Lipocalin-2 in Triple-Negative Breast Cancer: A Review of Its Pathophysiological Role in the Metastatic Cascade
Abstract
1. Introduction
1.1. Physiological Functions of LCN2
1.2. Pathological Functions of LCN2
1.3. Breast Cancer
2. Subtypes of Triple-Negative Breast Cancer
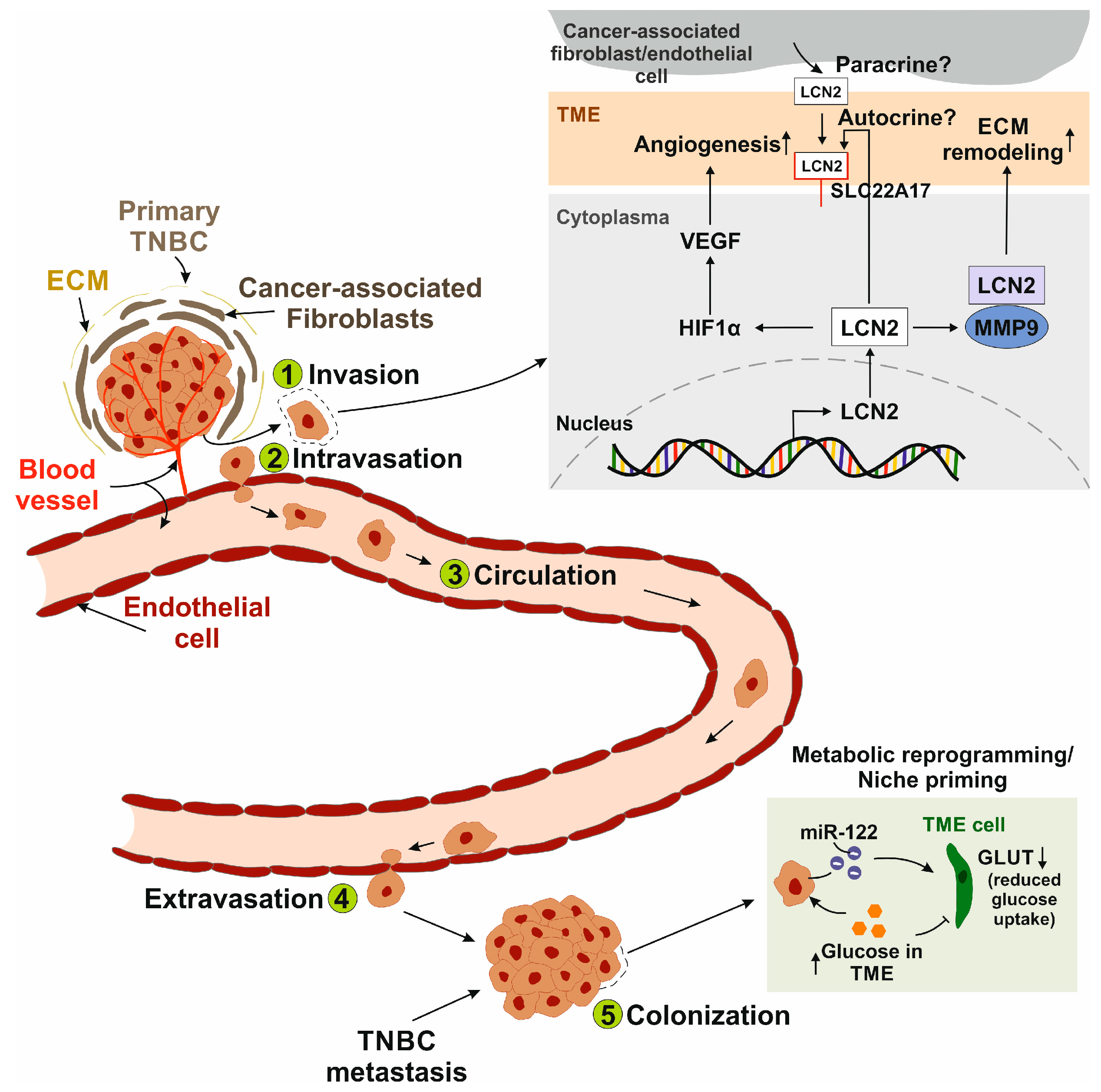
3. Metastasis of Triple-Negative Breast Cancer
3.1. Molecular Mechanisms
3.1.1. The Metastatic Cascade
3.1.2. Interaction with Tumor Microenvironment
3.2. Phenotypic Plasticity
4. LCN2 as a Prognostic and Therapeutic Target in Triple-Negative Breast Cancer
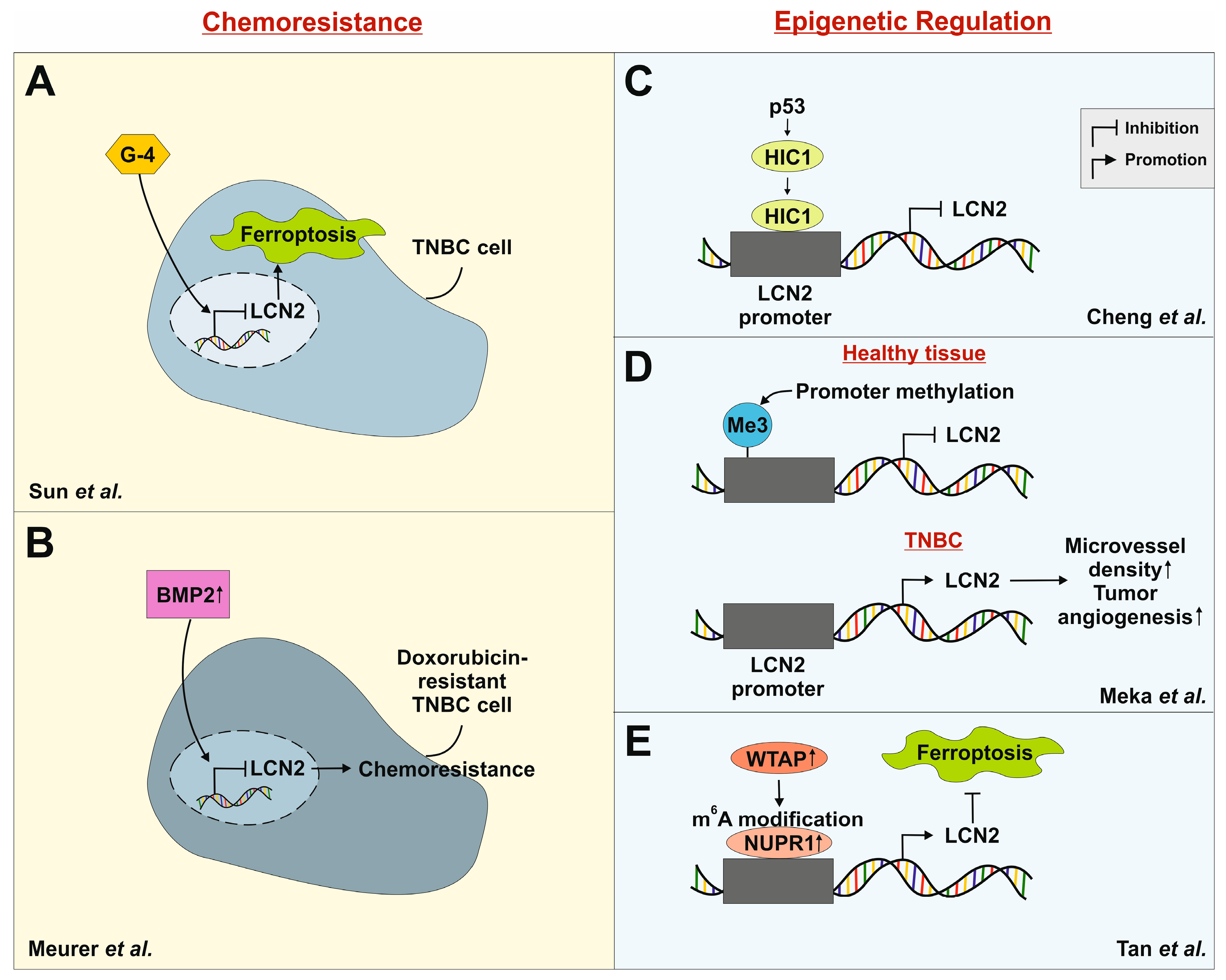
4.1. Chemoresistance in TNBC
4.2. Epigenetic Regulation of LCN2
5. Organ-Specific Role of LCN2 in Metastasis
5.1. Lung
5.2. Brain
5.3. Bone and Liver
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| CXCL1/2 | C-X-C chemokine motif 1/2 |
| EMT | Epithelial-to-mesenchymal transition |
| ER | Estrogen receptor |
| G-CSF | Granulocyte colony-stimulating factor |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| HIC1 | Hypermethylated in cancer 1 |
| HIF1α | Hypoxia-inducible factor 1 alpha |
| IL-8 | Interleukin 8 |
| LCN2 | Lipocalin-2 |
| LEC | Lymphatic endothelial cells |
| LRP2/5 | Low-density lipoprotein 2/5 |
| MC1R | Melanocortin receptor 1 |
| MEC | Microvascular endothelial cells |
| MET | Mesenchymal-to-epithelial transition |
| MMP-9 | Matrix-metalloproteinase |
| NGAL | Neutrophil-gelatinase associated protein |
| NUPR1 | Nuclear protein 1 |
| PR | Progesterone receptor |
| SLC22A17 | Solute carrier family 22 member 17 |
| TME | Tumor microenvironment |
| TNBC | Triple-negative breast cancer |
| TNFα | Tumor necrosis factor alpha |
| VEGF | Vascular endothelial growth factor |
| WTAP | Wt1-associated protein |
References
- Flower, D.R. The lipocalin protein family: Structure and function. Biochem. J. 1996, 318 (Pt 1), 1–14. [Google Scholar] [CrossRef]
- Kjeldsen, L.; Johnsen, A.H.; Sengeløv, H.; Borregaard, N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J. Biol. Chem. 1993, 268, 10425–10432. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Borregaard, N.; Kjeldsen, L.; Moses, M.A. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J. Biol. Chem. 2001, 276, 37258–37265. [Google Scholar] [CrossRef] [PubMed]
- Cramer, E.P.; Glenthøj, A.; Häger, M.; Juncker-Jensen, A.; Engelholm, L.H.; Santoni-Rugiu, E.; Lund, L.R.; Laerum, O.D.; Cowland, J.B.; Borregaard, N. No effect of NGAL/lipocalin-2 on aggressiveness of cancer in the MMTV-PyMT/FVB/N mouse model for breast cancer. PLoS ONE 2012, 7, e39646. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.A.; Paulsene, W.; Jide, X.; Ratledge, C.; Strong, R.K. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure 2005, 13, 29–41. [Google Scholar] [CrossRef]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef]
- Flo, T.H.; Smith, K.D.; Sato, S.; Rodriguez, D.J.; Holmes, M.A.; Strong, R.K.; Akira, S.; Aderem, A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [Google Scholar] [CrossRef]
- Bao, G.; Clifton, M.; Hoette, T.M.; Mori, K.; Deng, S.X.; Qiu, A.; Viltard, M.; Williams, D.; Paragas, N.; Leete, T.; et al. Iron traffics in circulation bound to a siderocalin (Ngal)-catechol complex. Nat. Chem. Biol. 2010, 6, 602–609. [Google Scholar] [CrossRef]
- Santiago-Sánchez, G.S.; Pita-Grisanti, V.; Quiñones-Díaz, B.; Gumpper, K.; Cruz-Monserrate, Z.; Vivas-Mejía, P.E. Biological Functions and Therapeutic Potential of Lipocalin 2 in Cancer. Int. J. Mol. Sci. 2020, 21, 4365. [Google Scholar] [CrossRef]
- Coles, M.; Diercks, T.; Muehlenweg, B.; Bartsch, S.; Zölzer, V.; Tschesche, H.; Kessler, H. The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin. J. Mol. Biol. 1999, 289, 139–157. [Google Scholar] [CrossRef]
- Devireddy, L.R.; Gazin, C.; Zhu, X.; Green, M.R. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 2005, 123, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.K.; Gasterich, N.; Weiskirchen, S.; Weiskirchen, R. Lipocalin 2 receptors: Facts, fictions, and myths. Front. Immunol. 2023, 14, 1229885. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025, 53, D20–D29. [Google Scholar] [CrossRef] [PubMed]
- Jani Kargar Moghaddam, S.; Mohammadi Roushandeh, A.; Hamidi, M.; Nemati, S.; Jahanian-Najafabadi, A.; Habibi Roudkenar, M. Lipocalin-2 Upregulation in Nasopharyngeal Carcinoma: A Novel Potential Diagnostic Biomarker. Iran. J. Med. Sci. 2023, 48, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Cowland, J.B.; Borregaard, N. Molecular characterization and pattern of tissue expression of the gene for neutrophil gelatinase-associated lipocalin from humans. Genomics 1997, 45, 17–23. [Google Scholar] [CrossRef]
- Soni, S.S.; Cruz, D.; Bobek, I.; Chionh, C.Y.; Nalesso, F.; Lentini, P.; de Cal, M.; Corradi, V.; Virzi, G.; Ronco, C. NGAL: A biomarker of acute kidney injury and other systemic conditions. Int. Urol. Nephrol. 2010, 42, 141–150. [Google Scholar] [CrossRef]
- Kessel, J.C.; Weiskirchen, R.; Schröder, S.K. Expression Analysis of Lipocalin 2 (LCN2) in Reproductive and Non-Reproductive Tissues of Esr1-Deficient Mice. Int. J. Mol. Sci. 2023, 24, 9280. [Google Scholar] [CrossRef]
- Chung, I.H.; Wu, T.I.; Liao, C.J.; Hu, J.Y.; Lin, Y.H.; Tai, P.J.; Lai, C.H.; Lin, K.H. Overexpression of lipocalin 2 in human cervical cancer enhances tumor invasion. Oncotarget 2016, 7, 11113–11126. [Google Scholar] [CrossRef]
- Yang, J.; Bielenberg, D.R.; Rodig, S.J.; Doiron, R.; Clifton, M.C.; Kung, A.L.; Strong, R.K.; Zurakowski, D.; Moses, M.A. Lipocalin 2 promotes breast cancer progression. Proc. Natl. Acad. Sci. USA 2009, 106, 3913–3918. [Google Scholar] [CrossRef]
- Moniaux, N.; Chakraborty, S.; Yalniz, M.; Gonzalez, J.; Shostrom, V.K.; Standop, J.; Lele, S.M.; Ouellette, M.; Pour, P.M.; Sasson, A.R.; et al. Early diagnosis of pancreatic cancer: Neutrophil gelatinase-associated lipocalin as a marker of pancreatic intraepithelial neoplasia. Br. J. Cancer 2008, 98, 1540–1547. [Google Scholar] [CrossRef]
- Hu, L.; Hittelman, W.; Lu, T.; Ji, P.; Arlinghaus, R.; Shmulevich, I.; Hamilton, S.R.; Zhang, W. NGAL decreases E-cadherin-mediated cell-cell adhesion and increases cell motility and invasion through Rac1 in colon carcinoma cells. Lab. Investig. 2009, 89, 531–548. [Google Scholar] [CrossRef]
- Cho, H.; Kim, J.-H. Lipocalin2 expressions correlate significantly with tumor differentiation in epithelial ovarian cancer. J. Histochem. Cytochem. 2009, 57, 513–521. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kaur, S.; Guha, S.; Batra, S.K. The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochim. Biophys. Acta 2012, 1826, 129–169. [Google Scholar] [CrossRef]
- Mertens, C.; Schnetz, M.; Rehwald, C.; Grein, S.; Elwakeel, E.; Weigert, A.; Brüne, B.; Jung, M. Iron-Bound Lipocalin-2 from Tumor-Associated Macrophages Drives Breast Cancer Progression Independent of Ferroportin. Metabolites 2021, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.S.; Wu, Q.; Yang, J.H.; Wang, H.Q.; Ding, X.D.; Yang, F.; Xu, X.C. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int. J. Cancer 2008, 122, 2050–2056. [Google Scholar] [CrossRef] [PubMed]
- Bahrun, U.; Wildana, W.; Rahmawati, H.; Kurniawan, L.B.; Hamdani, W. Lipocalin 2 could predict circulating MMP9 levels in patients with breast cancer. Breast Dis. 2021, 40, S115–S117. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer: Lyon, France. Available online: https://gco.iarc.who.int/today (accessed on 10 July 2025).
- Mohammed, A.A. Quantitative assessment of Ki67 expression in correlation with various breast cancer characteristics and survival rate; cross sectional study. Ann. Med. Surg. 2019, 48, 129–134. [Google Scholar] [CrossRef]
- Mohammed, A.A. The clinical behavior of different molecular subtypes of breast cancer. Cancer Treat. Res. Commun. 2021, 29, 100469. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, K.; Li, M.; Huang, W.; Zhang, F.; Wang, H. Lipocalin 2: A potential therapeutic target for breast cancer metastasis. Onco. Targets Ther. 2018, 11, 8099–8106. [Google Scholar] [CrossRef]
- Bao, Y.; Yan, Z.; Shi, N.; Tian, X.; Li, J.; Li, T.; Cheng, X.; Lv, J. LCN2: Versatile players in breast cancer. Biomed. Pharmacother. 2024, 171, 116091. [Google Scholar] [CrossRef]
- Sheppard, V.B.; Cavalli, L.R.; Dash, C.; Kanaan, Y.M.; Dilawari, A.A.; Horton, S.; Makambi, K.H. Correlates of Triple Negative Breast Cancer and Chemotherapy Patterns in Black and White Women with Breast Cancer. Clin. Breast Cancer 2017, 17, 232–238. [Google Scholar] [CrossRef]
- Schwentner, L.; Wolters, R.; Koretz, K.; Wischnewsky, M.B.; Kreienberg, R.; Rottscholl, R.; Wöckel, A. Triple-negative breast cancer: The impact of guideline-adherent adjuvant treatment on survival--a retrospective multi-centre cohort study. Breast Cancer Res. Treat. 2012, 132, 1073–1080. [Google Scholar] [CrossRef]
- Grosse, C.; Noack, P.; Grosse, A.; Preuss, C.I.; Schwarz, H.K.; Gitter, T.; Schrenk, P.; Frauchiger-Heuer, H.; Papassotiropoulos, B.; Tausch, C.; et al. Prognostic impact of histological subtyping in triple-negative breast cancer. Hum. Pathol. 2024, 152, 105640. [Google Scholar] [CrossRef]
- de Ruijter, T.C.; Veeck, J.; de Hoon, J.P.J.; van Engeland, M.; Tjan-Heijnen, V.C. Characteristics of triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 183–192. [Google Scholar] [CrossRef]
- Howard, F.M.; Olopade, O.I. Epidemiology of Triple-Negative Breast Cancer: A Review. Cancer J. 2021, 27, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Demere, Z.; Nair, K.; Ali, A.; Ferraro, G.B.; Natoli, T.; Deik, A.; Petronio, L.; Tang, A.A.; Zhu, C.; et al. A metastasis map of human cancer cell lines. Nature 2020, 588, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Hossain, F.; Danos, D.; Lassak, A.; Scribner, R.; Miele, L. Racial Disparities in Triple Negative Breast Cancer: A Review of the Role of Biologic and Non-biologic Factors. Front. Public Health 2020, 8, 576964. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Borri, F.; Granaglia, A. Pathology of triple negative breast cancer. Semin. Cancer Biol. 2021, 72, 136–145. [Google Scholar] [CrossRef]
- Wang, D.-Y.; Jiang, Z.; Ben-David, Y.; Woodgett, J.R.; Zacksenhaus, E. Molecular stratification within triple-negative breast cancer subtypes. Sci. Rep. 2019, 9, 19107. [Google Scholar] [CrossRef]
- Badowska-Kozakiewicz, A.M.; Budzik, M.P. Immunohistochemical characteristics of basal-like breast cancer. Contemp. Oncol./wspołcz. Onkol. 2016, 20, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qiao, X.; Liu, Z.; Zhang, W. The role of E2F2 in cancer progression and its value as a therapeutic target. Front. Immunol. 2024, 15, 1397303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, J.; Mao, K.; Deng, H.; Yang, Y.; Zhou, E.; Liu, J. Role of transforming growth factor-β1 in triple negative breast cancer patients. Int. J. Surg. 2017, 45, 72–76. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Pietenpol, J.A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 2014, 232, 142–150. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Gibson, G.R.; Qian, D.; Ku, J.K.; Lai, L.L. Metaplastic breast cancer: Clinical features and outcomes. Am. Surg. 2005, 71, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.J.; Mac Gabhann, F. Expression of VEGF and semaphorin genes define subgroups of triple negative breast cancer. PLoS ONE 2013, 8, e61788. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, Y.; Xue, J.; Li, J.; Yi, J.; Bu, J.; Zhang, Z.; Qiu, P.; Gu, X. Advances in immunotherapy for triple-negative breast cancer. Mol. Cancer 2023, 22, 145, Erratum in: Mol. Cancer 2023, 22, 154. [Google Scholar] [CrossRef]
- Dieci, M.V.; Tsvetkova, V.; Griguolo, G.; Miglietta, F.; Mantiero, M.; Tasca, G.; Cumerlato, E.; Giorgi, C.A.; Giarratano, T.; Faggioni, G.; et al. Androgen Receptor Expression and Association with Distant Disease-Free Survival in Triple Negative Breast Cancer: Analysis of 263 Patients Treated with Standard Therapy for Stage I-III Disease. Front. Oncol. 2019, 9, 452. [Google Scholar] [CrossRef]
- Vtorushin, S.; Dulesova, A.; Krakhmal, N. Luminal androgen receptor (LAR) subtype of triple-negative breast cancer: Molecular, morphological, and clinical features. J. Zhejiang Univ. Sci. B 2022, 23, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Bian, C.; Gu, K.; Wang, H.; Zhuang, S.; Tang, X.; Zhao, Y. Patterns of distant metastases in patients with triple-negative breast cancer—A population-based study. Precis. Med. Sci. 2023, 12, 182–195. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Tian, W.; Li, Y.; Qin, Q.; Mao, Y.; Liu, X.; Hong, J.; Hu, L.; Zeng, Q.; et al. Prognosis prediction and risk factors for triple-negative breast cancer patients with brain metastasis: A population-based study. Cancer Med. 2023, 12, 7951–7961. [Google Scholar] [CrossRef] [PubMed]
- Steward, L.; Conant, L.; Gao, F.; Margenthaler, J.A. Predictive factors and patterns of recurrence in patients with triple negative breast cancer. Ann. Surg. Oncol. 2014, 21, 2165–2171. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.-Y.; Chang, C.-J.; Cheng, J.-S. Survival, treatment regimens and medical costs of women newly diagnosed with metastatic triple-negative breast cancer. Sci. Rep. 2022, 12, 729. [Google Scholar] [CrossRef]
- Baser, O.; Wei, W.; Henk, H.J.; Teitelbaum, A.; Xie, L. Patient survival and healthcare utilization costs after diagnosis of triple-negative breast cancer in a United States managed care cancer registry. Curr. Med. Res. Opin. 2012, 28, 419–428. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Candido, S.; Abrams, S.L.; Steelman, L.S.; Lertpiriyapong, K.; Fitzgerald, T.L.; Martelli, A.M.; Cocco, L.; Montalto, G.; Cervello, M.; Polesel, J.; et al. Roles of NGAL and MMP-9 in the tumor microenvironment and sensitivity to targeted therapy. Biochim. Biophys. Acta 2016, 1863, 438–448. [Google Scholar] [CrossRef]
- Conn, E.M.; Botkjaer, K.A.; Kupriyanova, T.A.; Andreasen, P.A.; Deryugina, E.I.; Quigley, J.P. Comparative analysis of metastasis variants derived from human prostate carcinoma cells: Roles in intravasation of VEGF-mediated angiogenesis and uPA-mediated invasion. Am. J. Pathol. 2009, 175, 1638–1652. [Google Scholar] [CrossRef]
- Yang, J.; McNeish, B.; Butterfield, C.; Moses, M.A. Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. FASEB J. 2013, 27, 45–50. [Google Scholar] [CrossRef]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef]
- Boudreau, N.; Myers, C. Breast cancer-induced angiogenesis: Multiple mechanisms and the role of the microenvironment. Breast Cancer Res. 2003, 5, 140–146. [Google Scholar] [CrossRef]
- Banerjee, S.; Dowsett, M.; Ashworth, A.; Martin, L.-A. Mechanisms of disease: Angiogenesis and the management of breast cancer. Nat. Clin. Pract. Oncol. 2007, 4, 536–550. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed]
- Follain, G.; Osmani, N.; Azevedo, A.S.; Allio, G.; Mercier, L.; Karreman, M.A.; Solecki, G.; Garcia Leòn, M.J.; Lefebvre, O.; Fekonja, N.; et al. Hemodynamic Forces Tune the Arrest, Adhesion, and Extravasation of Circulating Tumor Cells. Dev. Cell 2018, 45, 33–52.e12. [Google Scholar] [CrossRef]
- Wang, S.; Ye, T.; Li, G.; Zhang, X.; Shi, H. Margination and adhesion dynamics of tumor cells in a real microvascular network. PLoS Comput. Biol. 2021, 17, e1008746. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.B.; Whisler, J.A.; Jeon, J.S.; Kamm, R.D. Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr. Biol. 2013, 5, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Roshanzamir, F.; Robinson, J.L.; Cook, D.; Karimi-Jafari, M.H.; Nielsen, J. Metastatic triple negative breast cancer adapts its metabolism to destination tissues while retaining key metabolic signatures. Proc. Natl. Acad. Sci. USA 2022, 119, e2205456119. [Google Scholar] [CrossRef]
- Kim, S.L.; Lee, S.T.; Min, I.S.; Park, Y.R.; Lee, J.H.; Kim, D.G.; Kim, S.W. Lipocalin 2 negatively regulates cell proliferation and epithelial to mesenchymal transition through changing metabolic gene expression in colorectal cancer. Cancer Sci. 2017, 108, 2176–2186. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, Q.; Dong, C. Metabolic reprogramming in triple-negative breast cancer. Cancer Biol. Med. 2020, 17, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, N.; Stearns, V.; Santa-Maria, C.A.; Popel, A.S. The tumor microenvironment and triple-negative breast cancer aggressiveness: Shedding light on mechanisms and targeting. Expert Opin. Ther. Targets 2022, 26, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Naba, A.; Clauser, K.R.; Lamar, J.M.; Carr, S.A.; Hynes, R.O. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 2014, 3, e01308. [Google Scholar] [CrossRef]
- Fraley, S.I.; Feng, Y.; Krishnamurthy, R.; Kim, D.-H.; Celedon, A.; Longmore, G.D.; Wirtz, D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010, 12, 598–604. [Google Scholar] [CrossRef]
- Han, J.; Chang, H.; Giricz, O.; Lee, G.Y.; Baehner, F.L.; Gray, J.W.; Bissell, M.J.; Kenny, P.A.; Parvin, B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput. Biol. 2010, 6, e1000684. [Google Scholar] [CrossRef]
- Bakal, C.; Aach, J.; Church, G.; Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 2007, 316, 1753–1756. [Google Scholar] [CrossRef]
- Baskaran, J.P.; Weldy, A.; Guarin, J.; Munoz, G.; Shpilker, P.H.; Kotlik, M.; Subbiah, N.; Wishart, A.; Peng, Y.; Miller, M.A.; et al. Cell shape, and not 2D migration, predicts extracellular matrix-driven 3D cell invasion in breast cancer. APL Bioeng. 2020, 4, 26105. [Google Scholar] [CrossRef] [PubMed]
- Mayorca-Guiliani, A.E.; Madsen, C.D.; Cox, T.R.; Horton, E.R.; Venning, F.A.; Erler, J.T. ISDoT: In situ decellularization of tissues for high-resolution imaging and proteomic analysis of native extracellular matrix. Nat. Med. 2017, 23, 890–898. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, abe6474. [Google Scholar] [CrossRef] [PubMed]
- Yuen, G.J.; Demissie, E.; Pillai, S. B lymphocytes and cancer: A love-hate relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef]
- Güç, E.; Pollard, J.W. Redefining macrophage and neutrophil biology in the metastatic cascade. Immunity 2021, 54, 885–902. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- de Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Ma, Q.; Dieterich, L.C.; Detmar, M. Multiple roles of lymphatic vessels in tumor progression. Curr. Opin. Immunol. 2018, 53, 7–12. [Google Scholar] [CrossRef]
- Restaino, A.C.; Vermeer, P.D. Neural regulations of the tumor microenvironment. FASEB Bioadv. 2022, 4, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Fertig, E.J.; Lee, E.; Pandey, N.B.; Popel, A.S. Analysis of gene expression of secreted factors associated with breast cancer metastases in breast cancer subtypes. Sci. Rep. 2015, 5, 12133. [Google Scholar] [CrossRef] [PubMed]
- Norton, K.-A.; Jin, K.; Popel, A.S. Modeling triple-negative breast cancer heterogeneity: Effects of stromal macrophages, fibroblasts and tumor vasculature. J. Theor. Biol. 2018, 452, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Malone, M.K.; Smrekar, K.; Park, S.; Blakely, B.; Walter, A.; Nasta, N.; Park, J.; Considine, M.; Danilova, L.V.; Pandey, N.B.; et al. Cytokines secreted by stromal cells in TNBC microenvironment as potential targets for cancer therapy. Cancer Biol. Ther. 2020, 21, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liu, Y.; Ali, N.M.; Zhang, B.; Cui, X. The role of innate immune cells in the tumor microenvironment and research progress in anti-tumor therapy. Front. Immunol. 2022, 13, 1039260. [Google Scholar] [CrossRef]
- Lei, X.; Lei, Y.; Li, J.-K.; Du, W.-X.; Li, R.-G.; Yang, J.; Li, J.; Li, F.; Tan, H.-B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef]
- Duan, X.; He, K.; Li, J.; Cheng, M.; Song, H.; Liu, J.; Liu, P. Tumor associated macrophages deliver iron to tumor cells via Lcn2. Int. J. Physiol. Pathophysiol. Pharmacol. 2018, 10, 105–114. [Google Scholar]
- Recalcati, S.; Locati, M.; Marini, A.; Santambrogio, P.; Zaninotto, F.; de Pizzol, M.; Zammataro, L.; Girelli, D.; Cairo, G. Differential regulation of iron homeostasis during human macrophage polarized activation. Eur. J. Immunol. 2010, 40, 824–835. [Google Scholar] [CrossRef]
- Jung, M.; Ören, B.; Mora, J.; Mertens, C.; Dziumbla, S.; Popp, R.; Weigert, A.; Grossmann, N.; Fleming, I.; Brüne, B. Lipocalin 2 from macrophages stimulated by tumor cell-derived sphingosine 1-phosphate promotes lymphangiogenesis and tumor metastasis. Sci. Signal. 2016, 9, ra64. [Google Scholar] [CrossRef]
- Jaberi, S.A.; Cohen, A.; D’Souza, C.; Abdulrazzaq, Y.M.; Ojha, S.; Bastaki, S.; Adeghate, E.A. Lipocalin-2: Structure, function, distribution and role in metabolic disorders. Biomed. Pharmacother. 2021, 142, 112002. [Google Scholar] [CrossRef]
- Xiong, S.; Dong, L.; Cheng, L. Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 2021, 14, 173. [Google Scholar] [CrossRef]
- Obeagu, E.I. N1 and N2 neutrophil subtypes in breast cancer: Functional implications and clinical perspectives: A narrative review. Ann. Med. Surg. 2025, 87, 5762–5769. [Google Scholar] [CrossRef]
- Wei, C.T.; Tsai, I.T.; Wu, C.C.; Hung, W.C.; Hsuan, C.F.; Yu, T.H.; Hsu, C.C.; Houng, J.Y.; Chung, F.M.; Lee, Y.J.; et al. Elevated plasma level of neutrophil gelatinase-associated lipocalin (NGAL) in patients with breast cancer. Int. J. Med. Sci. 2021, 18, 2689–2696. [Google Scholar] [CrossRef]
- Su, Y.; Leng, M.; Yang, Q.; Jiang, W.; Xiang, G.; Long, L.; Zhou, X. Targeting circulating tumor cell–neutrophil interactions: Nanoengineered strategies for inhibiting cancer metastasis. J. Nanobiotechnol. 2025, 23, 449. [Google Scholar] [CrossRef]
- Al Qutami, F.; AlHalabi, W.; Vijayakumar, A.; Rawat, S.S.; Mossa, A.H.; Jayakumar, M.N.; Samreen, B.; Hachim, M.Y. Characterizing the Inflammatory Profile of Neutrophil-Rich Triple-Negative Breast Cancer. Cancers 2024, 16, 747. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Wang, Q.; Li, M.; Shen, J.; Ji, J. Quantitative Proteomics of Tissue-Infiltrating T Cells from CRC Patients Identified Lipocalin-2 Induces T-Cell Apoptosis and Promotes Tumor Cell Proliferation by Iron Efflux. Mol. Cell. Proteom. 2024, 23, 100691. [Google Scholar] [CrossRef] [PubMed]
- Floderer, M.; Prchal-Murphy, M.; Vizzardelli, C. Dendritic cell-secreted lipocalin2 induces CD8+ T-cell apoptosis, contributes to T-cell priming and leads to a TH1 phenotype. PLoS ONE 2014, 9, e101881. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274. [Google Scholar] [CrossRef]
- Cheung, S.Y.; Boey, Y.J.Y.; Koh, V.C.Y.; Thike, A.A.; Lim, J.C.T.; Iqbal, J.; Tan, P.H. Role of epithelial-mesenchymal transition markers in triple-negative breast cancer. Breast Cancer Res. Treat. 2015, 152, 489–498. [Google Scholar] [CrossRef]
- Sarrió, D.; Rodriguez-Pinilla, S.M.; Hardisson, D.; Cano, A.; Moreno-Bueno, G.; Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008, 68, 989–997. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Córcoles, R.; Fabra, A.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef]
- Del Pozo Martin, Y.; Park, D.; Ramachandran, A.; Ombrato, L.; Calvo, F.; Chakravarty, P.; Spencer-Dene, B.; Derzsi, S.; Hill, C.S.; Sahai, E.; et al. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. Cell Rep. 2015, 13, 2456–2469. [Google Scholar] [CrossRef] [PubMed]
- Grasset, E.M.; Dunworth, M.; Sharma, G.; Loth, M.; Tandurella, J.; Cimino-Mathews, A.; Gentz, M.; Bracht, S.; Haynes, M.; Fertig, E.J.; et al. Triple-negative breast cancer metastasis involves complex epithelial-mesenchymal transition dynamics and requires vimentin. Sci. Transl. Med. 2022, 14, eabn7571. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Li, G.; Yuan, Y.; He, Y.; Wu, X.; Zhang, W.; Wu, Z.; Chen, T.; Wu, W.; Lobie, P.E.; et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE 2012, 7, e41523. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Seftor, R.E.; Trevor, K.T. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J. Pathol. 1997, 150, 483–495. [Google Scholar]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- Yang, Z.; Lian, X.; Luo, Y.; Ye, Q.; Guo, G.; Liu, G. Glutamine synthetase shields triple-negative breast cancer cells from ferroptosis in metastasis triggered by glutamine deprivation. Breast Cancer Res. 2025, 27, 165. [Google Scholar] [CrossRef]
- Kim, G.W.; Lee, D.H.; Jeon, Y.H.; Yoo, J.; Kim, S.Y.; Lee, S.W.; Cho, H.Y.; Kwon, S.H. Glutamine Synthetase as a Therapeutic Target for Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 1701. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Sun, G.; Wang, J.; Liu, F.; Zhao, C.; Cui, S.; Wang, Z.; Liu, Z.; Zhang, Q.; Xiang, C.; Zhang, Y.; et al. G-4 inhibits triple negative breast cancer by inducing cell apoptosis and promoting LCN2-dependent ferroptosis. Biochem. Pharmacol. 2024, 222, 116077. [Google Scholar] [CrossRef] [PubMed]
- Meurer, S.K.; Tezcan, O.; Lammers, T.; Weiskirchen, R. Differential regulation of Lipocalin 2 (LCN2) in doxorubicin-resistant 4T1 triple negative breast cancer cells. Cell. Signal. 2020, 74, 109731. [Google Scholar] [CrossRef]
- Cheng, G.; Sun, X.; Wang, J.; Xiao, G.; Wang, X.; Fan, X.; Zu, L.; Hao, M.; Qu, Q.; Mao, Y.; et al. HIC1 silencing in triple-negative breast cancer drives progression through misregulation of LCN2. Cancer Res. 2014, 74, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Meka, P.b.; Jarjapu, S.; Nanchari, S.R.; Vishwakarma, S.K.; Edathara, P.M.; Gorre, M.; Cingeetham, A.; Vuree, S.; Annamaneni, S.; Dunna, N.R.; et al. LCN2 Promoter Methylation Status as Novel Predictive Marker for Microvessel Density and Aggressive Tumor Phenotype in Breast Cancer Patients. Asian Pac. J. Cancer Prev. 2015, 16, 4965–4969. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; He, Y.; Yi, J.; Chen, J.; Guo, Q.; Liao, N.; Peng, L. WTAP Mediates NUPR1 Regulation of LCN2 Through m6A Modification to Influence Ferroptosis, Thereby Promoting Breast Cancer Proliferation, Migration and Invasion. Biochem. Genet. 2024, 62, 876–891. [Google Scholar] [CrossRef]
- Parrella, P.; Scintu, M.; Prencipe, M.; Poeta, M.L.; Gallo, A.P.; Rabitti, C.; Rinaldi, M.; Tommasi, S.; Paradiso, A.; Schittulli, F.; et al. HIC1 promoter methylation and 17p13.3 allelic loss in invasive ductal carcinoma of the breast. Cancer Lett. 2005, 222, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Wang, D.H.; Yen, R.C.; Luo, J.; Gu, W.; Baylin, S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005, 123, 437–448. [Google Scholar] [CrossRef]
- van Rechem, C.; Boulay, G.; Pinte, S.; Stankovic-Valentin, N.; Guérardel, C.; Leprince, D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol. Cell. Biol. 2010, 30, 4045–4059. [Google Scholar] [CrossRef]
- Hao, P.; Li, H.; Wu, A.; Zhang, J.; Wang, C.; Xian, X.; Ren, Q.; Hao, N.; Wang, Y.; Yue, F.; et al. Lipocalin2 promotes cell proliferation and migration in ovarian cancer through activation of the ERK/GSK3β/β-catenin signaling pathway. Life Sci. 2020, 262, 118492. [Google Scholar] [CrossRef]
- Monisha, J.; Roy, N.K.; Padmavathi, G.; Banik, K.; Bordoloi, D.; Khwairakpam, A.D.; Arfuso, F.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; et al. NGAL is Downregulated in Oral Squamous Cell Carcinoma and Leads to Increased Survival, Proliferation, Migration and Chemoresistance. Cancers 2018, 10, 228. [Google Scholar] [CrossRef]
- Feng, M.; Feng, J.; Chen, W.; Wang, W.; Wu, X.; Zhang, J.; Xu, F.; Lai, M. Lipocalin2 suppresses metastasis of colorectal cancer by attenuating NF-κB-dependent activation of snail and epithelial mesenchymal transition. Mol. Cancer 2016, 15, 77. [Google Scholar] [CrossRef]
- Xu, B.; Jin, D.Y.; Lou, W.H.; Wang, D.S. Lipocalin-2 is associated with a good prognosis and reversing epithelial-to-mesenchymal transition in pancreatic cancer. World J. Surg. 2013, 37, 1892–1900. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef]
- Piedra-Delgado, L.; Chambergo-Michilot, D.; Morante, Z.; Fairen, C.; Jerves-Coello, F.; Luque-Benavides, R.; Casas, F.; Bustamante, E.; Razuri-Bustamante, C.; Torres-Roman, J.S.; et al. Survival according to the site of metastasis in triple-negative breast cancer patients: The Peruvian experience. PLoS ONE 2024, 19, e0293833. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Gu, Y.; Yang, J.; Xu, L.; Mi, W.; Yu, W. Lipocalin 2 promotes lung metastasis of murine breast cancer cells. J. Exp. Clin. Cancer Res. 2008, 27, 83. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Ding, T.; Lin, H.; Wang, Y.; Hu, L.; Hu, J.; Feig, B.; Zhang, W.; Pusztai, L.; Symmans, W.F.; et al. Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 2009, 69, 8579–8584. [Google Scholar] [CrossRef]
- Chi, Y.; Remsik, J.; Kiseliovas, V.; Derderian, C.; Sener, U.; Alghader, M.; Saadeh, F.; Nikishina, K.; Bale, T.; Iacobuzio-Donahue, C.; et al. Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis. Science 2020, 369, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Smith, M.R.; Wang, Y.; D’Agostino, R.; Ruiz, J.; Lycan, T.; Kucera, G.L.; Miller, L.D.; Li, W.; Chan, M.D.; et al. c-Met Mediated Cytokine Network Promotes Brain Metastasis of Breast Cancer by Remodeling Neutrophil Activities. Cancers 2023, 15, 2626. [Google Scholar] [CrossRef]
- Berger, T.; Cheung, C.C.; Elia, A.J.; Mak, T.W. Disruption of the Lcn2 gene in mice suppresses primary mammary tumor formation but does not decrease lung metastasis. Proc. Natl. Acad. Sci. USA 2010, 107, 2995–3000. [Google Scholar] [CrossRef]
- Jin, J.; Gao, Y.; Zhang, J.; Wang, L.; Wang, B.; Cao, J.; Shao, Z.; Wang, Z. Incidence, pattern and prognosis of brain metastases in patients with metastatic triple negative breast cancer. BMC Cancer 2018, 18, 446. [Google Scholar] [CrossRef] [PubMed]
- Spector, R.; Robert Snodgrass, S.; Johanson, C.E. A balanced view of the cerebrospinal fluid composition and functions: Focus on adult humans. Exp. Neurol. 2015, 273, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Buschhaus, J.M.; Rajendran, S.; Humphries, B.A.; Cutter, A.C.; Muñiz, A.J.; Ciavattone, N.G.; Buschhaus, A.M.; Cañeque, T.; Nwosu, Z.C.; Sahoo, D.; et al. Effects of iron modulation on mesenchymal stem cell-induced drug resistance in estrogen receptor-positive breast cancer. Oncogene 2022, 41, 3705–3718. [Google Scholar] [CrossRef]
- Mackey, J.B.G.; Coffelt, S.B.; Carlin, L.M. Neutrophil Maturity in Cancer. Front. Immunol. 2019, 10, 1912. [Google Scholar] [CrossRef]
- Loh, J.J.; Ma, S. Hallmarks of cancer stemness. Cell Stem Cell 2024, 31, 617–639. [Google Scholar] [CrossRef]
- Lin, N.U.; Claus, E.; Sohl, J.; Razzak, A.R.; Arnaout, A.; Winer, E.P. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: High incidence of central nervous system metastases. Cancer 2008, 113, 2638–2645. [Google Scholar] [CrossRef]
- Luo, A.; Wu, F.; Han, R.; Huang, S.; Zhang, Y.; Jing, X.; Zhao, X. Clinicopathological features and prognostic evaluation of bone metastasis in triple-negative breast cancer. J. Cancer Res. Ther. 2017, 13, 778–784. [Google Scholar] [CrossRef]
- Yao, Y.; Chu, Y.; Xu, B.; Hu, Q.; Song, Q. Risk factors for distant metastasis of patients with primary triple-negative breast cancer. Biosci. Rep. 2019, 39, BSR20190288. [Google Scholar] [CrossRef]
- Bhatia, S.; Kramer, M.; Russo, S.; Naik, P.; Arun, G.; Brophy, K.; Andrews, P.; Fan, C.; Perou, C.M.; Preall, J.; et al. Patient-Derived Triple-Negative Breast Cancer Organoids Provide Robust Model Systems That Recapitulate Tumor Intrinsic Characteristics. Cancer Res. 2022, 82, 1174–1192. [Google Scholar] [CrossRef]

| TNBC Subtype [40] | TNBC Subtype [41] | Characteristics | Cell Line | LCN2 Expression (nTPM) * |
|---|---|---|---|---|
| Basal-like 1 | BL-1 | ↑ Ki67 [42] ↑ MYC activity (associated with worse survival) [43] ↑ RhoA activity (linked to poor prognosis) [43] ↑ basal cytokeratins (CK5/6, CK14, CK17) [44] Best overall survival among other TNBC subtypes [41] | HCC38 | 9.9 |
| HCC1143 | 284.2 | |||
| HCC1599 | 1225.9 | |||
| HCC1937 | 1796.1 | |||
| HCC2157 | 195.4 | |||
| MDA-MB-468 | 351.4 | |||
| Basal-like 2 | BL-2 | Basal-myoepithelial phenotype [42] ↑ Growth-factor signaling (EGFR, Wnt/β-catenin) [40] ↑ Glycolysis and gluconeogenesis ↑ E2F2 pathway (associated with poor overall survival) [45] ↑ TGF-β pathway (associated with worse overall and disease-free survival) [46] ↑ Basal cytokeratins (CK5/6, CK14, CK17) [44] | HCC70 | 270.5 |
| Mesenchymal | M | Genetic patterns responsible for cell motility and cell differentiation (Wnt, ALK pathway) [42] ↑ Extracellular matrix-receptor interactions [47] ↑ Genes involved in epithelial–mesenchymal transition [48] ↑ Resistance to chemotherapeutic agents [49] | BT-549 | 3.7 |
| Hs 578T | 0.4 | |||
| Mesenchymal stem-like | Genetic profiles associated with growth factor signaling pathways (EGFR, PDGF) [43] ↓ Proliferation-associated genes [47] ↑ Stem-cell-associated genes [47] ↓ Claudins [42] Pro-angiogenic gene expression (VEGFC, SEMA3G, SEMA5A) [50] | MDA-MB-231 | 16.4 | |
| MDA-MB-436 | 289.0 | |||
| MDA-MB-157 | 0.3 | |||
| Immunomodulatory | Gene ontologies related to immune cell processes (e.g., immune signal transduction) [42] ↑ TILs [51] ↑ IFN-α and IFN-γ pathways [43] Treatable with immune checkpoint inhibitors [51] More favorable prognosis compared to other subtypes [51] | DU4475 | 0.3 | |
| HCC1187 | 16.4 | |||
| HCC1806 | 137.6 | |||
| LAR | LAR | ↑ Androgen receptor ↑ Hormonal signaling pathways (steroid synthesis, androgen/estrogen metabolism) [48] ↓ Ki67 [52] ↑ Luminal cytokeratins (CK7/8, CK18, CK19) [53] | MDA-MB-453 | 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keller, D.T.; Weiskirchen, R.; Schröder-Lange, S.K. Lipocalin-2 in Triple-Negative Breast Cancer: A Review of Its Pathophysiological Role in the Metastatic Cascade. Int. J. Mol. Sci. 2025, 26, 10938. https://doi.org/10.3390/ijms262210938
Keller DT, Weiskirchen R, Schröder-Lange SK. Lipocalin-2 in Triple-Negative Breast Cancer: A Review of Its Pathophysiological Role in the Metastatic Cascade. International Journal of Molecular Sciences. 2025; 26(22):10938. https://doi.org/10.3390/ijms262210938
Chicago/Turabian StyleKeller, Diandra T., Ralf Weiskirchen, and Sarah K. Schröder-Lange. 2025. "Lipocalin-2 in Triple-Negative Breast Cancer: A Review of Its Pathophysiological Role in the Metastatic Cascade" International Journal of Molecular Sciences 26, no. 22: 10938. https://doi.org/10.3390/ijms262210938
APA StyleKeller, D. T., Weiskirchen, R., & Schröder-Lange, S. K. (2025). Lipocalin-2 in Triple-Negative Breast Cancer: A Review of Its Pathophysiological Role in the Metastatic Cascade. International Journal of Molecular Sciences, 26(22), 10938. https://doi.org/10.3390/ijms262210938








