Post-Translational Modifications of Huntingtin: Mechanistic Insights and Therapeutic Opportunities in Huntington’s Disease
Abstract
1. Introduction
2. Physiological Functions of Wild-Type HTT
3. Pathogenic Roles of mHTT
4. Post-Translational Modifications of HTT and Their Functional Implications
4.1. Phosphorylation Modifications (Summarized in Table 1)
4.1.1. Phosphorylations in the N17 Domain
4.1.2. Modulators of HTT Phosphorylation in the N17 Domain
4.1.3. Other Phosphorylation Sites on HTT Protein
4.1.4. Phosphorylations with Limited Information
| Residue | Function | Kinase | Phosphatase | Identification/Validation Method | References |
|---|---|---|---|---|---|
| T3 | Inhibits mHTT aggregation and decreases mHTT toxicity | IKKβ, GCK, HGK, TNIK | Not reported | MS/Ab/Phos-Tag SDS-PAGE | [46,87] |
| S13 | Inhibits mHTT aggregation, enhances mHTT degradation, and decreases mHTT toxicity | CK2, TBK1, HGK, TNIK, IKKβ | Not reported | MS/Ab | [46,60] |
| S16 | Inhibits mHTT aggregation, enhances mHTT degradation, and decreases mHTT toxicity | CK2, TBK1 | Not reported | MS/Ab | [46] |
| T107/S116 | Double phosphorylation enhances mHTT aggregation | MST3 | Not reported | ESI-MS/UPLC/Ab | [64] |
| S120 | Increases mHTT degradation and decreases aggregation and toxicity | NLK | Not reported | MS/in vitro kinase assays | [64,65] |
| S421 | Maintains mitochondrial potential and anterograde/retrograde vesicle transport. | AKT, SGK | PP1 and PP2A | MS/Ab | [67,68,78,82,88,89] |
| S434 | Decreases caspase-mediated mHTT cleavage and mHTT toxicity | CDK5 | Not reported | MS/Ab | [70,77,89] |
| S536 | Decreases calpain-mediated mHTT cleavage and cytotoxicity | Not reported | Not reported | MS | [14] |
| S1181/S1201 | Protects the cell from DNA damage-induced cell death, and dephosphorylation reduces mood disorders | CDK5 | Not reported | MS/Ab | [14,77,78,82] |
| S2114 | Enhances mHTT and PRC2 interaction | Not reported | Not reported | MS/Ab | [14,80] |
| S2116 | Enhances mHTT and PRC2 interaction, disrupts normal conformational stability. | Not reported | Not reported | MS/Ab | [14,80] |
| S2657 | Regulates axonal transport and mHTT-mediated neuronal cell death | ERK, GSK3β | Not reported | MS/in vitro kinase assay | [82] |
4.2. Acetylation
4.3. Ubiquitination
4.3.1. Ubiquitinations That Have Specific Functions
4.3.2. HTT Related E3 Ligase
4.4. SUMOylation
4.5. Palmitoylation Modification
4.6. Crosstalk Among PTMs
5. Pharmacological Modulation of PTMs
5.1. HDAC Inhibitors
5.2. Kinase Modulators
5.3. Palmitoylation Enhancer
6. Can Modulating Post-Translational Modifications Functionally Reverse mHTT Toxicity?
7. Research Limitations and Future Perspectives
7.1. Limitations of Experimental Models in Huntington’s Disease Research
7.2. Limitation of PTM-Mimetic or PTM-Null Mutants
7.3. Limited Understanding of the Interaction Between Striatum Metabolism and PTM
8. Summary and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ross, C.A.; Tabrizi, S.J. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011, 10, 83–98. [Google Scholar] [CrossRef]
- Gutekunst, C.-A.; Li, S.-H.; Yi, H.; Mulroy, J.S.; Kuemmerle, S.; Jones, R.; Rye, D.; Ferrante, R.J.; Hersch, S.M.; Li, X.-J. Nuclear and Neuropil Aggregates in Huntington’s Disease: Relationship to Neuropathology. J. Neurosci. 1999, 19, 2522–2534. [Google Scholar] [CrossRef]
- Takahashi, T.; Kikuchi, S.; Katada, S.; Nagai, Y.; Nishizawa, M.; Onodera, O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 2008, 17, 345–356. [Google Scholar] [CrossRef]
- Miller, J.; Arrasate, M.; Brooks, E.; Libeu, C.P.; Legleiter, J.; Hatters, D.; Curtis, J.; Cheung, K.; Krishnan, P.; Mitra, S.; et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 2011, 7, 925–934, Erratum in: Nat. Chem. Biol. 2012, 8, 318. [Google Scholar] [CrossRef]
- Zhai, W.; Jeong, H.; Cui, L.; Krainc, D.; Tjian, R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell 2005, 123, 1241–1253. [Google Scholar] [CrossRef]
- Milakovic, T.; Quintanilla, R.A.; Johnson, G.V. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: Functional consequences. J. Biol. Chem. 2006, 281, 34785–34795. [Google Scholar] [CrossRef]
- Vonsattel, J.P.; Myers, R.H.; Stevens, T.J.; Ferrante, R.J.; Bird, E.D.; Richardson, E.P., Jr. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985, 44, 559–577. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Ramdzan, Y.M.; Trubetskov, M.M.; Ormsby, A.R.; Newcombe, E.A.; Sui, X.; Tobin, M.J.; Bongiovanni, M.N.; Gras, S.L.; Dewson, G.; Miller, J.M.L.; et al. Huntingtin Inclusions Trigger Cellular Quiescence, Deactivate Apoptosis, and Lead to Delayed Necrosis. Cell Rep. 2017, 19, 919–927. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Scherzinger, E.; Lurz, R.; Turmaine, M.; Mangiarini, L.; Hollenbach, B.; Hasenbank, R.; Bates, G.P.; Davies, S.W.; Lehrach, H.; Wanker, E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 1997, 90, 549–558. [Google Scholar] [CrossRef]
- Cooper, J.K.; Schilling, G.; Peters, M.F.; Herring, W.J.; Sharp, A.H.; Kaminsky, Z.; Masone, J.; Khan, F.A.; Delanoy, M.; Borchelt, D.R.; et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 1998, 7, 783–790. [Google Scholar] [CrossRef]
- Jeong, H.; Then, F.; Melia, T.J., Jr.; Mazzulli, J.R.; Cui, L.; Savas, J.N.; Voisine, C.; Paganetti, P.; Tanese, N.; Hart, A.C.; et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 2009, 137, 60–72. [Google Scholar] [CrossRef]
- Schilling, B.; Gafni, J.; Torcassi, C.; Cong, X.; Row, R.H.; LaFevre-Bernt, M.A.; Cusack, M.P.; Ratovitski, T.; Hirschhorn, R.; Ross, C.A.; et al. Huntingtin Phosphorylation Sites Mapped by Mass Spectrometry: Modulation of Cleavage and Toxicity. J. Biol. Chem. 2006, 281, 23686–23697. [Google Scholar] [CrossRef]
- Sap, K.A.; Guler, A.T.; Bezstarosti, K.; Bury, A.E.; Juenemann, K.; Demmers, J.A.; Reits, E.A. Global Proteome and Ubiquitinome Changes in the Soluble and Insoluble Fractions of Q175 Huntington Mice Brains. Mol. Cell. Proteom. 2019, 18, 1705–1720. [Google Scholar] [CrossRef]
- Steffan, J.S.; Agrawal, N.; Pallos, J.; Rockabrand, E.; Trotman, L.C.; Slepko, N.; Illes, K.; Lukacsovich, T.; Zhu, Y.Z.; Cattaneo, E.; et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science 2004, 304, 100–104. [Google Scholar] [CrossRef]
- Yanai, A.; Huang, K.; Kang, R.; Singaraja, R.R.; Arstikaitis, P.; Gan, L.; Orban, P.C.; Mullard, A.; Cowan, C.M.; Raymond, L.A.; et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 2006, 9, 824–831. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Trottier, Y.; Devys, D.; Imbert, G.; Saudou, F.; An, I.; Lutz, Y.; Weber, C.; Agid, Y.; Hirsch, E.C.; Mandel, J.L. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nat. Genet. 1995, 10, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, S.; Liu, J.-P.; Chapman, D.L.; Papaioannou, V.E.; Efstratiadis, A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 1995, 11, 155–163. [Google Scholar] [CrossRef]
- Kozłowska, E.; Ciołak, A.; Adamek, G.; Szcześniak, J.; Fiszer, A. HTT loss-of-function contributes to RNA deregulation in developing Huntington’s disease neurons. Cell Biosci. 2025, 15, 100. [Google Scholar] [CrossRef]
- Carpentier, R.; Kim, J.; Capizzi, M.; Kim, H.; Fäßler, F.; Hansen, J.M.; Kim, M.J.; Denarier, E.; Blot, B.; Degennaro, M.; et al. Structure of the Huntingtin F-actin complex reveals its role in cytoskeleton organization. Sci. Adv. 2025, 11, eadw4124. [Google Scholar] [CrossRef]
- Louessard, M.; Cailleret, M.; Jarrige, M.; Bigarreau, J.; Lenoir, S.; Dufour, N.; Rey, M.; Saudou, F.; Deglon, N.; Perrier, A.L. Mono- and Biallelic Inactivation of Huntingtin Gene in Patient-Specific Induced Pluripotent Stem Cells Reveal HTT Roles in Striatal Development and Neuronal Functions. J. Huntington’s Dis. 2024, 13, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Prowse, E.N.P.; Chaudhary, A.R.; Sharon, D.; Hendricks, A.G. Huntingtin S421 phosphorylation increases kinesin and dynein engagement on early endosomes and lysosomes. Biophys. J. 2023, 122, 1168–1184. [Google Scholar] [CrossRef]
- Dietrich, P.; Johnson, I.M.; Alli, S.; Dragatsis, I. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLoS Genet. 2017, 13, e1006846. [Google Scholar] [CrossRef] [PubMed]
- Ehinger, Y.; Bruyère, J.; Panayotis, N.; Abada, Y.S.; Borloz, E.; Matagne, V.; Scaramuzzino, C.; Vitet, H.; Delatour, B.; Saidi, L. Huntingtin phosphorylation governs BDNF homeostasis and improves the phenotype of Mecp2 knockout mice. EMBO Mol. Med. 2020, 12, e10889. [Google Scholar] [CrossRef] [PubMed]
- Cason, S.E.; Carman, P.J.; Van Duyne, C.; Goldsmith, J.; Dominguez, R.; Holzbaur, E.L. Sequential dynein effectors regulate axonal autophagosome motility in a maturation-dependent pathway. J. Cell Biol. 2021, 220, e202010179. [Google Scholar] [CrossRef]
- Liot, G.; Zala, D.; Pla, P.; Mottet, G.; Piel, M.; Saudou, F. Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J. Neurosci. 2013, 33, 6298–6309. [Google Scholar] [CrossRef]
- Bruyere, J.; Abada, Y.-S.; Vitet, H.; Fontaine, G.; Deloulme, J.-C.; Cès, A.; Denarier, E.; Pernet-Gallay, K.; Andrieux, A.; Humbert, S. Presynaptic APP levels and synaptic homeostasis are regulated by Akt phosphorylation of huntingtin. eLife 2020, 9, e56371. [Google Scholar] [CrossRef]
- Twelvetrees, A.E.; Yuen, E.Y.; Arancibia-Carcamo, I.L.; MacAskill, A.F.; Rostaing, P.; Lumb, M.J.; Humbert, S.; Triller, A.; Saudou, F.; Yan, Z. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 2010, 65, 53–65. [Google Scholar] [CrossRef]
- Pearl, J.R.; Shetty, A.C.; Cantle, J.P.; Bergey, D.E.; Bragg, R.M.; Coffey, S.R.; Kordasiewicz, H.B.; Hood, L.E.; Price, N.D.; Ament, S.A.; et al. Altered huntingtin-chromatin interactions predict transcriptional and epigenetic changes in Huntington’s disease. Dis. Model. Mech. 2025, 18, dmm052282. [Google Scholar] [CrossRef]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef]
- Choi, Y.B.; Kadakkuzha, B.M.; Liu, X.A.; Akhmedov, K.; Kandel, E.R.; Puthanveettil, S.V. Huntingtin is critical both pre- and postsynaptically for long-term learning-related synaptic plasticity in Aplysia. PLoS ONE 2014, 9, e103004. [Google Scholar] [CrossRef]
- Gauthier, L.R.; Charrin, B.C.; Borrell-Pagès, M.; Dompierre, J.P.; Rangone, H.; Cordelières, F.P.; De Mey, J.; MacDonald, M.E.; Lessmann, V.; Humbert, S.; et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 2004, 118, 127–138. [Google Scholar] [CrossRef]
- Caviston, J.P.; Holzbaur, E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009, 19, 147–155. [Google Scholar] [CrossRef]
- Zuccato, C.; Ciammola, A.; Rigamonti, D.; Leavitt, B.R.; Goffredo, D.; Conti, L.; MacDonald, M.E.; Friedlander, R.M.; Silani, V.; Hayden, M.R.; et al. Loss of Huntingtin-Mediated BDNF Gene Transcription in Huntington’s Disease. Science 2001, 293, 493–498. [Google Scholar] [CrossRef]
- Rui, Y.N.; Xu, Z.; Patel, B.; Cuervo, A.M.; Zhang, S. HTT/Huntingtin in selective autophagy and Huntington disease: A foe or a friend within? Autophagy 2015, 11, 858–860. [Google Scholar] [CrossRef] [PubMed]
- Quintanilla, R.A.; Jin, Y.N.; von Bernhardi, R.; Johnson, G.V. Mitochondrial permeability transition pore induces mitochondria injury in Huntington disease. Mol. Neurodegener. 2013, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Yano, H.; Baranov, S.V.; Baranova, O.V.; Kim, J.; Pan, Y.; Yablonska, S.; Carlisle, D.L.; Ferrante, R.J.; Kim, A.H.; Friedlander, R.M. Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci. 2014, 17, 822–831. [Google Scholar] [CrossRef]
- Verhoef, L.G.G.C.; Lindsten, K.; Masucci, M.G.; Dantuma, N.P. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 2002, 11, 2689–2700. [Google Scholar] [CrossRef] [PubMed]
- Franco-Iborra, S.; Plaza-Zabala, A.; Montpeyo, M.; Sebastian, D.; Vila, M.; Martinez-Vicente, M. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy 2021, 17, 672–689. [Google Scholar] [CrossRef]
- Erie, C.; Sacino, M.; Houle, L.; Lu, M.L.; Wei, J. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur. J. Neurosci. 2015, 42, 1941–1951. [Google Scholar] [CrossRef]
- Sap, K.A.; Geijtenbeek, K.W.; Schipper-Krom, S.; Guler, A.T.; Reits, E.A. Ubiquitin-modifying enzymes in Huntington’s disease. Front. Mol. Biosci. 2023, 10, 1107323. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.M.; Aiken, C.T.; Kaltenbach, L.S.; Agrawal, N.; Illes, K.; Khoshnan, A.; Martinez-Vincente, M.; Arrasate, M.; O’Rourke, J.G.; Khashwji, H.; et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009, 187, 1083–1099. [Google Scholar] [CrossRef]
- O’Rourke, J.G.; Gareau, J.R.; Ochaba, J.; Song, W.; Raskó, T.; Reverter, D.; Lee, J.; Monteys, A.M.; Pallos, J.; Mee, L.; et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013, 4, 362–375. [Google Scholar] [CrossRef]
- Chiki, A.; Ricci, J.; Hegde, R.; Abriata, L.A.; Reif, A.; Boudeffa, D.; Lashuel, H.A. Site-Specific Phosphorylation of Huntingtin Exon 1 Recombinant Proteins Enabled by the Discovery of Novel Kinases. Chembiochem 2021, 22, 217–231. [Google Scholar] [CrossRef]
- Tao, M.; Pandey, N.K.; Barnes, R.; Han, S.; Langen, R. Structure of Membrane-Bound Huntingtin Exon 1 Reveals Membrane Interaction and Aggregation Mechanisms. Structure 2019, 27, 1570–1580.E4. [Google Scholar] [CrossRef]
- Gu, X.; Cantle, J.P.; Greiner, E.R.; Lee, C.Y.; Barth, A.M.; Gao, F.; Park, C.S.; Zhang, Z.; Sandoval-Miller, S.; Zhang, R.L.; et al. N17 Modifies mutant Huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron 2015, 85, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Palidwor, G.A.; Shcherbinin, S.; Huska, M.R.; Rasko, T.; Stelzl, U.; Arumughan, A.; Foulle, R.; Porras, P.; Sanchez-Pulido, L.; Wanker, E.E.; et al. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput. Biol. 2009, 5, e1000304. [Google Scholar] [CrossRef] [PubMed]
- Aiken, C.T.; Steffan, J.S.; Guerrero, C.M.; Khashwji, H.; Lukacsovich, T.; Simmons, D.; Purcell, J.M.; Menhaji, K.; Zhu, Y.-Z.; Green, K.; et al. Phosphorylation of Threonine 3: Implications for Huntingtin Aggregation and Neurotoxicity*. J. Biol. Chem. 2009, 284, 29427–29436. [Google Scholar] [CrossRef]
- Branco-Santos, J.; Herrera, F.; Poças, G.M.; Pires-Afonso, Y.; Giorgini, F.; Domingos, P.M.; Outeiro, T.F. Protein phosphatase 1 regulates huntingtin exon 1 aggregation and toxicity. Hum. Mol. Genet. 2017, 26, 3763–3775. [Google Scholar] [CrossRef]
- Chiki, A.; DeGuire, S.M.; Ruggeri, F.S.; Sanfelice, D.; Ansaloni, A.; Wang, Z.M.; Cendrowska, U.; Burai, R.; Vieweg, S.; Pastore, A.; et al. Mutant Exon1 Huntingtin Aggregation is Regulated by T3 Phosphorylation-Induced Structural Changes and Crosstalk between T3 Phosphorylation and Acetylation at K6. Angew. Chem. Int. Ed. Engl. 2017, 56, 5202–5207. [Google Scholar] [CrossRef]
- Zhang, Z.; Gehin, C.; Abriata, L.A.; Dal Peraro, M.; Lashuel, H. Differential Effects of Post-translational Modifications on the Membrane Interaction of Huntingtin Protein. ACS Chem. Neurosci. 2024, 15, 2408–2419. [Google Scholar] [CrossRef]
- Gu, X.; Greiner, E.R.; Mishra, R.; Kodali, R.; Osmand, A.; Finkbeiner, S.; Steffan, J.S.; Thompson, L.M.; Wetzel, R.; Yang, X.W. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron 2009, 64, 828–840. [Google Scholar] [CrossRef]
- Hegde, R.N.; Chiki, A.; Petricca, L.; Martufi, P.; Arbez, N.; Mouchiroud, L.; Auwerx, J.; Landles, C.; Bates, G.P.; Singh-Bains, M.K.; et al. TBK1 phosphorylates mutant Huntingtin and suppresses its aggregation and toxicity in Huntington’s disease models. EMBO J. 2020, 39, e104671. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Y.; Bäuerlein, F.J.B.; Saha, I.; Hartl, F.U.; Baumeister, W.; Wilfling, F. Autophagy preferentially degrades non-fibrillar polyQ aggregates. Mol. Cell 2024, 84, 1980–1994.E8. [Google Scholar] [CrossRef]
- Atwal, R.S.; Desmond, C.R.; Caron, N.; Maiuri, T.; Xia, J.; Sipione, S.; Truant, R. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat. Chem. Biol. 2011, 7, 453–460. [Google Scholar] [CrossRef]
- Bowie, L.E.; Maiuri, T.; Alpaugh, M.; Gabriel, M.; Arbez, N.; Galleguillos, D.; Hung, C.L.K.; Patel, S.; Xia, J.; Hertz, N.T.; et al. N6-Furfuryladenine is protective in Huntington’s disease models by signaling huntingtin phosphorylation. Proc. Natl. Acad. Sci. USA 2018, 115, E7081–E7090. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; Ruzzene, M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 2019, 38, 287. [Google Scholar] [CrossRef] [PubMed]
- Cariulo, C.; Martufi, P.; Verani, M.; Toledo-Sherman, L.; Lee, R.; Dominguez, C.; Petricca, L.; Caricasole, A. IKBKB reduces huntingtin aggregation by phosphorylating serine 13 via a non-canonical IKK pathway. Life Sci. Alliance 2023, 6, e202302006. [Google Scholar] [CrossRef]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- da Cruz e Silva, E.F.; Fox, C.A.; Ouimet, C.C.; Gustafson, E.; Watson, S.J.; Greengard, P. Differential expression of protein phosphatase 1 isoforms in mammalian brain. J. Neurosci. 1995, 15, 3375–3389. [Google Scholar] [CrossRef]
- Arbez, N.; Ratovitski, T.; Roby, E.; Chighladze, E.; Stewart, J.C.; Ren, M.; Wang, X.; Lavery, D.J.; Ross, C.A. Post-translational modifications clustering within proteolytic domains decrease mutant huntingtin toxicity. J. Biol. Chem. 2017, 292, 19238–19249. [Google Scholar] [CrossRef]
- Kolla, R.; Gopinath, P.; Ricci, J.; Reif, A.; Rostami, I.; Lashuel, H.A. A New Chemoenzymatic Semisynthetic Approach Provides Insight into the Role of Phosphorylation beyond Exon1 of Huntingtin and Reveals N-Terminal Fragment Length-Dependent Distinct Mechanisms of Aggregation. J. Am. Chem. Soc. 2021, 143, 9798–9812. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, X.; Liu, H.; LeBron, J.; Alexandris, A.; Peng, Q.; Gu, H.; Yang, F.; Li, Y.; Wang, R.; et al. Nemo-like kinase reduces mutant huntingtin levels and mitigates Huntington’s disease. Hum. Mol. Genet. 2020, 29, 1340–1352. [Google Scholar] [CrossRef]
- Humbert, S.; Bryson, E.A.; Cordelières, F.P.; Connors, N.C.; Datta, S.R.; Finkbeiner, S.; Greenberg, M.E.; Saudou, F. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev. Cell 2002, 2, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Zala, D.; Colin, E.; Rangone, H.; Liot, G.; Humbert, S.; Saudou, F. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum. Mol. Genet. 2008, 17, 3837–3846. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ng, B.; Sim, B.; Radulescu, C.I.; Yusof, N.; Goh, W.I.; Lin, S.; Lim, J.S.Y.; Cha, Y.; Kusko, R.; et al. pS421 huntingtin modulates mitochondrial phenotypes and confers neuroprotection in an HD hiPSC model. Cell Death Dis. 2020, 11, 809. [Google Scholar] [CrossRef]
- Rangone, H.; Poizat, G.; Troncoso, J.; Ross, C.A.; MacDonald, M.E.; Saudou, F.; Humbert, S. The serum- and glucocorticoid-induced kinase SGK inhibits mutant huntingtin-induced toxicity by phosphorylating serine 421 of huntingtin. Eur. J. Neurosci. 2004, 19, 273–279. [Google Scholar] [CrossRef]
- Luo, S.; Vacher, C.; Davies, J.E.; Rubinsztein, D.C. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: Implications for mutant huntingtin toxicity. J. Cell Biol. 2005, 169, 647–656. [Google Scholar] [CrossRef]
- Brito, V.; Giralt, A.; Masana, M.; Royes, A.; Espina, M.; Sieiro, E.; Alberch, J.; Castañé, A.; Girault, J.-A.; Ginés, S. Cyclin-Dependent Kinase 5 Dysfunction Contributes to Depressive-like Behaviors in Huntington’s Disease by Altering the DARPP-32 Phosphorylation Status in the Nucleus Accumbens. Biol. Psychiatry 2019, 86, 196–207. [Google Scholar] [CrossRef]
- Gafni, J.; Hermel, E.; Young, J.E.; Wellington, C.L.; Hayden, M.R.; Ellerby, L.M. Inhibition of Calpain Cleavage of Huntingtin Reduces Toxicity: Accumulation of Calpain/Caspase Fragments in the Nucleus*. J. Biol. Chem. 2004, 279, 20211–20220. [Google Scholar] [CrossRef]
- Barbaro, B.A.; Lukacsovich, T.; Agrawal, N.; Burke, J.; Bornemann, D.J.; Purcell, J.M.; Worthge, S.A.; Caricasole, A.; Weiss, A.; Song, W.; et al. Comparative study of naturally occurring huntingtin fragments in Drosophila points to exon 1 as the most pathogenic species in Huntington’s disease. Hum. Mol. Genet. 2015, 24, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Landles, C.; Sathasivam, K.; Weiss, A.; Woodman, B.; Moffitt, H.; Finkbeiner, S.; Sun, B.; Gafni, J.; Ellerby, L.M.; Trottier, Y.; et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 2010, 285, 8808–8823. [Google Scholar] [CrossRef] [PubMed]
- Southwell, A.L.; Bugg, C.W.; Kaltenbach, L.S.; Dunn, D.; Butland, S.; Weiss, A.; Paganetti, P.; Lo, D.C.; Patterson, P.H. Perturbation with intrabodies reveals that calpain cleavage is required for degradation of huntingtin exon 1. PLoS ONE 2011, 6, e16676. [Google Scholar] [CrossRef] [PubMed]
- Mauthe, M.; van de Beek, N.; Mari, M.; Korsten, G.; Nobari, P.; Castelino, K.B.; de Mattos, E.P.; Ouhida, I.; Dijkstra, J.L.; Schipper-Krom, S.; et al. A chaperone-proteasome-based fragmentation machinery is essential for aggrephagy. Nat. Cell Biol. 2025, 27, 1448–1464. [Google Scholar] [CrossRef]
- Anne, S.L.; Saudou, F.; Humbert, S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 2007, 27, 7318–7328. [Google Scholar] [CrossRef]
- Ratovitski, T.; O’Meally, R.N.; Jiang, M.; Chaerkady, R.; Chighladze, E.; Stewart, J.C.; Wang, X.; Arbez, N.; Roby, E.; Alexandris, A.; et al. Post-Translational Modifications (PTMs), Identified on Endogenous Huntingtin, Cluster within Proteolytic Domains between HEAT Repeats. J. Proteome Res. 2017, 16, 2692–2708. [Google Scholar] [CrossRef]
- Ben M’Barek, K.; Pla, P.; Orvoen, S.; Benstaali, C.; Godin, J.D.; Gardier, A.M.; Saudou, F.; David, D.J.; Humbert, S. Huntingtin Mediates Anxiety/Depression-Related Behaviors and Hippocampal Neurogenesis. J. Neurosci. 2013, 33, 8608–8620. [Google Scholar] [CrossRef]
- Jung, T.; Shin, B.; Tamo, G.; Kim, H.; Vijayvargia, R.; Leitner, A.; Marcaida, M.J.; Astorga-Wells, J.; Jung, R.; Aebersold, R.; et al. The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain. Structure 2020, 28, 1035–1050.E8. [Google Scholar] [CrossRef]
- Carmichael, J.; Sugars, K.L.; Bao, Y.P.; Rubinsztein, D.C. Glycogen synthase kinase-3beta inhibitors prevent cellular polyglutamine toxicity caused by the Huntington’s disease mutation. J. Biol. Chem. 2002, 277, 33791–33798. [Google Scholar] [CrossRef]
- Krzystek, T.J.; Rathnayake, R.; Zeng, J.; Huang, J.; Iacobucci, G.; Yu, M.C.; Gunawardena, S. Opposing roles for GSK3β and ERK1-dependent phosphorylation of huntingtin during neuronal dysfunction and cell death in Huntington’s disease. Cell Death Dis. 2025, 16, 328. [Google Scholar] [CrossRef]
- Frame, S.; Cohen, P.; Biondi, R.M. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 2001, 7, 1321–1327. [Google Scholar] [CrossRef]
- Watkin, E.E.; Arbez, N.; Waldron-Roby, E.; O’Meally, R.; Ratovitski, T.; Cole, R.N.; Ross, C.A. Phosphorylation of mutant huntingtin at serine 116 modulates neuronal toxicity. PLoS ONE 2014, 9, e88284. [Google Scholar] [CrossRef]
- Huang, B.; Lucas, T.; Kueppers, C.; Dong, X.; Krause, M.; Bepperling, A.; Buchner, J.; Voshol, H.; Weiss, A.; Gerrits, B.; et al. Scalable production in human cells and biochemical characterization of full-length normal and mutant huntingtin. PLoS ONE 2015, 10, e0121055. [Google Scholar] [CrossRef]
- Rrustemi, T.; Meyer, K.; Roske, Y.; Uyar, B.; Akalin, A.; Imami, K.; Ishihama, Y.; Daumke, O.; Selbach, M. Pathogenic mutations of human phosphorylation sites affect protein-protein interactions. Nat. Commun. 2024, 15, 3146. [Google Scholar] [CrossRef]
- Bustamante, M.B.; Ansaloni, A.; Pedersen, J.F.; Azzollini, L.; Cariulo, C.; Wang, Z.-M.; Petricca, L.; Verani, M.; Puglisi, F.; Park, H.; et al. Detection of huntingtin exon 1 phosphorylation by Phos-Tag SDS-PAGE: Predominant phosphorylation on threonine 3 and regulation by IKKβ. Biochem. Biophys. Res. Commun. 2015, 463, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Metzler, M.; Gan, L.; Mazarei, G.; Graham, R.K.; Liu, L.; Bissada, N.; Lu, G.; Leavitt, B.R.; Hayden, M.R. Phosphorylation of Huntingtin at Ser421 in YAC128 Neurons Is Associated with Protection of YAC128 Neurons from NMDA-Mediated Excitotoxicity and Is Modulated by PP1 and PP2A. J. Neurosci. 2010, 30, 14318–14329. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Callegari, E.; Gloeckner, C.J.; Ueffing, M.; Wang, H. Mass spectrometric identification of novel posttranslational modification sites in Huntingtin. Proteomics 2012, 12, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Held, J.M.; DeGiacomo, F.; Bonner, A.; Chen, J.M.; Schilling, B.; Czerwieniec, G.A.; Gibson, B.W.; Ellerby, L.M. Mass spectrometric identification of novel lysine acetylation sites in huntingtin. Mol. Cell Proteom. 2011, 10, M111.009829. [Google Scholar] [CrossRef]
- Chaibva, M.; Jawahery, S.; Pilkington, A.W.; Arndt, J.R.; Sarver, O.; Valentine, S.; Matysiak, S.; Legleiter, J. Acetylation within the First 17 Residues of Huntingtin Exon 1 Alters Aggregation and Lipid Binding. Biophys. J. 2016, 111, 349–362. [Google Scholar] [CrossRef]
- Hockly, E.; Richon, V.M.; Woodman, B.; Smith, D.L.; Zhou, X.; Rosa, E.; Sathasivam, K.; Ghazi-Noori, S.; Mahal, A.; Lowden, P.A.; et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, R.J.; Kubilus, J.K.; Lee, J.; Ryu, H.; Beesen, A.; Zucker, B.; Smith, K.; Kowall, N.W.; Ratan, R.R.; Luthi-Carter, R.; et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J. Neurosci. 2003, 23, 9418–9427. [Google Scholar] [CrossRef]
- Yang, X.; Duckhorn, J.; Marshall, J.; Huang, Y.A. Interlinked destinies: How ubiquitin-proteasome and autophagy systems underpin neurocognitive outcomes. Exp. Neurol. 2024, 379, 114869. [Google Scholar] [CrossRef]
- Swanson, M.E.V.; Tan, A.Y.S.; Tippett, L.J.; Turner, C.P.; Curtis, M.A.; Scotter, E.L.; Lashuel, H.A.; Dragunow, M.; Faull, R.L.M.; Murray, H.C.; et al. Huntingtin inclusion bodies have distinct immunophenotypes and ubiquitination profiles in the Huntington’s disease human cerebral cortex. Sci. Rep. 2025, 15, 15546. [Google Scholar] [CrossRef] [PubMed]
- Hakim-Eshed, V.; Boulos, A.; Cohen-Rosenzweig, C.; Yu-Taeger, L.; Ziv, T.; Kwon, Y.T.; Riess, O.; Phuc Nguyen, H.H.; Ziv, N.E.; Ciechanover, A. Site-specific ubiquitination of pathogenic huntingtin attenuates its deleterious effects. Proc. Natl. Acad. Sci. USA 2020, 117, 18661–18669. [Google Scholar] [CrossRef]
- Boulos, A.; Maroun, D.; Ciechanover, A.; Ziv, N.E. Peripheral sequestration of huntingtin delays neuronal death and depends on N-terminal ubiquitination. Commun. Biol. 2024, 7, 1014. [Google Scholar] [CrossRef]
- Jana, N.R.; Dikshit, P.; Goswami, A.; Kotliarova, S.; Murata, S.; Tanaka, K.; Nukina, N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005, 280, 11635–11640. [Google Scholar] [CrossRef]
- He, B.; Bai, S.; Hnat, A.T.; Kalman, R.I.; Minges, J.T.; Patterson, C.; Wilson, E.M. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J. Biol. Chem. 2004, 279, 30643–30653. [Google Scholar] [CrossRef] [PubMed]
- Miller, V.M.; Nelson, R.F.; Gouvion, C.M.; Williams, A.; Rodriguez-Lebron, E.; Harper, S.Q.; Davidson, B.L.; Rebagliati, M.R.; Paulson, H.L. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 2005, 25, 9152–9161. [Google Scholar] [CrossRef]
- Omura, T.; Kaneko, M.; Okuma, Y.; Orba, Y.; Nagashima, K.; Takahashi, R.; Fujitani, N.; Matsumura, S.; Hata, A.; Kubota, K.; et al. A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem. 2006, 99, 1456–1469. [Google Scholar] [CrossRef]
- Yang, H.; Zhong, X.; Ballar, P.; Luo, S.; Shen, Y.; Rubinsztein, D.C.; Monteiro, M.J.; Fang, S. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res. 2007, 313, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Fishman, P.S.; Thakor, N.V.; Oyler, G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003, 278, 22044–22055. [Google Scholar] [CrossRef]
- Campesan, S.; del Popolo, I.; Marcou, K.; Straatman-Iwanowska, A.; Repici, M.; Boytcheva, K.V.; Cotton, V.E.; Allcock, N.; Rosato, E.; Kyriacou, C.P.; et al. Bypassing mitochondrial defects rescues Huntington’s phenotypes in Drosophila. Neurobiol. Dis. 2023, 185, 106236. [Google Scholar] [CrossRef]
- Bhutani, S.; Das, A.; Maheshwari, M.; Lakhotia, S.C.; Jana, N.R. Dysregulation of core components of SCF complex in poly-glutamine disorders. Cell Death Dis. 2012, 3, e428. [Google Scholar] [CrossRef]
- Fiorillo, A.; Morea, V.; Colotti, G.; Ilari, A. Huntingtin Ubiquitination Mechanisms and Novel Possible Therapies to Decrease the Toxic Effects of Mutated Huntingtin. J. Pers. Med. 2021, 11, 1309. [Google Scholar] [CrossRef]
- Mishra, A.; Dikshit, P.; Purkayastha, S.; Sharma, J.; Nukina, N.; Jana, N.R. E6-AP Promotes Misfolded Polyglutamine Proteins for Proteasomal Degradation and Suppresses Polyglutamine Protein Aggregation and Toxicity. J. Biol. Chem. 2008, 283, 7648–7656. [Google Scholar] [CrossRef]
- Maheshwari, M.; Shekhar, S.; Singh, B.K.; Jamal, I.; Vatsa, N.; Kumar, V.; Sharma, A.; Jana, N.R. Deficiency of Ube3a in Huntington’s disease mice brain increases aggregate load and accelerates disease pathology. Hum. Mol. Genet. 2014, 23, 6235–6245. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Yan, S.; Wang, C.E.; Li, S.; Li, X.J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. USA 2014, 111, 5706–5711. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, M.; Samanta, A.; Godavarthi, S.K.; Mukherjee, R.; Jana, N.R. Dysfunction of the ubiquitin ligase Ube3a may be associated with synaptic pathophysiology in a mouse model of Huntington disease. J. Biol. Chem. 2012, 287, 29949–29957. [Google Scholar] [CrossRef]
- Subramaniam, S.; Mealer, R.G.; Sixt, K.M.; Barrow, R.K.; Usiello, A.; Snyder, S.H. Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 2010, 285, 20428–20432. [Google Scholar] [CrossRef]
- Soares, E.S.; Prediger, R.D.; Brocardo, P.S.; Cimarosti, H.I. SUMO-modifying Huntington’s disease. IBRO Neurosci. Rep. 2022, 12, 203–209. [Google Scholar] [CrossRef]
- Lemarié, F.L.; Caron, N.S.; Sanders, S.S.; Schmidt, M.E.; Nguyen, Y.T.N.; Ko, S.; Xu, X.; Pouladi, M.A.; Martin, D.D.O.; Hayden, M.R. Rescue of aberrant huntingtin palmitoylation ameliorates mutant huntingtin-induced toxicity. Neurobiol. Dis. 2021, 158, 105479. [Google Scholar] [CrossRef]
- Virlogeux, A.; Scaramuzzino, C.; Lenoir, S.; Carpentier, R.; Louessard, M.; Genoux, A.; Lino, P.; Hinckelmann, M.-V.; Perrier, A.L.; Humbert, S.; et al. Increasing brain palmitoylation rescues behavior and neuropathology in Huntington disease mice. Sci. Adv. 2021, 7, eabb0799. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef]
- Liu, J.P.; Zeitlin, S.O. Is Huntingtin Dispensable in the Adult Brain? J. Huntingt. Dis. 2017, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gardian, G.; Browne, S.E.; Choi, D.K.; Klivenyi, P.; Gregorio, J.; Kubilus, J.K.; Ryu, H.; Langley, B.; Ratan, R.R.; Ferrante, R.J.; et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J. Biol. Chem. 2005, 280, 556–563. [Google Scholar] [CrossRef]
- Mielcarek, M.; Benn, C.L.; Franklin, S.A.; Smith, D.L.; Woodman, B.; Marks, P.A.; Bates, G.P. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLoS ONE 2011, 6, e27746. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, P.; Lovrecic, L.; Krainc, D. Sodium phenylbutyrate in Huntington’s disease: A dose-finding study. Mov. Disord. 2007, 22, 1962–1964. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Parise, R.A.; Ramanathan, R.K.; Lagattuta, T.F.; Musguire, L.A.; Stoller, R.G.; Potter, D.M.; Argiris, A.E.; Zwiebel, J.A.; Egorin, M.J.; et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin. Cancer Res. 2007, 13, 3605–3610. [Google Scholar] [CrossRef]
- Apostol, B.L.; Simmons, D.A.; Zuccato, C.; Illes, K.; Pallos, J.; Casale, M.; Conforti, P.; Ramos, C.; Roarke, M.; Kathuria, S.; et al. CEP-1347 reduces mutant huntingtin-associated neurotoxicity and restores BDNF levels in R6/2 mice. Mol. Cell. Neurosci. 2008, 39, 8–20. [Google Scholar] [CrossRef]
- Parkinson Study Group Precert Investigators. Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease. Neurology 2007, 69, 1480–1490. [Google Scholar] [CrossRef]
- Huang, Z.N.; Chen, J.M.; Huang, L.C.; Fang, Y.H.; Her, L.S. Inhibition of p38 Mitogen-Activated Protein Kinase Ameliorates HAP40 Depletion-Induced Toxicity and Proteasomal Defect in Huntington’s Disease Model. Mol. Neurobiol. 2021, 58, 2704–2723. [Google Scholar] [CrossRef] [PubMed]
- Bowles, K.R.; Jones, L. Kinase signalling in Huntington’s disease. J. Huntingt. Dis. 2014, 3, 89–123. [Google Scholar] [CrossRef]
- Garcia, M.; Charvin, D.; Caboche, J. Expanded huntingtin activates the c-Jun terminal kinase/c-Jun pathway prior to aggregate formation in striatal neurons in culture. Neuroscience 2004, 127, 859–870. [Google Scholar] [CrossRef]
- Perrin, V.; Dufour, N.; Raoul, C.; Hassig, R.; Brouillet, E.; Aebischer, P.; Luthi-Carter, R.; Déglon, N. Implication of the JNK pathway in a rat model of Huntington’s disease. Exp. Neurol. 2009, 215, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Fawcett, J.R.; Thorne, R.G.; DeFor, T.A.; Frey, W.H., 2nd. Intranasal administration of insulin-like growth factor-I bypasses the blood-brain barrier and protects against focal cerebral ischemic damage. J. Neurol. Sci. 2001, 187, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Werner, H. The IGF1 Signaling Pathway: From Basic Concepts to Therapeutic Opportunities. Int. J. Mol. Sci. 2023, 24, 14882. [Google Scholar] [CrossRef]
- Lin, D.T.S.; Conibear, E. Enzymatic protein depalmitoylation by acyl protein thioesterases. Biochem. Soc. Trans. 2015, 43, 193–198. [Google Scholar] [CrossRef]
- Huang, K.; Sanders, S.; Singaraja, R.; Orban, P.; Cijsouw, T.; Arstikaitis, P.; Yanai, A.; Hayden, M.R.; El-Husseini, A. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009, 23, 2605–2615. [Google Scholar] [CrossRef]
- El-Husseini, A.E.-D.; Schnell, E.; Dakoji, S.; Sweeney, N.; Zhou, Q.; Prange, O.; Gauthier-Campbell, C.; Aguilera-Moreno, A.; Nicoll, R.A.; Bredt, D.S. Synaptic Strength Regulated by Palmitate Cycling on PSD-95. Cell 2002, 108, 849–863. [Google Scholar] [CrossRef]
- Mätlik, K.; Baffuto, M.; Kus, L.; Deshmukh, A.L.; Davis, D.A.; Paul, M.R.; Carroll, T.S.; Caron, M.-C.; Masson, J.-Y.; Pearson, C.E.; et al. Cell-type-specific CAG repeat expansions and toxicity of mutant Huntingtin in human striatum and cerebellum. Nat. Genet. 2024, 56, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, K.; Jadhav, B.; Zanovello, M.; Gagliardi, D.; Clarkson, C.; Facchini, S.; Garg, P.; Martin-Trujillo, A.; Gies, S.J.; Deforie, V.G.; et al. Increased frequency of repeat expansion mutations across different populations. Nat. Med. 2024, 30, 3357–3368. [Google Scholar] [CrossRef]
- Handsaker, R.E.; Kashin, S.; Reed, N.M.; Tan, S.; Lee, W.S.; McDonald, T.M.; Morris, K.; Kamitaki, N.; Mullally, C.D.; Morakabati, N.R.; et al. Long somatic DNA-repeat expansion drives neurodegeneration in Huntington’s disease. Cell 2025, 188, 623–639.E19. [Google Scholar] [CrossRef]
- Scahill, R.I.; Farag, M.; Murphy, M.J.; Hobbs, N.Z.; Leocadi, M.; Langley, C.; Knights, H.; Ciosi, M.; Fayer, K.; Nakajima, M.; et al. Somatic CAG repeat expansion in blood associates with biomarkers of neurodegeneration in Huntington’s disease decades before clinical motor diagnosis. Nat. Med. 2025, 31, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Cubo, E.; Martinez-Horta, S.I.; Santalo, F.S.; Descalls, A.M.; Calvo, S.; Gil-Polo, C.; Muñoz, I.; Llano, K.; Mariscal, N.; Diaz, D.; et al. Clinical manifestations of homozygote allele carriers in Huntington disease. Neurology 2019, 92, e2101–e2108. [Google Scholar] [CrossRef] [PubMed]
- Louçã, M.; El Akrouti, D.; Lemesle, A.; Louessard, M.; Dufour, N.; Baroin, C.; de la Fouchardière, A.; Cotter, L.; Jean-Jacques, H.; Redeker, V.; et al. Huntingtin lowering impairs the maturation and synchronized synaptic activity of human cortical neuronal networks derived from induced pluripotent stem cells. Neurobiol. Dis. 2024, 200, 106630, Erratum in Neurobiol. Dis. 2024, 202, 106703. [Google Scholar] [CrossRef]
- Spronck, E.A.; Vallès, A.; Lampen, M.H.; Montenegro-Miranda, P.S.; Keskin, S.; Heijink, L.; Evers, M.M.; Petry, H.; Deventer, S.J.V.; Konstantinova, P.; et al. Intrastriatal Administration of AAV5-miHTT in Non-Human Primates and Rats Is Well Tolerated and Results in miHTT Transgene Expression in Key Areas of Huntington Disease Pathology. Brain Sci. 2021, 11, 129. [Google Scholar] [CrossRef]
- Gao, L.; Bhattacharyya, A.; Beers, B.; Kaushik, D.; Bredlau, A.L.; Kristensen, A.; Abd-Elaziz, K.; Grant, R.; Golden, L.; Kong, R. Pharmacokinetics and pharmacodynamics of PTC518, an oral huntingtin lowering splicing modifier: A first-in-human study. Br. J. Clin. Pharmacol. 2024, 90, 3242–3251. [Google Scholar] [CrossRef]
- Kordasiewicz, H.B.; Stanek, L.M.; Wancewicz, E.V.; Mazur, C.; McAlonis, M.M.; Pytel, K.A.; Artates, J.W.; Weiss, A.; Cheng, S.H.; Shihabuddin, L.S.; et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 2012, 74, 1031–1044. [Google Scholar] [CrossRef]
- Southwell, A.L.; Kordasiewicz, H.B.; Langbehn, D.; Skotte, N.H.; Parsons, M.P.; Villanueva, E.B.; Caron, N.S.; Østergaard, M.E.; Anderson, L.M.; Xie, Y.; et al. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci. Transl. Med. 2018, 10, eaar3959. [Google Scholar] [CrossRef] [PubMed]
- Wennagel, D.; Braz, B.Y.; Capizzi, M.; Barnat, M.; Humbert, S. Huntingtin coordinates dendritic spine morphology and function through cofilin-mediated control of the actin cytoskeleton. Cell Rep. 2022, 40, 111261. [Google Scholar] [CrossRef]
- Ochaba, J.; Lukacsovich, T.; Csikos, G.; Zheng, S.; Margulis, J.; Salazar, L.; Mao, K.; Lau, A.L.; Yeung, S.Y.; Humbert, S.; et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 16889–16894. [Google Scholar] [CrossRef]
- Burrus, C.J.; McKinstry, S.U.; Kim, N.; Ozlu, M.I.; Santoki, A.V.; Fang, F.Y.; Ma, A.; Karadeniz, Y.B.; Worthington, A.K.; Dragatsis, I.; et al. Striatal Projection Neurons Require Huntingtin for Synaptic Connectivity and Survival. Cell Rep. 2020, 30, 642–657.E6. [Google Scholar] [CrossRef]
- Bragg, R.M.; Coffey, S.R.; Cantle, J.P.; Hu, S.; Singh, S.; Legg, S.R.W.; McHugh, C.A.; Toor, A.; Zeitlin, S.O.; Kwak, S.; et al. Huntingtin loss in hepatocytes is associated with altered metabolism, adhesion, and liver zonation. Life Sci. Alliance 2023, 6, e202302098. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C.; Wang, Z.; Zhu, C.; Li, J.; Sha, T.; Ma, L.; Gao, C.; Yang, Y.; Sun, Y.; et al. Allele-selective lowering of mutant HTT protein by HTT–LC3 linker compounds. Nature 2019, 575, 203–209. [Google Scholar] [CrossRef]
- Li, E.; Choi, J.; Sim, H.-R.; Kim, J.; Jun, J.H.; Kyung, J.; Ha, N.; Kim, S.; Ryu, K.H.; Chung, S.S.; et al. A novel HDAC6 inhibitor, CKD-504, is effective in treating preclinical models of Huntington’s disease. BMB Rep. 2023, 56, 178–183. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, L.; Zhu, W.; Song, J.; Lin, J.; Li, Y.; Zhang, J.; Song, X.; Xing, T.; Guo, T.; et al. TRIM37 is a primate-specific E3 ligase for Huntingtin and accounts for the striatal degeneration in Huntington’s disease. Sci. Adv. 2024, 10, eadl2036. [Google Scholar] [CrossRef] [PubMed]
- Bagherpoor Helabad, M.; Matlahov, I.; Kumar, R.; Daldrop, J.O.; Jain, G.; Weingarth, M.; van der Wel, P.C.A.; Miettinen, M.S. Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease. Nat. Commun. 2024, 15, 10793. [Google Scholar] [CrossRef] [PubMed]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Kay, C.; Leavitt, B.R.; Nance, M.; Ross, C.A.; Scahill, R.I.; Wetzel, R.; et al. Huntington disease. Nat. Rev. Dis. Primers 2015, 1, 15005. [Google Scholar] [CrossRef]
- Ochaba, J.; Fote, G.; Kachemov, M.; Thein, S.; Yeung, S.Y.; Lau, A.L.; Hernandez, S.; Lim, R.G.; Casale, M.; Neel, M.J.; et al. IKKβ slows Huntington’s disease progression in R6/1 mice. Proc. Natl. Acad. Sci. USA 2019, 116, 10952–10961. [Google Scholar] [CrossRef] [PubMed]
- Chiki, A.; Zhang, Z.; Rajasekhar, K.; Abriata, L.A.; Rostami, I.; Krapp, L.F.; Boudeffa, D.; Dal Peraro, M.; Lashuel, H.A. Investigating Crosstalk Among PTMs Provides Novel Insight Into the Structural Basis Underlying the Differential Effects of Nt17 PTMs on Mutant Httex1 Aggregation. Front. Mol. Biosci. 2021, 8, 686086. [Google Scholar] [CrossRef]
- Han, I.; You, Y.; Kordower, J.H.; Brady, S.T.; Morfini, G.A. Differential vulnerability of neurons in Huntington’s disease: The role of cell type-specific features. J. Neurochem. 2010, 113, 1073–1091. [Google Scholar] [CrossRef]
- Zeron, M.M.; Hansson, O.; Chen, N.; Wellington, C.L.; Leavitt, B.R.; Brundin, P.; Hayden, M.R.; Raymond, L.A. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron 2002, 33, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Panov, A.V.; Gutekunst, C.A.; Leavitt, B.R.; Hayden, M.R.; Burke, J.R.; Strittmatter, W.J.; Greenamyre, J.T. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002, 5, 731–736. [Google Scholar] [CrossRef] [PubMed]
 ”) denote inhibitory regulation, and question marks (“?”) indicate unclear or yet-to-be-determined roles.
”) denote inhibitory regulation, and question marks (“?”) indicate unclear or yet-to-be-determined roles.
 ”) denote inhibitory regulation, and question marks (“?”) indicate unclear or yet-to-be-determined roles.
”) denote inhibitory regulation, and question marks (“?”) indicate unclear or yet-to-be-determined roles.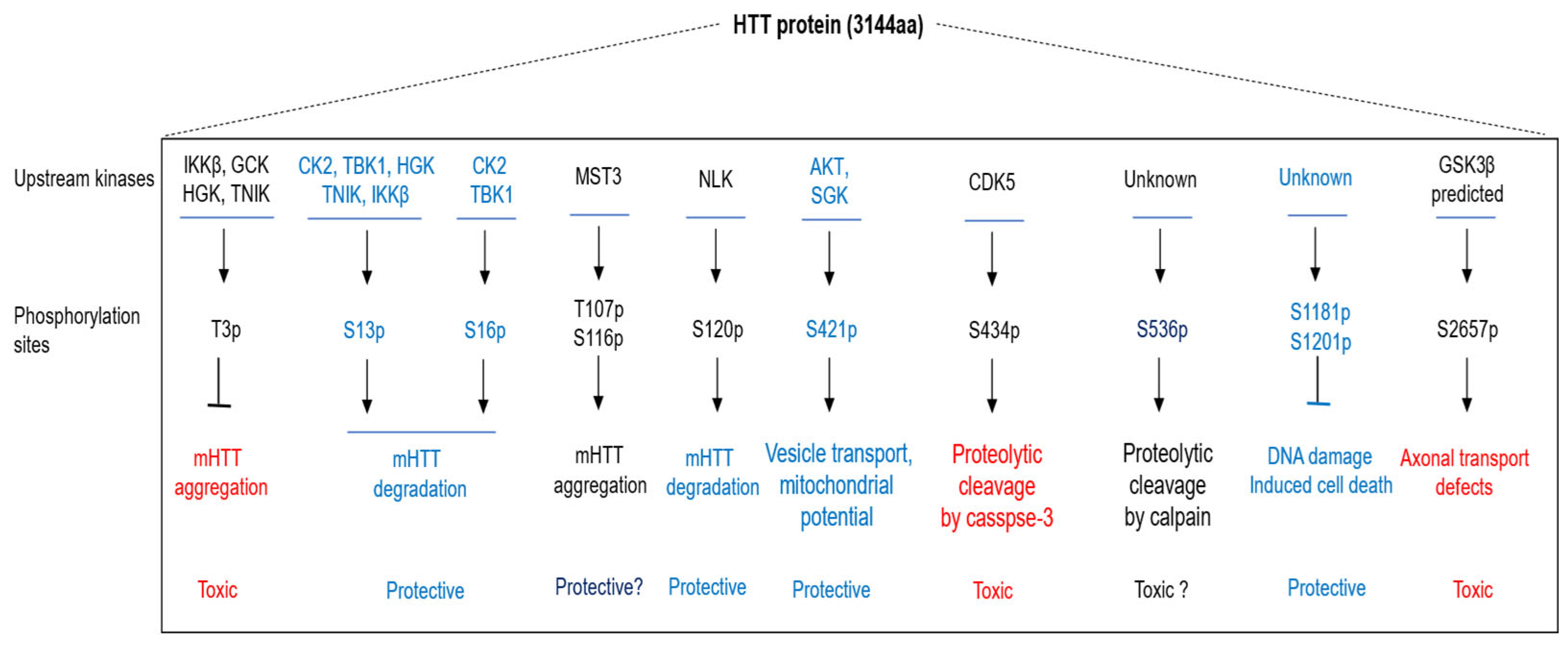
 ”) denote inhibitory regulation.
”) denote inhibitory regulation.
 ”) denote inhibitory regulation.
”) denote inhibitory regulation.
 ” represents inhibitory regulation, and “?” denotes unclear or yet-to-be-determined regulatory mechanisms or functions.
” represents inhibitory regulation, and “?” denotes unclear or yet-to-be-determined regulatory mechanisms or functions.
 ” represents inhibitory regulation, and “?” denotes unclear or yet-to-be-determined regulatory mechanisms or functions.
” represents inhibitory regulation, and “?” denotes unclear or yet-to-be-determined regulatory mechanisms or functions.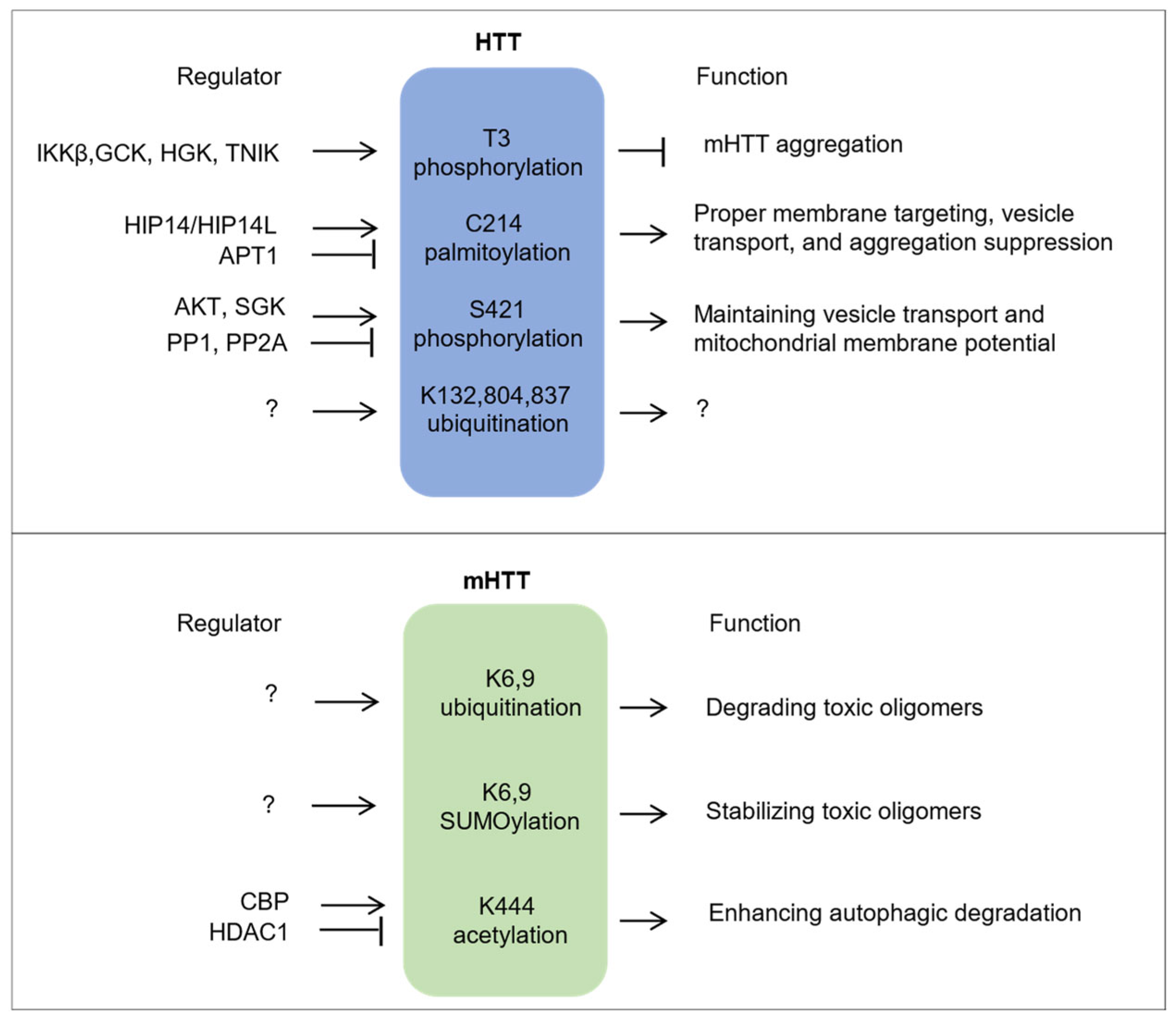
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, S.; Wang, C. Post-Translational Modifications of Huntingtin: Mechanistic Insights and Therapeutic Opportunities in Huntington’s Disease. Int. J. Mol. Sci. 2025, 26, 10907. https://doi.org/10.3390/ijms262210907
Zhang X, Zhang S, Wang C. Post-Translational Modifications of Huntingtin: Mechanistic Insights and Therapeutic Opportunities in Huntington’s Disease. International Journal of Molecular Sciences. 2025; 26(22):10907. https://doi.org/10.3390/ijms262210907
Chicago/Turabian StyleZhang, Xiaoxia, Shengping Zhang, and Chuangui Wang. 2025. "Post-Translational Modifications of Huntingtin: Mechanistic Insights and Therapeutic Opportunities in Huntington’s Disease" International Journal of Molecular Sciences 26, no. 22: 10907. https://doi.org/10.3390/ijms262210907
APA StyleZhang, X., Zhang, S., & Wang, C. (2025). Post-Translational Modifications of Huntingtin: Mechanistic Insights and Therapeutic Opportunities in Huntington’s Disease. International Journal of Molecular Sciences, 26(22), 10907. https://doi.org/10.3390/ijms262210907





