CrossRef]
- Wang, M.L.; Morris, B.; Tonnis, B.; Davis, J.; Pederson, G.A. Assessment of Oil Content and Fatty Acid Composition Variability in Two Economically Important Hibiscus Species. J. Agric. Food Chem. 2012, 60, 6620–6626. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of Whole Chloroplast Genome Sequences to Choose Noncoding Regions for Phylogenetic Studies in Angiosperms: The Tortoise and the Hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, Z.; Qi, F.; Fang, Y.; Lin, K.; Pavan, S.; Huang, B.; Dong, W.; Du, P.; Tian, M.; et al. Chloroplast and Whole-Genome Sequencing Shed Light on the Evolutionary History and Phenotypic Diversification of Peanuts. Nat. Genet. 2024, 56, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Shi, W.; Wang, H.; Zhang, Z.; Tao, R.; Liu, J.; Wang, S.; Engel, M.S.; Shi, C. Comparative Analysis of 12 Water Lily Plastid Genomes Reveals Genomic Divergence and Evolutionary Relationships in Early Flowering Plants. Mar. Life Sci. Technol. 2024, 6, 425–441. [Google Scholar] [CrossRef]
- Da-Costa-Rocha, I.; Bonnlaender, B.; Sievers, H.; Pischel, I.; Heinrich, M. Hibiscus sabdariffa L.—A Phytochemical and Pharmacological Review. Food Chem. 2014, 165, 424–443. [Google Scholar] [CrossRef]
- Tao, A.; Li, Y.; Chen, J.; Li, J.; Xu, J.; Lin, L.; Zhang, L.; Fang, P. Development of Roselle (Hibiscus sabdariffa L.) Transcriptome-Based Simple Sequence Repeat Markers and Their Application in Roselle. Plants 2024, 13, 3517. [Google Scholar] [CrossRef]
- Kwon, S.-H.; Park, Y.; Jang, Y.L.; Kwon, H.-Y. The Complete Chloroplast Genome Sequence of Hibiscus sabdariffa (Malvaceae). Korean J. Plant Taxon. 2022, 52, 123–126. [Google Scholar] [CrossRef]
- Patel, S. Hibiscus Sabdariffa: An Ideal yet under-Exploited Candidate for Nutraceutical Applications. Biomed. Prev. Nutr. 2014, 4, 23–27. [Google Scholar] [CrossRef]
- Shruthi, V.H.; Ramachandra, C.T.; Udaykumar Nidoni, U.N.; Sharanagouda Hiregoudar, S.H.; Nagaraj Naik, N.N.; Kurubar, A.R. Roselle (Hibiscus sabdariffa L.) as a Source of Natural Colour: A Review. Plant Arch. 2016, 16, 515–522. [Google Scholar]
- Kwon, S.-H.; Kwon, H.-Y.; Choi, Y.-I.; Shin, H. Comprehensive Analysis of Chloroplast Genome of Hibiscus Sinosyriacus: Evolutionary Studies in Related Species and Genera. Forests 2023, 14, 2221. [Google Scholar] [CrossRef]
- Palmer, J.D.; Stein, D.B. Conservation of Chloroplast Genome Structure among Vascular Plants. Curr. Genet. 1986, 10, 823–833. [Google Scholar] [CrossRef]
- Huang, X.; Coulibaly, D.; Tan, W.; Ni, Z.; Shi, T.; Li, H.; Hayat, F.; Gao, Z. The Analysis of Genetic Structure and Characteristics of the Chloroplast Genome in Different Japanese Apricot Germplasm Populations. BMC Plant Biol. 2022, 22, 354. [Google Scholar] [CrossRef]
- Koo, H.; Shin, A.-Y.; Hong, S.; Kim, Y.-M. The Complete Chloroplast Genome of Hibiscus syriacus Using Long-Read Sequencing: Comparative Analysis to Examine the Evolution of the Tribe Hibisceae. Front. Plant Sci. 2023, 14, 1111968. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic Simple Sequence Repeat Regions in Chloroplast Genomes: Applications to the Population Genetics of Pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef]
- Abdullah; Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast Genome of Hibiscus rosa-Sinensis (Malvaceae): Comparative Analyses and Identification of Mutational Hotspots. Genomics 2020, 112, 581–591. [Google Scholar] [CrossRef]
- Abdullah; Chen, C.; Yan, R.; Sammad, A.; Heidari, P.; Hussain, T.; Mohammed, I.H.; Waheed, M.T.; Ahmed, I.; Vallada, A.; et al. Comparative Chloroplast Genomics of Hibiscus (Hibisceae, Malvoideae, Malvaceae) and Its Phylogenetic Implications. Res. Sq. 2025. [Google Scholar] [CrossRef]
- Wang, D.; Liu, F.; Wang, L.; Huang, S.; Yu, J. Nonsynonymous Substitution Rate (Ka) Is a Relatively Consistent Parameter for Defining Fast-Evolving and Slow-Evolving Protein-Coding Genes. Biol. Direct 2011, 6, 13. [Google Scholar] [CrossRef]
- Drescher, A.; Ruf, S.; Calsa, T.; Carrer, H.; Bock, R. The Two Largest Chloroplast Genome—Encoded Open Reading Frames of Higher Plants Are Essential Genes. Plant J. 2000, 22, 97–104. [Google Scholar] [CrossRef]
- De Vries, J.; Archibald, J.M.; Gould, S.B. The Carboxy Terminus of YCF1 Contains a Motif Conserved throughout >500 Myr of Streptophyte Evolution. Genome Biol. Evol. 2017, 9, 473–479. [Google Scholar] [CrossRef]
- Goldbecker, E.S.; De Vries, J. Systems Biology of Streptophyte Cell Evolution. Annu. Rev. Plant Biol. 2025, 76, 493–522. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 Genes from 64 Plastid Genomes Resolves Relationships in Angiosperms and Identifies Genome-Scale Evolutionary Patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, L.; Qi, J.; Zhang, L. Complete Chloroplast Genome Sequence of Hibiscus Cannabinus and Comparative Analysis of the Malvaceae Family. Front. Genet. 2020, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.-H.; Kwon, H.-Y.; Shin, H. Genetic Insights into the Extremely Dwarf Hibiscus Syriacus Var. Micranthus: Complete Chloroplast Genome Analysis and Development of a Novel dCAPS Marker. Curr. Issues Mol. Biol. 2024, 46, 2757–2771. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward Almost Closed Genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Kurtz, S. The Vmatch Large Scale Sequence Analysis Software. Available online: http://vmatch.de/vmweb.pdf (accessed on 9 November 2025).
- Katoh, K.; Kuma, K.-i.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a Program to Detect tRNA Genes and tmRNA Genes in Nucleotide Sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. raxmlGUI: A Graphical Front-End for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
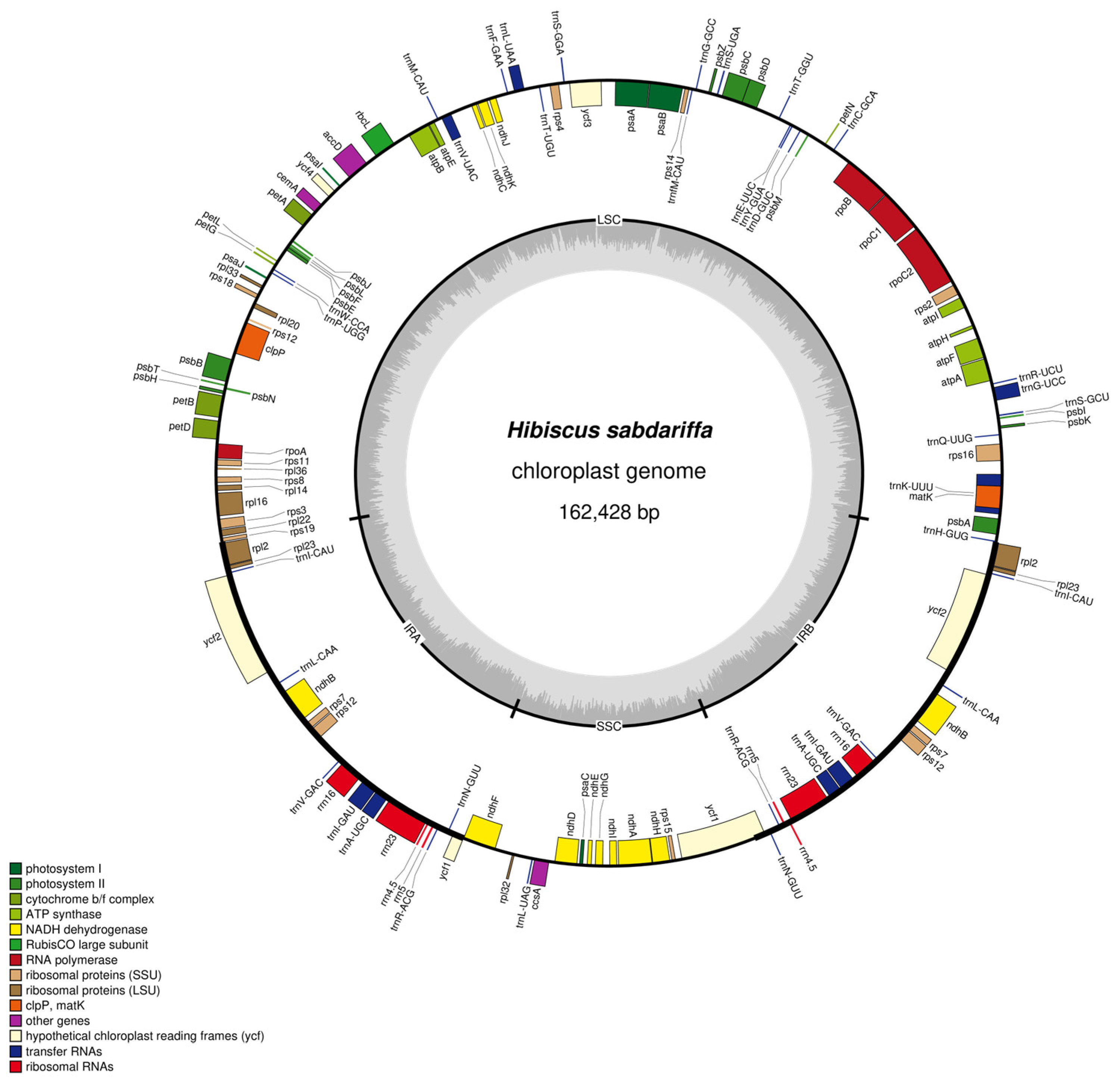



| Sequence Type Characteristics | Base Type | Number | % |
|---|---|---|---|
| Large-Scale Copy Region (LSC) Feature | A | 28,931 | 32.03% |
| C | 15,998 | 17.71% | |
| G | 15,132 | 16.75% | |
| T | 30,266 | 33.51% | |
| GC | 31,130 | 34.46% | |
| All | 90,327 | 100.00% | |
| Small-Scale Copy Region (SSC) Feature | A | 6603 | 33.66% |
| C | 3232 | 16.48% | |
| G | 2922 | 14.90% | |
| T | 6860 | 34.97% | |
| GC | 6154 | 31.37% | |
| All | 19,617 | 100.00% | |
| Inverse Repeat Sequence a (IRa) Feature | A | 7493 | 28.55% |
| C | 5393 | 20.55% | |
| G | 5802 | 22.11% | |
| T | 7554 | 28.79% | |
| GC | 11,195 | 42.66% | |
| All | 26,242 | 100.00% | |
| Inverse Repeat Sequence b (IRb) Feature | A | 7554 | 28.79% |
| C | 5802 | 22.11% | |
| G | 5393 | 20.55% | |
| T | 7493 | 28.55% | |
| GC | 11,195 | 42.66% | |
| All | 26,242 | 100.00% |
| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB *(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of the cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Subunits of photochlorophyllide reductase | - | |
| Self-replication | Proteins of the large ribosomal subunit | rpl14, rpl16 *, rpl2 *(2), rpl20, rpl22, rpl23 (2), rpl32, rpl33, rpl36 |
| Proteins of the small ribosomal subunit | rps11, rps12 **(2), rps14, rps15, rps16 *, rps18, rps19, rps2, rps3, rps4, rps7 (2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Ribosomal RNAs | rrn16 (2), rrn23 (2), rrn4.5 (2), rrn5 (2) | |
| Transfer RNAs | trnA-UGC *(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU (2), trnI-GAU *(2), trnK-UUU *, trnL-CAA (2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU (2), trnP-UGG, trnQ-UUG, trnR-ACG (2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (2), trnV-UAC *, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP ** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | - | |
| other | - | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1(2), ycf2(2), ycf3 **, ycf4 |
| Symbol | Codon | No. | RSCU | Symbol | Codon | No. | RSCU | Symbol | Codon | No. | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ter * | UAA | 43 | 1.6539 | Arg | AGA | 406 | 1.7862 | Lys | AAA | 893 | 1.5174 |
| Ter * | UAG | 18 | 0.6924 | Arg | AGG | 146 | 0.642 | Lys | AAG | 284 | 0.4826 |
| Ter * | UGA | 17 | 0.6537 | Arg | CGA | 315 | 1.3854 | Leu | CUA | 316 | 0.7896 |
| Ala | GCA | 340 | 1.046 | Arg | CGC | 108 | 0.4752 | Leu | CUC | 152 | 0.3798 |
| Ala | GCC | 213 | 0.6552 | Arg | CGG | 93 | 0.4092 | Leu | CUG | 154 | 0.3846 |
| Ala | GCG | 169 | 0.52 | Arg | CGU | 296 | 1.302 | Leu | CUU | 495 | 1.2366 |
| Ala | GCU | 578 | 1.7784 | Ser | AGC | 101 | 0.3588 | Leu | UUA | 794 | 1.9836 |
| Cys | UGC | 58 | 0.4604 | Ser | AGU | 343 | 1.2192 | Leu | UUG | 491 | 1.2264 |
| Cys | UGU | 194 | 1.5396 | Ser | UCA | 359 | 1.2762 | Met | AUA | 0 | 0 |
| Asp | GAC | 184 | 0.4058 | Ser | UCC | 259 | 0.9204 | Met | AUC | 0 | 0 |
| Asp | GAU | 723 | 1.5942 | Ser | UCG | 143 | 0.5082 | Met | AUG | 531 | 7 |
| Glu | GAA | 897 | 1.5012 | Ser | UCU | 483 | 1.7166 | Met | AUU | 0 | 0 |
| Glu | GAG | 298 | 0.4988 | Thr | ACA | 351 | 1.2072 | Met | CUG | 0 | 0 |
| Phe | UUC | 429 | 0.6646 | Thr | ACC | 228 | 0.784 | Met | GUG | 0 | 0 |
| Phe | UUU | 862 | 1.3354 | Thr | ACG | 136 | 0.4676 | Met | UUG | 0 | 0 |
| Gly | GGA | 622 | 1.5452 | Thr | ACU | 448 | 1.5408 | Asn | AAC | 255 | 0.467 |
| Gly | GGC | 177 | 0.4396 | Val | GUA | 476 | 1.5244 | Asn | AAU | 837 | 1.533 |
| Gly | GGG | 288 | 0.7156 | Val | GUC | 149 | 0.4772 | Pro | CCA | 265 | 1.1276 |
| Gly | GGU | 523 | 1.2992 | Val | GUG | 168 | 0.538 | Pro | CCC | 180 | 0.766 |
| His | CAC | 140 | 0.5036 | Val | GUU | 456 | 1.4604 | Pro | CCG | 129 | 0.5488 |
| His | CAU | 416 | 1.4964 | Trp | UGG | 399 | 1 | Pro | CCU | 366 | 1.5576 |
| Ile | AUA | 622 | 0.9468 | Tyr | UAC | 171 | 0.3972 | Gln | CAA | 632 | 1.551 |
| Ile | AUC | 370 | 0.5631 | Tyr | UAU | 690 | 1.6028 | Gln | CAG | 183 | 0.449 |
| Ile | AUU | 979 | 1.4901 |
| Length | F | P | R | C | Total |
|---|---|---|---|---|---|
| 30 | 10 | 7 | 6 | 1 | 24 |
| 31 | 6 | 4 | 2 | 0 | 12 |
| 32 | 4 | 0 | 3 | 1 | 8 |
| 33 | 2 | 1 | 2 | 1 | 6 |
| 34 | 4 | 4 | 1 | 0 | 9 |
| 35 | 0 | 0 | 1 | 0 | 1 |
| 36 | 1 | 1 | 0 | 0 | 2 |
| 38 | 2 | 1 | 0 | 0 | 3 |
| 39 | 1 | 0 | 1 | 0 | 2 |
| 40 | 1 | 2 | 0 | 0 | 3 |
| 41 | 2 | 0 | 0 | 0 | 2 |
| 46 | 0 | 1 | 0 | 0 | 1 |
| 52 | 1 | 0 | 0 | 0 | 1 |
| 59 | 0 | 0 | 1 | 0 | 1 |
| 26,242 | 0 | 1 | 0 | 0 | 1 |
| Total | 34 | 22 | 17 | 3 | 76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, J.; Ji, Q.; An, X.; Luo, X.; Chen, C.; Liu, T.; Zou, L.; Li, S.; Du, G.; Chen, J.; et al. Characteristics and Phylogenetic Analysis of the Complete Chloroplast Genome of Hibiscus sabdariffa L. Int. J. Mol. Sci. 2025, 26, 11001. https://doi.org/10.3390/ijms262211001
Dong J, Ji Q, An X, Luo X, Chen C, Liu T, Zou L, Li S, Du G, Chen J, et al. Characteristics and Phylogenetic Analysis of the Complete Chloroplast Genome of Hibiscus sabdariffa L. International Journal of Molecular Sciences. 2025; 26(22):11001. https://doi.org/10.3390/ijms262211001
Chicago/Turabian StyleDong, Junyuan, Qingqing Ji, Xingcai An, Xiahong Luo, Changli Chen, Tingting Liu, Lina Zou, Shaocui Li, Guanghui Du, Jikang Chen, and et al. 2025. "Characteristics and Phylogenetic Analysis of the Complete Chloroplast Genome of Hibiscus sabdariffa L." International Journal of Molecular Sciences 26, no. 22: 11001. https://doi.org/10.3390/ijms262211001
APA StyleDong, J., Ji, Q., An, X., Luo, X., Chen, C., Liu, T., Zou, L., Li, S., Du, G., Chen, J., & An, X. (2025). Characteristics and Phylogenetic Analysis of the Complete Chloroplast Genome of Hibiscus sabdariffa L. International Journal of Molecular Sciences, 26(22), 11001. https://doi.org/10.3390/ijms262211001






