Abstract
Two-component systems (TCSs) in bacteria are often involved in the global regulation of various physiological activities and behaviours. This study investigated the GacSA TCS and DJ41_1407 transcriptional sensor adjacent to GacA in Acinetobacter baumannii ATCC 19606. The relationship between GacS, GacA, and DJ41_1407 and their functions and signal transduction mechanisms are described. A. baumannii ATCC 19606 mutants, ∆gacS, ∆gacA, and ∆DJ41_1407, were generated using markerless mutation and cultured in LB medium, then collected for RNA sequencing. It was found that GacS, GacA, and DJ41_1407 regulate a series of genes involved in carbon metabolism. Quantitative reverse transcription PCR (qRT-PCR) results showed that DJ41_1407 and GacA may regulate the expression of adh4, ipdC, iacH, and paa. Phos-tag™ results revealed that GacS plays a more significant role in GacA phosphorylation. GacA regulated colony size and growth conditions in rich medium. Compared to the wild-type strain, the ∆gacA and ∆gacSA mutants exhibited smaller colony sizes, and mutation of the gacS, gacA, and DJ41_1407 genes also reduced bacterial virulence as determined by the Galleria mellonella infection assay. GacA also plays a crucial role in modulating antibiotic resistance, and the ∆gacA∆DJ41_1407 mutant demonstrated greater susceptibility to antibiotics. These results highlight the multiple functions regulated by the GacSA global TCS in A. baumannii ATCC 19606.
1. Introduction
Acinetobacter baumannii, a Gram-negative bacterium, acts as an opportunistic human pathogen [1] that has garnered attention for its role in causing various severe nosocomial infections, including skin and soft tissue infections, wound infections, urinary tract infections, and secondary meningitis, among others [2,3,4]. Of these, ventilator-associated pneumonia and bloodstream infections are among the most significant, as they are linked to higher mortality rates [5]. A. baumannii is classified as an ESKAPE (an acronym of the scientific names for six highly virulent and antibiotic-resistant pathogens, including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) pathogen due to its high levels of antibiotic resistance [6], and its ability to form biofilms, which enhances its capacity to persist on artificial surfaces within hospital environments for extended periods [7]. A. baumannii generally utilises two-component systems (TCSs) to regulate adaptive responses and traits associated with virulence.
TCSs are one of the largest signal transduction pathways in living organisms and play an important role in sensing external signals, regulating response and promoting survival in changing environments. Most TCSs in bacteria can regulate gene expression related to antibiotic resistance [8], virulence, biofilm formation [9], motility [10], and ethanol metabolism [11,12]. Moreover, some of these have been identified as global TCSs, due to their capacity to regulate various physiological activities and behaviours. For example, GacS/GacA in Pseudomonas fluorescens MFE01 control antimicrobial activities against phytopathogens and emissions of volatile organic compounds (VOCs) [13]. In Legionella pneumophila, PmrAB was found to exert a broad influence on genes encoding eukaryotic-like proteins, the Dot/Icm apparatus, secreted effectors, type II-secreted proteins, regulators of the post-exponential phase, stress response genes, flagellar biosynthesis genes, metabolic genes, and genes of unknown function [14]. It has also been shown to be essential for the intracellular proliferation of L. pneumophila within human macrophages and protozoa [14]. Disparities in gene regulation and intracellular growth patterns observed between the pmrA and pmrB mutants suggest potential crosstalk with other TCSs [14]. Additionally, CovSR (CsrSR) in group B streptococcus has been identified as a global TCS implicated in virulence. Expression profiling analysis revealed that CovSR regulates a substantial array of genes that encode proteins potentially involved in secretion or associated with the cell surface [15]. AirSR is a redox-dependent global regulatory system in Staphylococcus aureus that plays essential roles in gene regulation through a redox-active Fe–S cluster under O2-limited conditions, and research has shown that an AirSR-mutant strain demonstrated GacSA heightened resistance to H2O2, vancomycin, norfloxacin, and ciprofloxacin in anaerobic environments [16].
GacSA has been identified as a global regulator in Gram-negative bacteria. In P. aeruginosa, GacSA (GacS-PA0928, GacA-PA2586) is one of the crucial TCSs and plays a central role in regulating the expression of virulence factors, secondary metabolites, biofilm formation, and quorum sensing [17,18]. Additionally, GacSA acts as a key switch that determines the transition between acute and chronic infections [19]. In P. aeruginosa, the phosphorylation of GacS is regulated by two hybrid sensor kinases, RetS (PA4856) [20] and LadS (PA3974) [21]. RetS directly interacts with GacS, inhibiting its phosphorylation [21,22], while LadS phosphorylates GacS [21]. The phosphorylated GacS then promotes the transcription of two small regulatory RNAs, rgRsmZ (PA3621.1) and rgRsmY (PA0527.1). Both rgRsmZ and rgRsmY function as inhibitors of the negative regulator, the RNA-binding protein RsmA (PA0905). RsmA, in turn, positively regulates genes associated with the Type 3 secretion system, type IV pili formation, and iron homeostasis while repressing quorum sensing, Type 6 secretion, and potentially other transcription factors [23,24,25]. In K. pneumoniae, the GacSA homologue protein is known as the BarA/UvrY TCS. Studies have demonstrated that inactivation of the barA and uvrY genes is linked to reduced capsule polysaccharide (CPS) production [26]. Transcriptome and proteome analyses further showed that cefiderocol stress downregulates UvrY transcription and expression levels, indicating that UvrY may play a role in the response to cefiderocol pressure. However, the regulated genes and activation signals associated with the BarA/UvrY TCS system still require further investigation [27].
GacSA is known to control the expression of 674 genes associated with virulence, biofilm formation, pili production, resistance to human serum, motility, metabolism of aromatic compounds, and more in A. baumannii ATCC 17978 [28,29]. Unlike other well-characterized TCSs such as AdeRS [30], BaeSR [31], PmrAB [32], and BfmRS [33], GacSA does not exist as a contiguous operon in A. baumannii ATCC 17978, and the GacS and GacA genes are positioned differently on the chromosome. However, the response regulator GacA was found to be transcribed in the same direction as the adjacent sensor kinase of unknown function, namely DJ41_1407 in A. baumannii ATCC 19606, indicating potential crosstalk between these two TCSs, as GacA may have co-opted this sensor kinase. Given the differences between strains, the signal transduction and function of GacSA in other strains, such as A. baumannii ATCC 19606, remain unclear.
Considering that GacSA functions as an important global regulator in A. baumannii, a better understanding of its role in physiological mechanisms could potentially lead to the development of novel strategies that can address issues such as virulence and antibiotic resistance in this critical nosocomial pathogen. In this study, we aim to confirm the roles of GacSA in A. baumannii ATCC 19606 and to investigate the relationship between GacA and its adjacent sensor kinase DJ41_1407.
2. Results
2.1. GacSA Is a TCS in A. baumannii ATCC 19606
The gacS gene in A. baumannii ATCC 19606 is designated as DJ41_1085 and annotated as a hybrid sensor histidine kinase/response regulator. The conserved and functional domains of GacS were analysed using CDvist (http://cdvist.joulinelab.org/ (accessed on 28 August 2025); Figure S1A), which revealed the presence of a phosphatidate phosphatase domain, a Hamp domain, a histidine kinase domain for phosphorylation, an ATPase domain, a FleQ domain for flagellar regulation, a receiver domain for phosphorylation, and an Hpt domain for phosphate transfer. The potential phosphorylation sites include a histidine residue at position 299 and an aspartic acid residue at position 719. The gacA gene in A. baumannii ATCC 19606 is designated as DJ41_1406 and annotated as a response regulator. The conserved and functional domains of GacA were analysed using CDvist (Figure S1B), revealing a receiver domain for phosphorylation and a helix-turn-helix domain for DNA binding. The potential phosphorylation site is the aspartic acid residue at position 54.
Phos-tagTM was used to investigate whether GacS phosphorylates GacA, in order to determine if GacS and GacA constitute a TCS in A. baumannii ATCC 19606. Wild-type and ΔgacS strains were cultured under the same conditions, specifically LB medium or M9 medium supplemented with 5 mM citrate, indole-3-acetic acid (IAA), or tryptophan (Figure S2). The results indicated that when gacS was mutated, there was decreased expression of phosphorylated GacA, even across different culture conditions. This finding suggests that GacA is phosphorylated by GacS and that they form a TCS.
2.2. GacSA Transcription Analysis
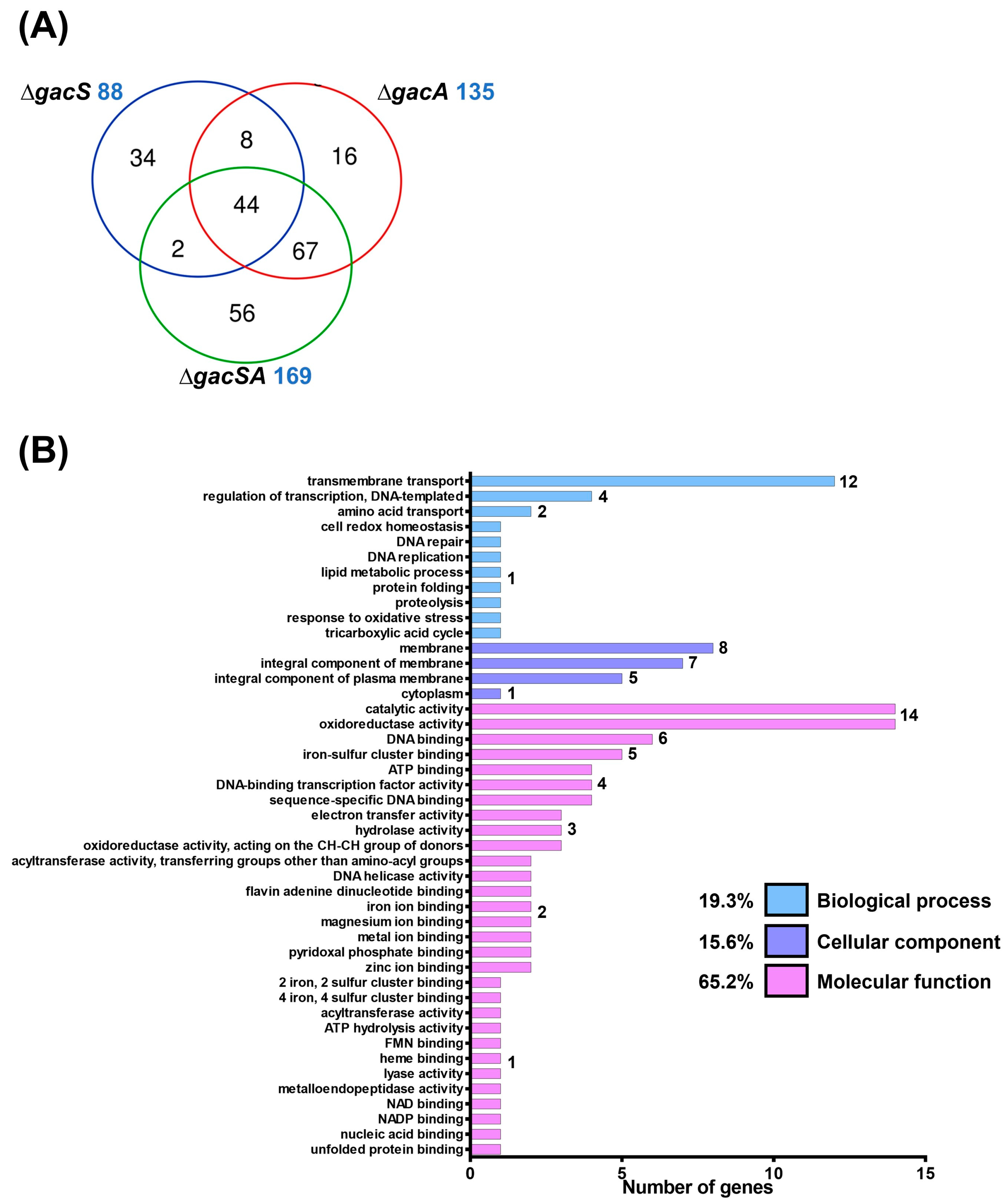
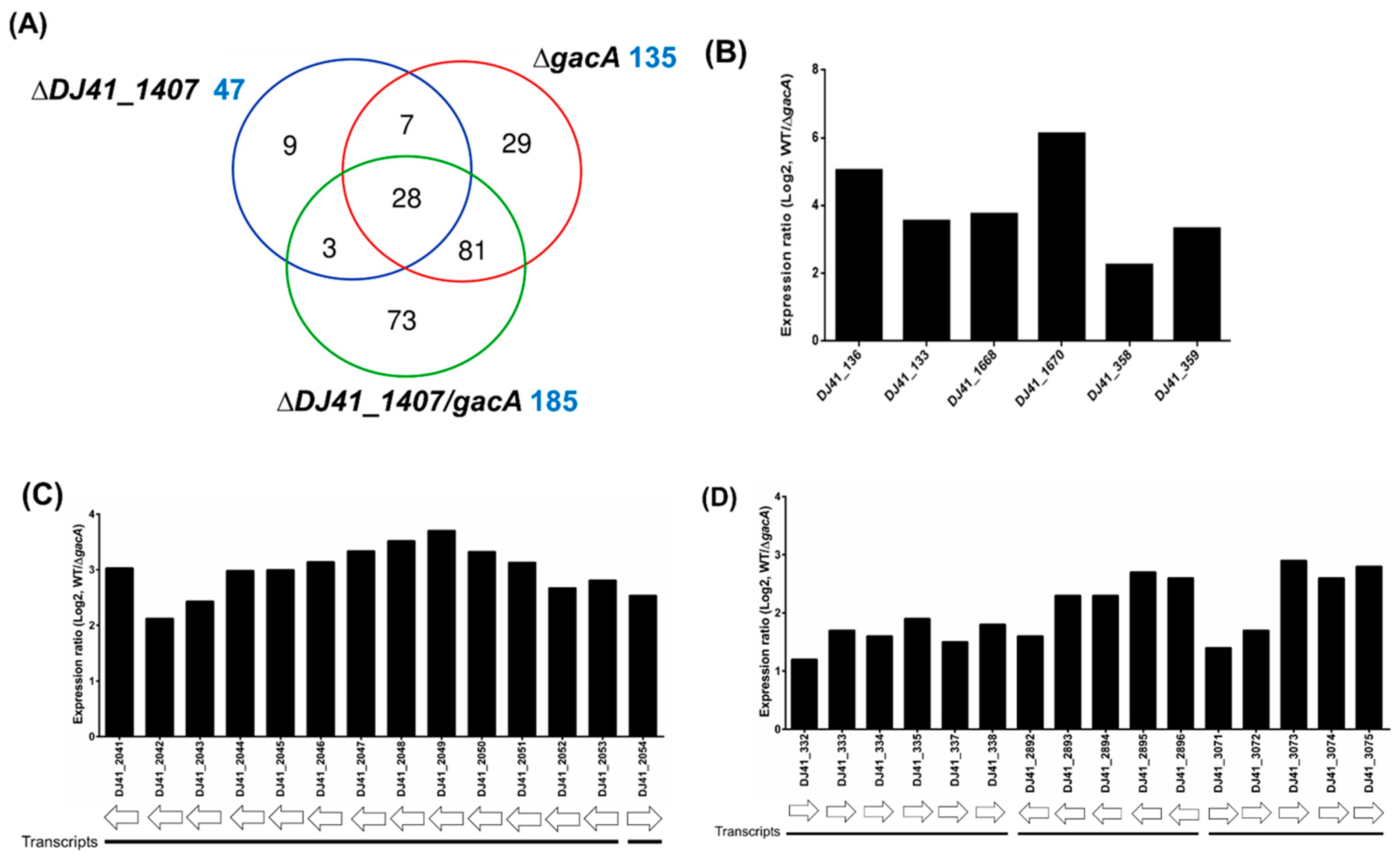
To analyse the functions of GacS and GacA, wild-type, ∆gacS, ∆gacA, and ∆gacSA strains were cultured in LB medium and subsequently sent to Welgene Biotech Co., Ltd. (Taipei, Taiwan) for transcriptome analysis. RNA expression levels of the mutants were compared against wild-type. The fold-change in each mutant relative to the wild-type was presented as a log2 expression ratio (wild-type/mutant). A total of 88, 135, and 169 genes exhibited differences in expression activity compared with the wild-type for the ∆gacS, ∆gacA, and ∆gacSA mutants, respectively. Among the regulated genes in each group, 44 genes were found to be co-regulated by GacS, GacA, and GacSA, indicating that the combination of GacS and GacA plays a significant role in key regulatory processes. Additionally, 67 genes were co-regulated solely by GacA and GacSA, suggesting that GacA may exclusively regulate the expression of these genes in response to phosphorylation by other sensor kinases (Figure 1A; Tables S1–S7).
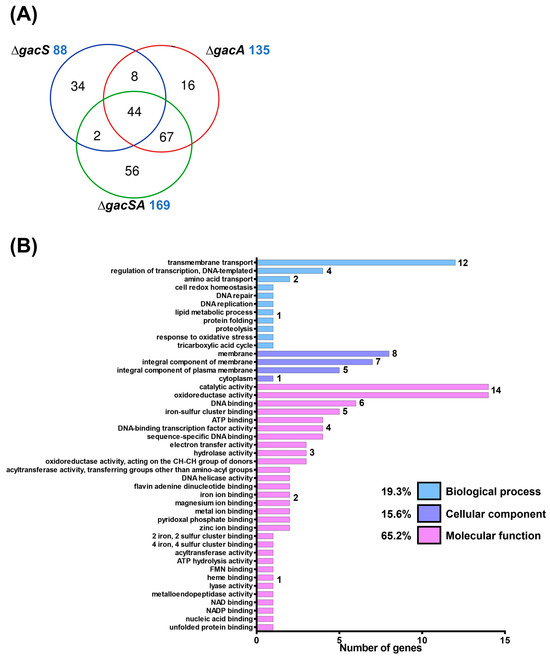
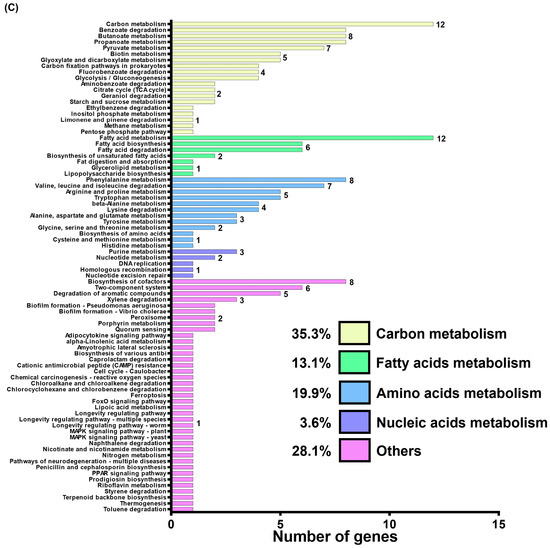
Figure 1.
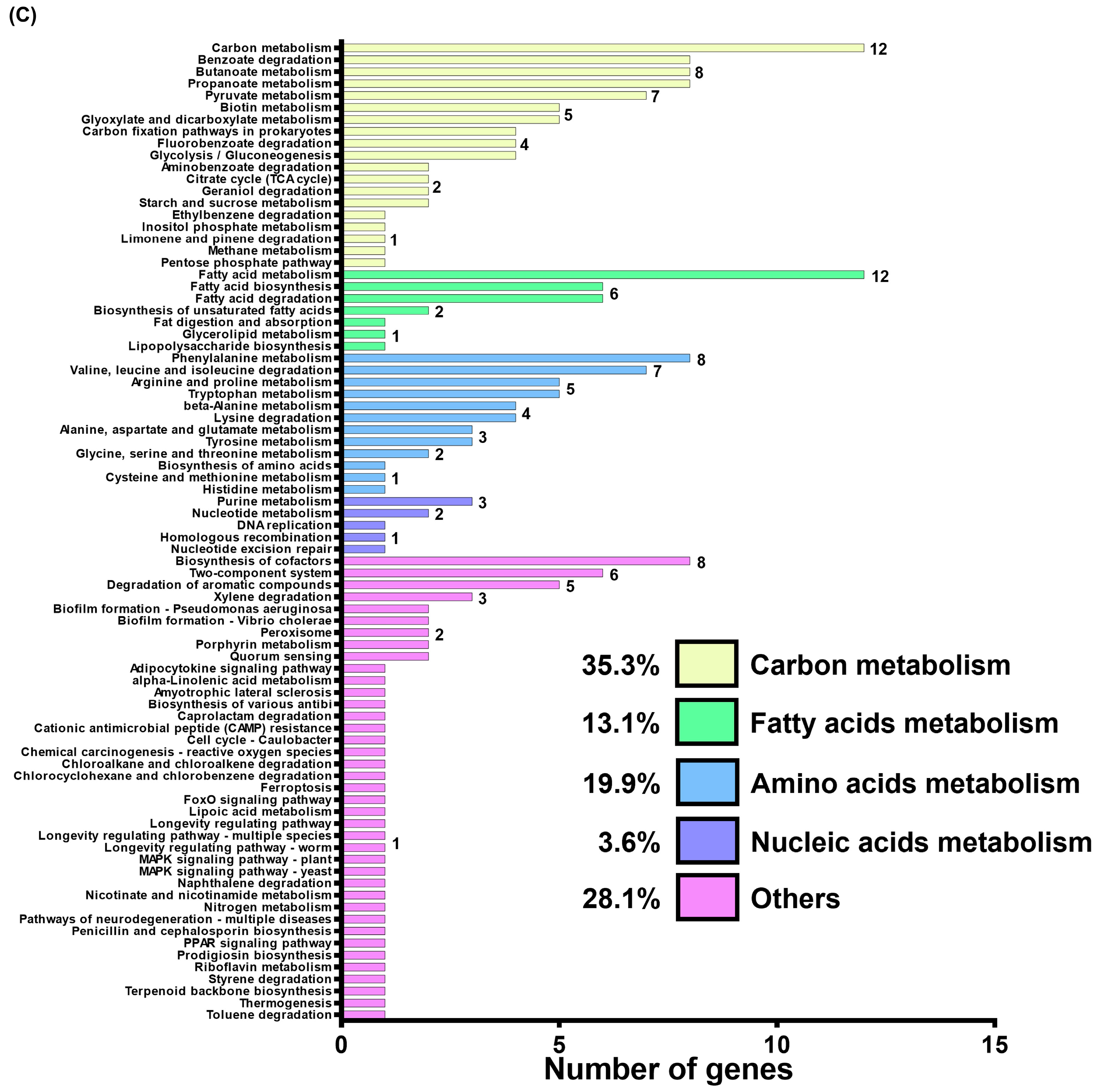
Transcriptome analysis of the GacSA regulon. (A) Comparison between the total number of genes regulated by GacS (blue), GacA (red) and GacSA (green). Genes with log2 expression ratio (wild-type/mutant) > 1.0 or < −1.0 in ∆gacS, ∆gacA and ∆gacSA were compared. A total of 88, 135 and 169 genes respectively showed difference in expression activity for the ∆gacS, ∆gacA and ∆gacSA mutants, compared with wild type. (B) Metabolic classification of genes regulated by GacSA through gene ontology classification. The indicated gene function groups were divided into three categories based on their roles in biological processes (blue), cellular components (purple), and molecular functions (pink). The number of genes found in these gene function groups ranged from 1 to 17, and these numbers are labelled on the right of the graph bar. (C) Metabolic classification of genes regulated by GacSA through KEGG classification. The indicated gene function groups were divided into five categories based on their roles in carbon metabolism (yellow), fatty acid metabolism (green), amino acid metabolism (blue), nucleic acid metabolism (purple), and others (pink). The number of genes found in these gene function groups ranged from 1 to 18, and these numbers are labelled on the right of the graph bar.
To gain a deeper understanding of GacSA regulation, genes with a log2 expression ratio (wild-type/∆gacSA) greater than 1.0 and less than −1.0 were analysed and classified through gene ontology and KEGG pathway classification. In the gene ontology classification, the genes regulated by GacSA were found to be involved in molecular functions (59.2%), cellular components (23.2%), and biological processes (17.6%) (Figure 1B). Most of the genes regulated by GacSA were associated with molecular functions; for instance, 16 genes were linked to oxidoreductase activity, and 14 genes were related to catalytic activity. KEGG pathway classification revealed that, of the genes regulated by GacSA, 35.8%, 30.6%, 23.3%, 6.7%, and 3.6% were respectively involved in carbon metabolism, other metabolic processes, amino acid metabolism, fatty acid metabolism, and nucleic acid metabolism. GacSA primarily regulated genes associated with carbon metabolism and amino acid metabolism; specifically, 18 genes were related to benzoate degradation, while 13 genes were linked to phenylalanine metabolism (Figure 1C).
2.3. GacA and DJ41_1407 Are Expressed in the Same Transcript
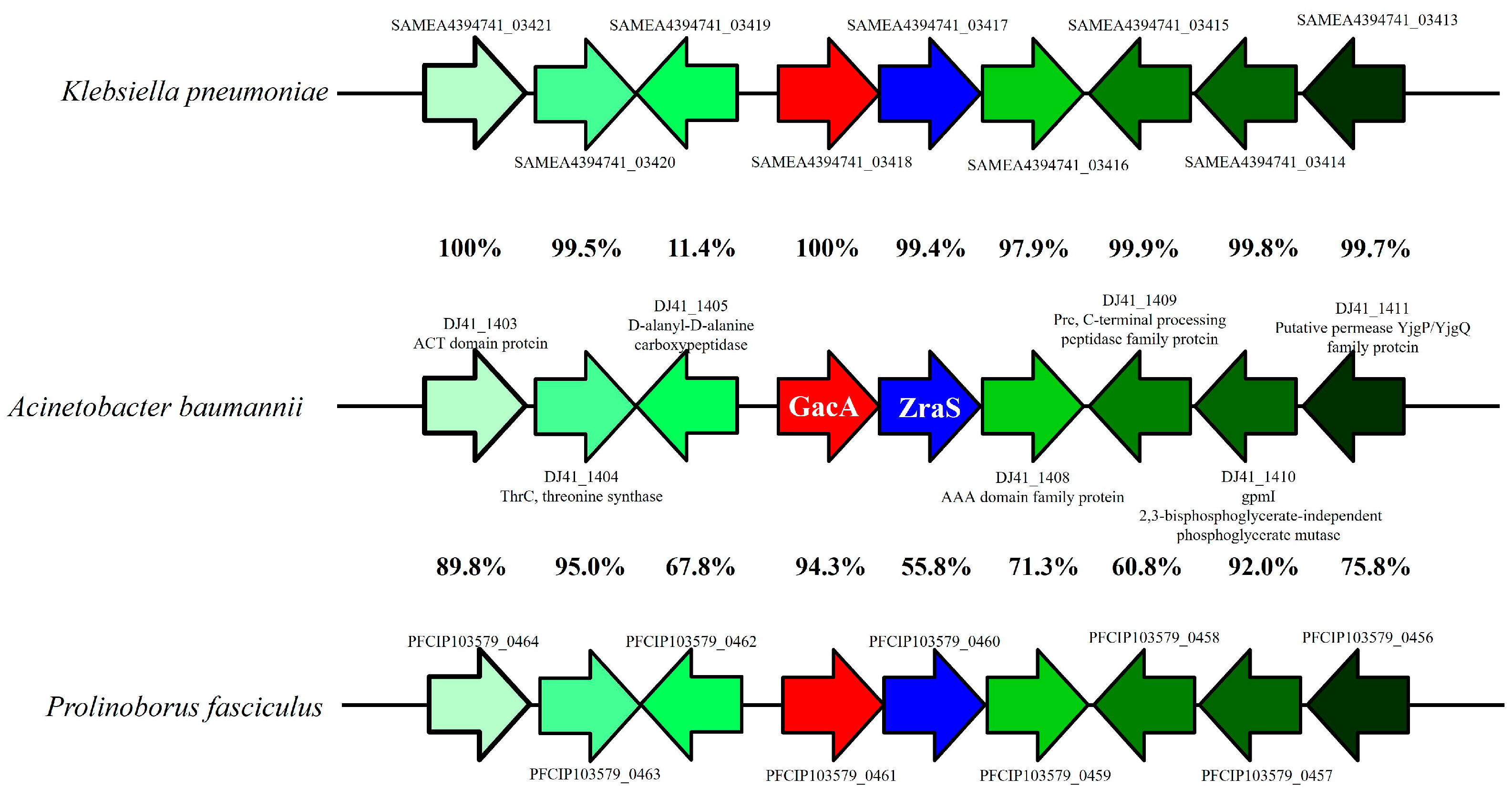
Amino acid identity analysis revealed that DJ41_1407 and GacA can be found in most Acinetobacter spp. Genome structure analysis of the neighbour regions of the gacA and DJ41_1407 gene clusters revealed that this gene cluster is also found in Klebsiella pneumonia and Prolinoborus fasciculus, with high amino acid identity among homologues. The gene organization and gene orientation are also identical between different strains, which shows that this gene cluster is highly conserved among bacterial species (Figure 2).
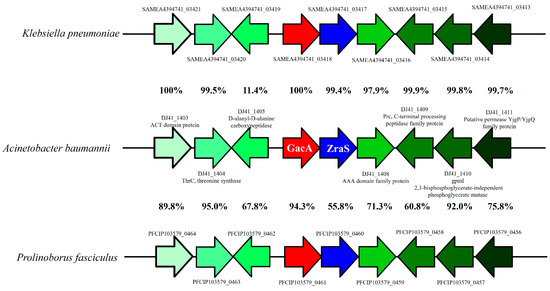
Figure 2.
Comparative genomic analysis of the gacA and DJ41_1407 gene clusters and neighbouring regions. The percentages between two gene clusters represent the amino acid similarity between homologues of two different species.
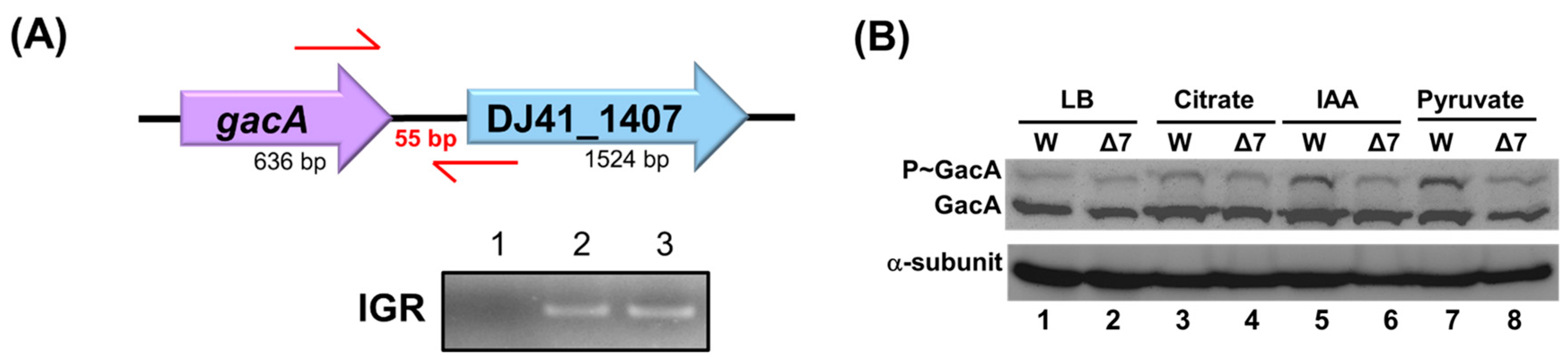
TCS genes in bacterial chromosomes are often located adjacent to one another. In the A. baumannii ATCC 19606 genome, the gacA and DJ41_1407 genes are transcribed in the same direction. Their intergenic region is only 55 base pairs long, suggesting that they may constitute a TCS. RNA was extracted from the wild-type strain, and reverse transcription PCR (RT-PCR) was performed to analyze the transcripts of these genes. The cDNA was used to amplify the intergenic region between gacA and DJ41_1407. If gacA and DJ41_1407 are expressed from the same transcript, a product should be successfully amplified; otherwise, no product would be obtained. The results demonstrated that the intergenic region between gacA and DJ41_1407 was successfully amplified (Figure 3A, lane 3), indicating that these two genes are transcribed from the same mRNA.
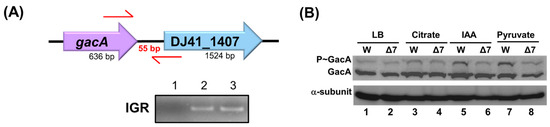
Figure 3.
gacA and DJ41_1407 expression in the same transcript. (A) Analysis of polymerase chain reaction (PCR) products from the gacA and DJ41_1407 intergenic region. 1: Negative control, 2: A. baumannii chromosome as template (positive control), 3: A. baumannii complementary DNA (cDNA) as template. IGR: gacA and DJ41_1407 intergenic region amplification. (B) Analysing the intensity of phosphorylated GacA protein with Phos-tagTM. W: A. baumannii ATCC 19606; Δ7: A. baumannii ATCC 19606 ΔDJ41_1407; LB: strains cultured in LB medium; Citrate: strains cultured in M9 medium with 5 mM citrate; IAA: 5 mM IAA added; Pyruvate: 5 mM pyruvate added.
2.4. GacA and DJ41_1407 Constitute a TCS
DJ41_1407 has been identified as a sensor kinase. The original amino acid sequences of DJ41_1407 were acquired from GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 28 August 2025)), and the conserved and functional domains were analysed using Cdvist (Figure S1C), which revealed a per-Arnt sim domain for signal sensing, a histidine kinase domain for phosphorylation, and an ATPase domain. The potential phosphorylation site is the histidine residue at position 308. To investigate the interaction between GacA and DJ41_1407 in A. baumannii ATCC 19606, Phos-tagTM was used to determine whether DJ41_1407 phosphorylates GacA. The wild-type and ΔDJ41_1407 strains were cultured under the same conditions (Figure 3B). The results indicated that when DJ41_1407 was mutated, a decrease in phosphorylated GacA was observed in cultures in M9 medium with 5 mM citrate, IAA, or pyruvate. This suggests that GacA is phosphorylated by DJ41_1407 and that they function as a TCS. However, the level of GacA phosphorylation by DJ41_1407 was lower compared to that of GacS on GacA (Figure S2). In summary, although GacS plays a more significant role in GacA phosphorylation, DJ41_1407 can also phosphorylate GacA under culture conditions (Figure 3B).
2.5. Transcriptome Analysis of GacA-Regulated Genes
To analyse the function of GacS and GacA, wild-type and ∆DJ41_1407, ∆gacA, and ∆DJ41_1407∆gacA mutant strains were cultured in LB medium and were sent to Welgene Biotech Co., Ltd. for transcriptome analysis. A total of 47, 135, and 185 genes respectively showed differences in expression activity compared to wild-type for the ∆DJ41_1407, ∆gacA, and ∆DJ41_1407∆gacA mutants (Figure 4A; Tables S1–S7). Considering that the gene ontology classification results showed that GacA regulates many genes that may be associated with bacterial metabolism, genes and gene clusters with expression differences greater than 2-fold (log2 greater than 1.0 or less than −1.0) were independently analysed to investigate this further. Analysis revealed a 5-fold decrease in gene expression for DJ14_136 in ΔgacA (Figure 4B), for which the translated protein is alcohol dehydrogenase (Adh4, DJ41_136). Previous studies have shown it to be primarily involved in the alcohol metabolism of A. baumannii, converting ethanol to aldehyde [34]. DJ14_133 showed a 3.5-fold decrease in gene expression in ΔgacA, and its translated protein is annotated as aldehyde dehydrogenase (ALDH), responsible for converting aldehyde to less toxic acetate. DJ41_1668, also annotated as an ALDH, exhibited similar down-regulation in ΔgacA (Figure 4B). In the ΔgacA transcriptome analysis, DJ41_1670 showed a 6-fold decrease in gene expression, and its translated protein, indole-3-pyruvate decarboxylase (IpdC), has been demonstrated to participate in the production of the plant hormone IAA in A. baumannii [35]. Genes DJ41_358 and DJ41_359, with respectively 2.2- and 3.3-fold decreased expression in ΔgacA, encode IacH (indole-3-acetic acid catabolism) and IacA, both previously linked to the breakdown of the plant hormone IAA [24,25] (Figure 4B).
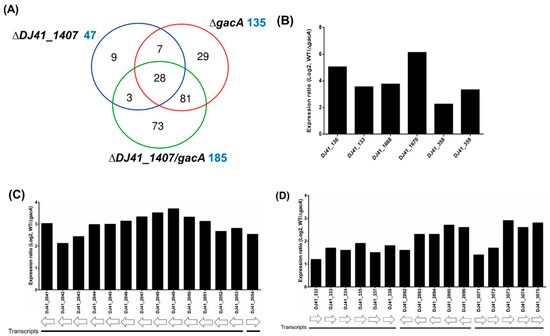
Figure 4.
Transcriptome analysis of the DJ41_1407 and GacA regulon. (A) Comparison between the total number of genes regulated by DJ41_1407 (blue), GacA (red) and DJ41_1407/GacA (green). Genes with log2 expression ratio (wild-type/mutant) > 1.0 or < −1.0 in ∆DJ41_1407, ∆gacA, and ∆DJ41_1407/gacA were compared. A total of 47, 135 and 185 genes, respectively, showed differences in expression activity for the ∆DJ41_1407, ∆gacA, and ∆DJ41_1407/gacA mutant strains compared with the wild-type. (B) Gene clusters regulated by GacA with an expression fold-change greater than 2. (C,D) Gene clusters regulated by GacA are involved in benzoate degradation and the phenylalanine pathway. Gene cluster (C) DJ41_2041-DJ41_2054 was found to be related to phenylalanine metabolism through KEGG analysis. Gene clusters (D) DJ41_332-DJ4_338, DJ41_2892-DJ41_2896 and DJ41_3071-DJ41_3075 were found to be related to benzoate degradation and amino acid metabolism.
Furthermore, ΔgacA transcriptome analysis revealed that the gene cluster DJ41_2041-DJ41_2054 exhibited a greater than 2-fold decrease in gene expression in ΔgacA (Figure 4C). The proteins translated from DJ41_2041-DJ41_2054 were previously shown to be involved in the degradation of phenylacetic acid (PAA), indicating that GacA regulates the metabolisation of alcohols into acetic acid. Acetic acid is then converted to IAA, which, through the PAA degradation pathway, is further degraded into acetyl-CoA and succinyl-CoA, ultimately entering the citric acid cycle for energy production (Figure S3). In addition, three gene clusters, DJ41_332-DJ4_338, DJ41_2892-DJ41_2896, and DJ41_3071-DJ41_3075, exhibited more than a 2-fold increase (log2 of 1.0) in gene expression in ΔgacA (Figure 4D). The functions of the translated proteins from these clusters were analysed using the KEGG enzyme database (https://www.genome.jp/kegg/annotation/enzyme.html (accessed on 28 August 2025)). DJ41_332-DJ4_338 is involved in benzoate metabolism, DJ41_2892-DJ41_2896 is associated with arginine and proline metabolism, and DJ41_3071-DJ41_3075 is involved in alanine, aspartate, and glutamate metabolism. These clusters convert amino acids into succinate and oxaloacetate, which also enter the citric acid cycle for energy production (Figure S3).
2.6. GacA Regulates Genes Involved in Alcohol, IAA, and Phenylacetic Acid Metabolism
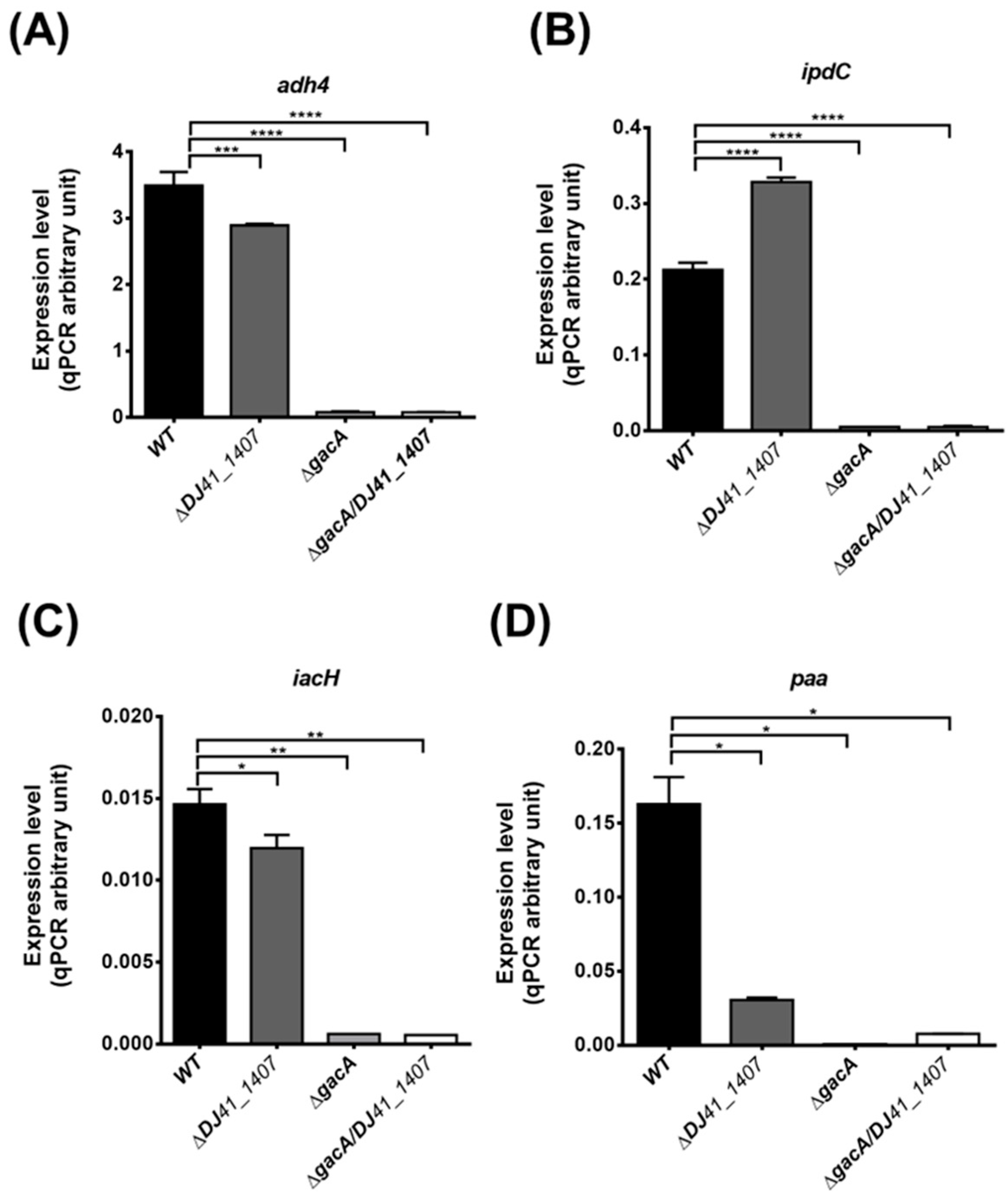
To validate the transcriptome analysis results, real-time quantitative polymerase chain reaction (qRT-PCR) was conducted to measure gene expression levels of adh4, ipdC, iacH, and paa in each strain. The results showed a significant reduction in adh4 expression in the ΔDJ41_1407 strain, suggesting that DJ41_1407 modulates the expression of the adh4 gene. In both ΔgacA and ΔDJ41_1407ΔgacA strains, the expression of the adh4 gene is nearly absent, implying that in A. baumannii, transcription of the adh4 gene is primarily regulated by GacA (Figure 5A). In comparison to the wild-type, ipdC expression is nearly absent in both ΔgacA and ΔDJ41_1407ΔgacA strains (Figure 5B). The results for iacH gene expression also indicate that in the ΔgacA and ΔDJ41_1407ΔgacA strains, iacH gene expression is almost negligible (Figure 5C). An examination of the expression levels of the paa gene revealed a reduction in paa expression in the ΔDJ41_1407 strain, while in the ΔgacA and ΔDJ41_1407ΔgacA strains, paa gene expression is almost absent (Figure 5D). The qRT-PCR results are consistent with the transcriptomic analysis results, indicating that DJ41_1407/GacA not only participates in amino acid metabolism but also regulates bacterial metabolism in alcohol, IAA and phenylacetic acid. This underscores the crucial role of DJ41_1407/GacA in bacterial energy acquisition.
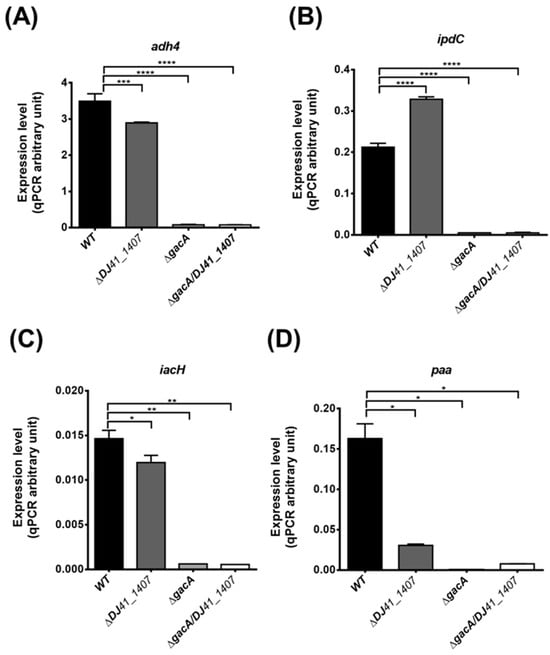
Figure 5.
Real-time quantitative PCR analysis of gene expression in wild-type and ∆DJ41_1407, ∆gacA and ∆DJ41_1407/gacA mutants. Expression differences in (A) adh4, (B) ipdC, (C) iacH and (D) paa genes. One-way analysis of variance (ANOVA) was adopted to evaluate the significance of differences. * p value < 0.05; ** p value < 0.01; *** p value < 0.001; **** p value < 0.0001.
2.7. Possible Binding Region of GacA
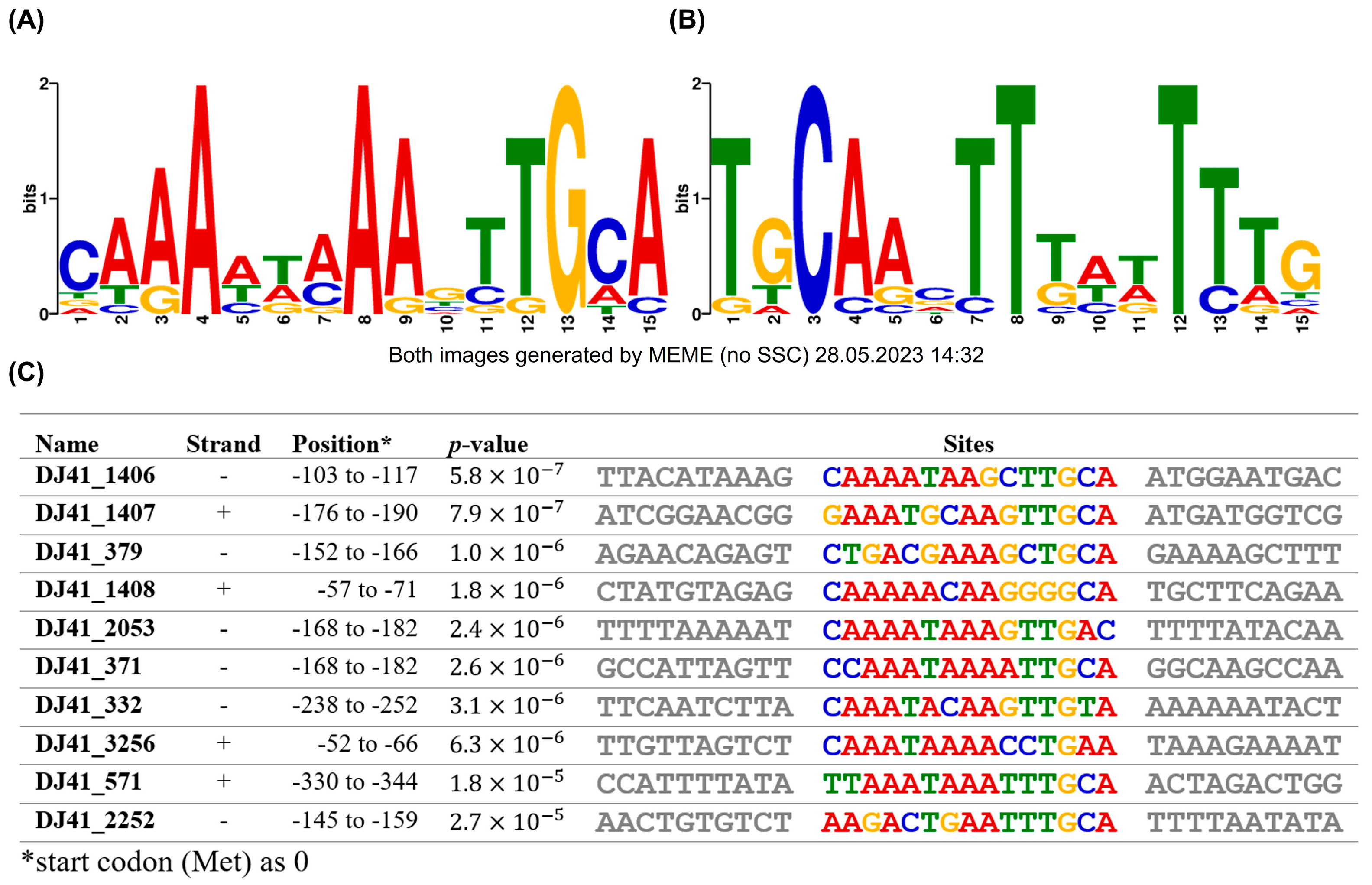
The promoter regions of gene clusters regulated by GacSA were analysed using MEME to identify potential GacA binding boxes. The GacA binding box is within a 15 bp region, with a 5′ CAAAWWAAAGTTGCA 3′ sequence on the positive strand (Figure 6A). These sequences were also consistent with those in the upstream region of genes exhibiting opposite transcriptional orientations (Figure 6B). Previous studies have shown that the regulator of a TCS binds to its promoter to self-regulate its expression, a phenomenon also observed in GacA. The GacA binding box was identified in the upstream region from −103 to −117 of DJ41_1406, with the 5′ CAAAATAAGCTTGCA 3′ sequence. This finding suggests that GacA regulates its own expression, a characteristic commonly observed in most bacterial TCS (Figure 6C).
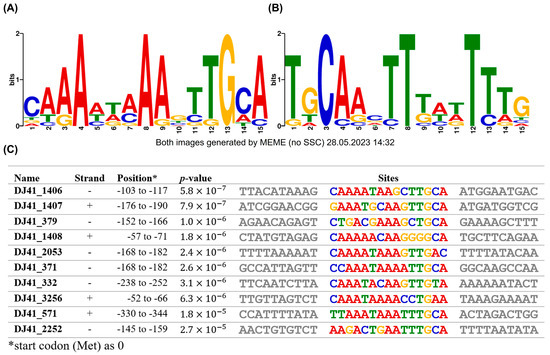
Figure 6.
Putative GacA binding box analysis by MEME Suite. Conservation of the GacA binding box in (A) positive strand and (B) negative strand. (C) Exact sequence of the GacA binding box in the promoter region of different genes. The positions represent the location of the conserved sequence upstream of analysed genes, while the first base pair of the start codon was considered as +1. The highly conserved sequence location is likely to be the binding box of GacA.
2.8. GacA Regulates Bacterial Growth
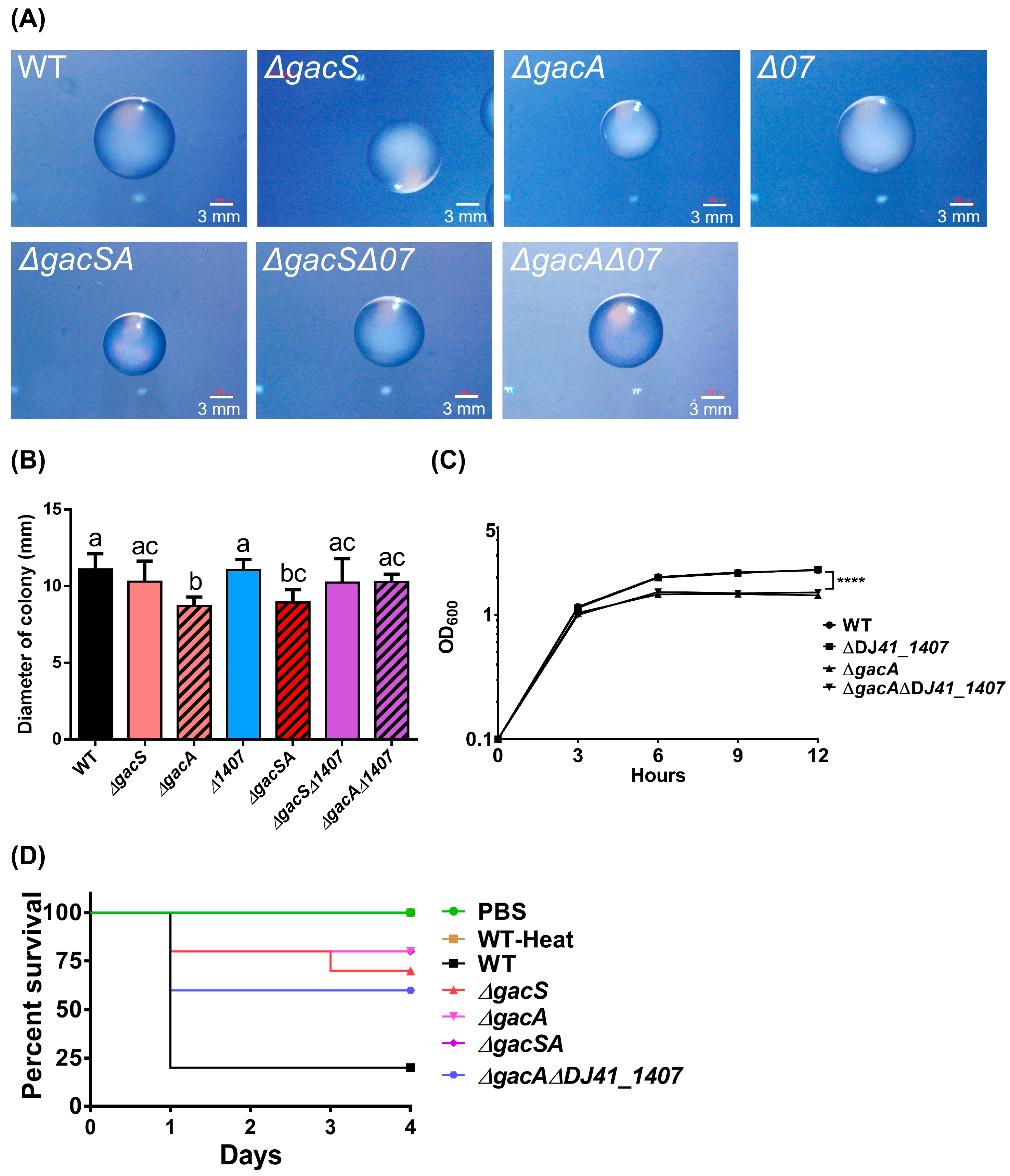
To understand the biological role of GacA and its corresponding sensor, colonies of A. baumannii ATCC 19606 and its mutants were observed on LB agar plates using a stereoscopic microscope. The results indicated that the colony sizes of the ∆gacA and ∆gacSA mutants were significantly smaller than those of the other strains (Figure 7A). ImageJ (https://imagej.net/ij/ (accessed on 28 August 2025)) was utilised to analyse colony diameter and revealed that the colony sizes of ∆gacA were 1.28-fold smaller than those of all other strains, except for ∆gacSA. Additionally, ∆gacSA exhibited colony sizes that were 1.21-fold smaller than those of A. baumannii ATCC 19606, ∆gacS, and ∆DJ41_1407. No significant differences were observed among the wild type, ∆gacS, ∆DJ41_1407, ∆gacS∆DJ41_1407, and ∆gacA∆DJ41_1407 strains (Figure 7B). The growth curves of the wild-type and mutant strains were assessed in LB medium over 12 h. The results indicated a significantly reduced optical density (OD) in the ∆gacA and ∆gacA∆DJ41_1407 mutants (Figure 7C). These results suggest that GacA can play a role in colony size and growth conditions in rich medium for A. baumannii ATCC 19606.
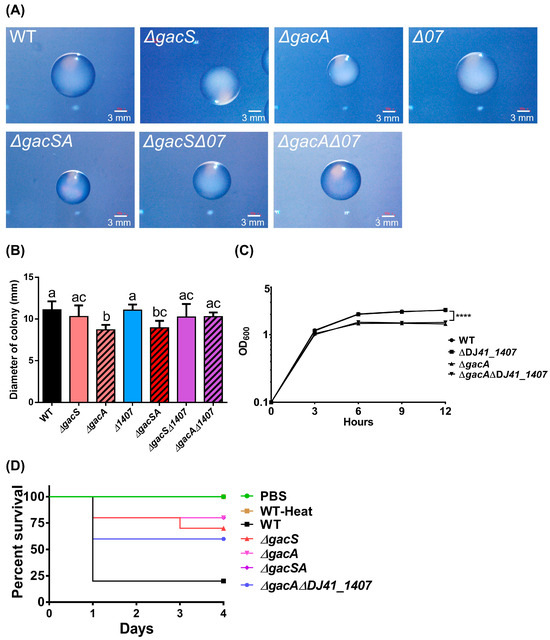
Figure 7.
Biological properties of A. baumannii ATCC 19606 wild-type and mutant strains. (A) Images of colonies were taken at 12 h post-cultivation. The length of the white scale bar represents 3 mm. (B) ImageJ was used to analyse colony diameter, using the scale bar as a standard. Each strain was measured in 10 colony replicates. A graph bar with the same alphabet indicates that there were no significant differences between each bar (p < 0.05). (C) The growth curve of different strains cultured in LB medium, including wild-type (WT), ΔDJ41_1407, ΔgacA and ΔgacAΔDJ41_1407. One-way ANOVA was adopted to evaluate the significance of differences. **** p value < 0.0001. (D) Survival rate of G. mellonella larvae after bacterial infection. The x-axis indicates the time from infection of G. mellonella larvae by different bacterial strains, while the y-axis represents the survival rate of G. mellonella larvae in each group. G. mellonella larvae were infected with heat-treated wild-type (WT-Heat), wild-type (WT), ΔgacS, ΔgacA, ΔgacSA and ΔgacAΔDJ41_1407 strains at 5 × 106 CFU/larvae for 4 days (n = 10), or injected with Phosphate-Buffered Saline (PBS).
2.9. GacSA Regulates Virulence
To determine whether GacS, GacA, and DJ41_1407 can influence the virulence of A. baumannii, we infected Galleria mellonella larvae with A. baumannii ATCC 19606 and its mutant strains. The survival rates of the larvae were monitored over 96 h. Results indicated that larvae injected with Phosphate-Buffered Saline (PBS) survived throughout the 96 h, demonstrating health and no mortality. In contrast, larvae injected with the wild-type strain exhibited a survival rate of only 20% after 24 h, indicating high bacterial virulence. Conversely, larvae infected with the ΔgacA and ΔgacSA strains showed a survival rate of 80%, while those infected with ΔgacS had a survival rate of 70%. This suggests that bacterial virulence was reduced after the gacS and gacA genes were mutated. Additionally, larvae infected with the ΔgacAΔDJ41_1407 strain demonstrated a 60% survival rate after 96 h, indicating that bacterial virulence was also reduced when both the gacA and DJ41_1407 genes were mutated. These results suggest that the gacS, gacA, and DJ41_1407 genes play a crucial role in regulating the expression of bacterial virulence genes (Figure 7D).
2.10. GacA Regulates Antibiotic Resistance
Previous studies have demonstrated that TCSs can significantly influence bacterial resistance to antibiotics (8). Therefore, we aimed to determine whether GacS, GacA, and DJ41_1407 might also affect the antibiotic resistance of A. baumannii ATCC 19606. Wild-type and mutant strains were cultured in media containing varying concentrations of apramycin, gentamicin, kanamycin, chloramphenicol, polymyxin B, and colistin, to assess the minimum inhibitory concentration (MIC) of these antibiotics for each strain (Table 1). For the ∆gacA∆DJ41_1407 strain, there was a 4-fold decrease in the MIC for apramycin and a 2-fold reduction in the MICs for gentamicin, kanamycin, and chloramphenicol, compared to the wild-type strain. The MICs of all tested antibiotics for the ∆gacS, ∆DJ41_1407, and ∆gacS∆DJ41_1407 strains remained unchanged from those of the wild-type (Table 1). These findings indicate that GacA plays a crucial role in modulating the antibiotic resistance of A. baumannii ATCC 19606.

Table 1.
Minimum inhibitory concentrations (MICs) of wild-type and mutant bacterial strains against different antibiotics.
3. Discussion
The GacSA TCS has been identified as a global regulator in A. baumannii ATCC17978, controlling the expression of 674 genes associated with virulence, biofilm formation, pili production, resistance against human serum, motility, and metabolism of aromatic compounds [28,29]. In this study, GacSA was found to regulate virulence in A. baumannii ATCC 19606, exhibiting certain similarities with GacSA in A. baumannii ATCC 17978. In both A. baumannii ATCC 19606 and A. baumannii ATCC 17978, GacSA was observed to affect the expression of genes involved in virulence and biofilm formation, as well as the metabolism of aromatic compounds, such as the catabolic pathway of aromatic compounds known as the PAA pathway, which was also investigated in this study.
In this study, we discovered that GacA was phosphorylated by both GacS (Figure S2A) and DJ41_1407 (Figure 3B), indicating crosstalk between GacSA, DJ41_1407, and GacA. This phenomenon is also observed in other bacterial species. In Serratia marcescens, QseBC is involved in quorum sensing, while RssAB plays a role in swarming and biofilm formation [36]. QseC can dephosphorylate RssB ∼ P, deactivating RssAB signaling and facilitating bacterial surface migration initiation during swarming development. These findings highlight the crosstalk between two TCSs, which cooperatively regulate flagellar biosynthesis in a stage-specific manner during swarming development [36]. Another unique crosstalk system between TCSs has been observed in E. coli, involving the HprR and CusR response regulators [37,38]. HprSR is involved in the stress response to hydrogen peroxide, while CusSR is responsible for the response to Cu(II) [37,38]. Interestingly, HprR and CusR recognize the same binding sequence, suggesting that they can regulate the same target genes, but cannot bind to these targets simultaneously. Both regulators recognize and transcribe the hiuH and cusC promoters, albeit with different efficiencies, indicating a collaborative mechanism [37,38]. However, as protein concentrations increase, HprR and CusR compete to transcribe common targets, demonstrating a competitive interplay [37,38]. Further research is needed to ascertain whether GacSA and DJ41_1407 have similarly competitive crosstalk mechanisms in place.
We further found that GacSA in A. baumannii enhanced resistance against apramycin, gentamicin, and kanamycin (Table 1). In comparison, GacSA in P. aeruginosa has also been implicated in antibiotic resistance against three different antibiotic families, tobramycin, ciprofloxacin, and tetracycline [39], likely through the RsmA/rgRsmZ pathway. The BfiRS system potentially collaborates with GacSA and contributes to the regulation loop involving Rsm, thereby exerting control over biofilm formation [40]. Compared to GacSA in P. aeruginosa, fewer functions of GacSA in A. baumannii ATCC 19606 have been identified for now, but key similarities can be noted and may point the way for future research.
This study observed that virulence of A. baumannii ATCC 19606 against G. mellonella larvae was significantly reduced after the deletion of gacS and/or gacA genes (Figure 7D). It has been previously reported in A. baumannii ATCC 17978 [29] that mutation of the gacS gene results in avirulence of the mutated strains against Candida albicans and mice. The study further noted that virulence was restored by complementation [29]. Previous research showed that dissemination from infection sites to other organs in murine models was much reduced with gacS deletion mutant strains compared to wild type, and the mutant strains were also more susceptible to the killing effects of human serum [29]. This has been linked to the PAA pathway, as the paa operon is completely repressed in gacS deletion mutant strains, thus preventing them from utilising L-phenylalanine as a carbon source [29]. This study also observed that deletion of the gacS and/or gacA genes downregulates the expression of key genes involved in alcohol and amino acid metabolism, which can limit energy sources available to the mutant strains (Figure 4; Tables S1–S7). Moreover, ∆gacSA mutant strains exhibited smaller colony sizes (Figure 7B). These results suggest that disruption of the GacSA TCS has profound effects on bacterial metabolism, growth, dissemination, and virulence, and may have implications for the development of novel antibacterial strategies.
Together, the findings in this study show that GacSA as well as DJ41_1407/GacA are TCSs and further indicate crosstalk between these two TCSs. GacA can be phosphorylated by both GacS and DJ41_1407, with GacS playing a more significant role in GacA phosphorylation than DJ41_1407. Together, GacS, GacA, and DJ41_1407 were found to regulate key functions and genes involved in benzoate degradation, phenylalanine metabolism, virulence, and antibiotic resistance of A. baumannii ATCC 19606. These findings shed light on the GacSA TCS in A. baumannii ATCC 19606 and offer important insights regarding the regulation of energy production, virulence, and resistance.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Culture Media, and Markerless Mutation
The bacterial strains used in this study are detailed in Table 2. The plasmids and primers employed in constructing the mutants are respectively listed in Table 3 and Table 4. LB medium (0.5% yeast extract, 1% tryptone, and 1% NaCl [41]; Becton Dickinson (BD), Franklin Lakes, NJ, USA) and agar (BD) were utilised to culture the bacterial strains statically or with shaking at 200 rpm at 37 °C. PCR-amplified 1 kb fragments of the upstream and downstream regions of GacS, GacA, and DJ41_1407 genes were inserted into the plasmid pK18mobsacB and transformed into the donor strain E. coli S17-1λπ. The recombinant plasmid containing the donor was then introduced into A. baumannii through conjugation. The transconjugants were cultured on LB plates supplemented with ampicillin and kanamycin to select for A. baumannii that harboured the integrated recombinant pK18mobsacB, indicating the occurrence of the first homologous recombination. Colony PCR confirmed successful recombination. The positive colony was subsequently cultured in LB with 20% sucrose, resulting in a second homologous recombination with a 50% probability of deleting the desired gene fragment [42].

Table 2.
Bacterial strains used in this study.

Table 3.
Plasmids used in this study.

Table 4.
Primers used in this study.
4.2. Transcriptome Sequencing and Analysis
Bacterial strains were cultured in LB medium containing 50 μg/mL of ampicillin at 37 °C for 12 to 14 h and then subcultured with an initial optical density (OD) at 600 nm (OD600) of 0.1 for 3 h. After this incubation period, the bacterial solution was aliquoted to achieve an OD600 of 0.6 in each microcentrifuge tube. Subsequently, 1/10 of the volume of acid phenol [5% acid phenol, 95% ethanol] was added to the microcentrifuge tubes and mixed thoroughly to fix the samples. The microcentrifuge tubes were centrifuged at 4 °C and 17,000× g for 10 min. Following centrifugation, the supernatant was carefully removed, and the pellet was resuspended in 1 mL Thermo Fisher TRIzol™ Reagent (Invitrogen, Waltham, MA, USA). After resuspension, the samples were stored at −80 °C and were sent to Welgene Biotech Co., Ltd. for transcriptome analysis [8]. RNA sequence reads were generated using Illumina HiSeq 2000 (San Diego, CA, USA) and then underwent selection to remove reads containing adaptors, reads containing over 10% of unknown sequences, and reads of low quality (defined as more than half of all bases having a quality score < 5). The curated set of reads was then mapped to the A. baumannii ATCC 19606 genome published on GenBank (accession numbers: SRX3312085 and SRX3312086) using SOAP2 software (https://bioinformaticshome.com/db/tool/SOAP2 (accessed on 28 August 2025)) [44]. We subsequently calculated gene expression levels as reads per kilobase of genes per million reads (RPKMs) [45,46], and false discovery rates (FDRs) [47] and the RPKM ratios of two samples were utilized to discern differentially expressed genes (DEGs). We considered DEGs to be genes with FDR ≤ 0.01 and an absolute log2 value ratio of greater than 1.
4.3. RNA Extraction and Reverse Transcription (RT)
Bacterial strains were cultured at 37 °C with agitation overnight and then subcultured in 50 mL of LB medium for 3 h after OD600 reached 0.3. A total of 0.6 of OD600 samples were then collected and mixed with 0.1 volume of fixing solution (5% acid phenol, 95% ethanol). After centrifugation at 17,000× g at 4 °C, the cell pellets were stored at −80 °C for RNA extraction. Cell pellets were thawed on ice and resuspended in 1 mL of NucleoZOL (MACHEREY-NAGEL, Düren, Germany), then mixed thoroughly with 400 μL of diethyl pyrocarbonate (DEPC)-treated H2O, and incubated at room temperature for 15 min. The supernatant was recovered after centrifugation at 17,000× g at 4 °C for 20 min and then mixed with 5 μL of 100% 4-bromoanisole and incubated at room temperature for 10 min. Excess protein was then removed by centrifugation at 17,000× g at 4 °C for 20 min. The resulting RNA suspension was mixed with an equal volume of isopropanol for 15 min to induce RNA precipitation. The derived RNA pellet was washed twice with ice-cold 75% ethanol and resuspended in 30 μL of DEPC-treated H2O for analysis [34].
A total of 2 μg of RNA was used to prepare cDNA. The RT-PCR mixture contained 10× reaction buffer, 200 U of MMLV high-performance reverse transcriptase (Epicentre, Madison, WI, USA), 100 mM of dithiothreitol (DTT), 2.5 mM dNTP, and 1 nM of hexamer. The reaction was conducted in a Biometra TADVANCED Thermal Cycler (Analytik Jena, Jena, Germany). The gyrase gene served as an internal control, and the other gene-specific primers used to determine the presence and expression levels of the respective genes are listed in Table 4 [35].
4.4. Phos-Tagtm Acrylamide Gel Electrophoresis
Wild-type, ΔgacS, and ΔDJ41_1407 strains were cultured in LB broth with shaking at 37 °C overnight. Bacteria were subcultured in M9 medium containing 5 mM citrate, IAA, or tryptophan. Bacteria were collected after OD600 reached 0.6 and then centrifuged and resuspended in 130 μL of 1 M formic acid, 54 μL of sample buffer [130 mM Tris-Cl (pH 6.8), 6% SDS, 15% β-mercaptonethanol, 3% glycerol, 15% bromophenol blue], and 24 μL of 5 N NaOH. Phos-tagTM containing 12% sodium dodecyl sulfate (SDS)-acrylamide resolving gel was prepared with 50 μL of 50 mM Phos-tagTM, 1.25 mL of 1.5 M Tris-Cl (pH 8.8), 2.12 mL of 30% acrylamide solution, 50 μL of 10 mM MnCl2, 50 μL of 10% SDS, 1.25 mL of ddH2O, 25 μL of 10% ammonium persulfate (APS), and 5 μL of N, N,N’,N’-tetramethylethylenediamine (TEMED). A 6% staking gel was prepared with 1 mL of 0.5 M Tris-Cl (pH 6.8), 800 μL of 30% acrylamide solution, 40 μL of 10% SDS, 2.12 mL of ddH2O, 40 μL of 10% APS, and 4 μL of TEMED. Electrophoresis of prepared samples was conducted at 120 V at room temperature for 90 min [48].
4.5. G. mellonella Larvae Infection Assay
Bacterial strains were cultured in LB medium at 37 °C for 12–14 h. The overnight bacterial solution was washed twice with PBS buffer (0.14 M NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) to remove LB medium. The bacterial solution was then diluted to 5 × 108 CFU/mL. Larvae were infected with only PBS and heat-killed wild-type bacteria as the control group (n = 10). Heat-killed wild-type bacteria were prepared by heating the bacterial solution at 100 °C for 5 min. Using a Hamilton syringe [Hamilton® syringe, 700 series, 701N, Reno, NV, USA], 10 μL of diluted bacterial solutions was injected into G. mellonella larvae through the last left pro-leg of the abdominal region. The final bacterial concentration was 5 × 106 CFU/larva. Infected larvae were incubated at 37 °C. Survival (alive/dead) and melanisation scores were measured every 24 h for 96 h [49].
4.6. MIC Test
The MIC test protocol is based on a modified version of the broth dilution method established by the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [50], and the concentrations of the antibiotics used were based on results derived from previous studies [8,51]. Bacteria were incubated in a shaking incubator at 37 °C for 12–16 h. A 96-well culture plate was prepared, with 270 μL of LB medium added to the first well and 135 μL of LB medium added to each subsequent well. Using kanamycin (Sigma-Aldrich, St. Louis, MO, USA) as an example, a final concentration of 200 μg/mL kanamycin was added to the first well. Half of the volume (135 μL) was transferred to the second well (½ dilution) thoroughly, and then ½ was serially diluted to each subsequent well. After dilution, 135 μL of LB medium was added to each well. The first well had a final concentration of 100 μg/mL kanamycin. Bacterial strains were respectively treated with the following antibiotics (Sigma-Aldrich): apramycin, gentamycin, kanamycin, tetracycline, polymyxin B, and colistin. Each desired antibiotic experiment group was serially performed with six replicates. Overnight bacterial solution was adjusted to OD600 = 1.0, and 30 μL was added into each well to make the initial OD600 0.1. After incubating at 37 °C for 12 h, the OD600 value was measured to determine the MIC of the antibiotic. The MIC was defined as the antibiotic concentration that was twice the concentration at which there was no bacterial growth [8].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms262110620/s1.
Author Contributions
Conceptualization, G.-H.L.; methodology, M.-Y.W.; software, G.-H.L.; validation, Y.-H.T. and M.-Y.W.; formal analysis, M.-Y.W. and Y.-H.T.; investigation, M.-Y.W. and Y.-H.T.; resources, G.-H.L.; data curation, Y.-H.T. and M.-Y.W.; writing—original draft preparation, G.-H.L. and Y.-H.T.; writing—review and editing, G.-H.L.; visualization, G.-H.L. and Y.-H.T.; supervision, G.-H.L.; project administration, G.-H.L.; funding acquisition, G.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Council under grant number MOST111-2635B320-001-MY2. The funder had no role in study design, data collection, interpretation, or the decision to submit the work for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Acknowledgments
We are grateful for the support from the Core Facility Center, Tzu Chi University.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lastoria, L.C.; Caldeira, S.M.; Moreira, R.G.; Akazawa, R.T.; Maion, J.C.; Fortaleza, C.M.C.B. Ecological competition and the incidence of Acinetobacter baumannii bloodstream infections in a teaching hospital in Southeastern Brazil. Rev. Soc. Bras. Med. Trop. 2014, 47, 583–588. [Google Scholar] [CrossRef][Green Version]
- Bergogne-Berezin, E.; Towner, K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Actis, L.; Pachon, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Espinal, P.; Vila-Farres, X.; Vila, J. The Acinetobacter baumannii Oxymoron: Commensal Hospital Dweller Turned Pan-Drug-Resistant Menace. Front. Microbiol. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Espinal, P.; Marti, S.; Vila, J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J. Hosp. Infect. 2012, 80, 56–60. [Google Scholar] [CrossRef]
- Toh, H.Y.; Lin, G.H. Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC 19606 Virulence and Antibiotic Response. Int. J. Mol. Sci. 2024, 25, 3862. [Google Scholar] [CrossRef]
- Roy, S.; Chowdhury, G.; Mukhopadhyay, A.K.; Dutta, S.; Basu, S. Convergence of Biofilm Formation and Antibiotic Resistance in Acinetobacter baumannii Infection. Front. Med. 2022, 9, 793615. [Google Scholar] [CrossRef]
- Shan, W.; Kan, J.; Cai, X.; Yin, M. Insights into mucoid Acinetobacter baumannii: A review of microbiological characteristics, virulence, and pathogenic mechanisms in a threatening nosocomial pathogen. Microbiol. Res. 2022, 261, 127057. [Google Scholar] [CrossRef]
- Shu, H.Y.; Huang, Y.W.; Tsai, P.Y.; Hsieh, K.S.; Lin, G.H. Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii. Int. J. Mol. Sci. 2022, 23, 12606. [Google Scholar] [CrossRef]
- Huang, Y.W.; Shu, H.Y.; Lin, G.H. Gene Expression of Ethanol and Acetate Metabolic Pathways in the Acinetobacter baumannii EmaSR Regulon. Microorganisms 2024, 12, 331. [Google Scholar] [CrossRef]
- Dupont, C.A.; Bourigault, Y.; Biziere-Maco, H.; Boukerb, A.M.; Latour, X.; Barbey, C.; Verdon, J.; Merieau, A. The GacS/GacA two-component system strongly regulates antimicrobial competition mechanisms of Pseudomonas fluorescens MFE01 strain. J. Bacteriol. 2025, 207, e0038824. [Google Scholar] [CrossRef]
- Al-Khodor, S.; Kalachikov, S.; Morozova, I.; Price, C.T.; Abu Kwaik, Y. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect. Immun. 2009, 77, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Lamy, M.C.; Zouine, M.; Fert, J.; Vergassola, M.; Couve, E.; Pellegrini, E.; Glaser, P.; Kunst, F.; Msadek, T.; Trieu-Cuot, P.; et al. CovS/CovR of group B streptococcus: A two-component global regulatory system involved in virulence. Mol. Microbiol. 2004, 54, 1250–1268. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lin, K.; Liu, Y.; Zhang, H.; Lei, L. Two-component signaling pathways modulate drug resistance of Staphylococcus aureus (Review). Biomed. Rep. 2020, 13, 5. [Google Scholar]
- Kitten, T.; Kinscherf, T.G.; McEvoy, J.L.; Willis, D.K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 1998, 28, 917–929. [Google Scholar] [CrossRef]
- Pessi, G.; Williams, F.; Hindle, Z.; Heurlier, K.; Holden, M.T.; Camara, M.; Haas, D.; Williams, P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6676–6683. [Google Scholar] [CrossRef]
- Coleman, F.T.; Mueschenborn, S.; Meluleni, G.; Ray, C.; Carey, V.J.; Vargas, S.O.; Cannon, C.L.; Ausubel, F.M.; Pier, G.B. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl. Acad. Sci. USA 2003, 100, 1949–1954. [Google Scholar] [CrossRef]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef]
- Brencic, A.; Lory, S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009, 72, 612–632. [Google Scholar] [CrossRef]
- Wolfgang, M.C.; Lee, V.T.; Gilmore, M.E.; Lory, S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 2003, 4, 253–263. [Google Scholar] [CrossRef]
- Heeb, S.; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe. Interact. 2001, 14, 1351–1363. [Google Scholar] [CrossRef]
- Dorman, M.J.; Feltwell, T.; Goulding, D.A.; Parkhill, J.; Short, F.L. The Capsule Regulatory Network of Klebsiella pneumoniae Defined by density-TraDISort. mBio 2018, 9, e01863-18. [Google Scholar] [CrossRef]
- Bao, J.; Xie, L.; Ma, Y.; An, R.; Gu, B.; Wang, C. Proteomic and Transcriptomic Analyses Indicate Reduced Biofilm-Forming Abilities in Cefiderocol-Resistant Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 778190. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.S.; Ellett, F.; Murray, G.L.; Kostoulias, X.; Cerqueira, G.M.; Schulze, K.E.; Maifiah, M.H.M.; Li, J.; Creek, D.J.; Lieschke, G.J.; et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA 2016, 113, 9599–9604. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, G.M.; Kostoulias, X.; Khoo, C.; Aibinu, I.; Qu, Y.; Traven, A.; Peleg, A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014, 210, 46–55. [Google Scholar] [CrossRef]
- De Silva, P.M.; Kumar, A. Effect of Sodium Chloride on Surface-Associated Motility of Acinetobacter baumannii and the Role of AdeRS Two-Component System. J. Membr. Biol. 2018, 251, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lin, Y.Y.; Yeh, H.W.; Lan, C.Y. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 2014, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacob, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef] [PubMed]
- Liou, M.L.; Soo, P.C.; Ling, S.R.; Kuo, H.Y.; Tang, C.Y.; Chang, K.C. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J. Microbiol. Immunol. Infect. 2014, 47, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.H.; Hsieh, M.C.; Shu, H.Y. Role of Iron-Containing Alcohol Dehydrogenases in Acinetobacter baumannii ATCC 19606 Stress Resistance and Virulence. Int. J. Mol. Sci. 2021, 22, 9921. [Google Scholar] [CrossRef]
- Lin, H.R.; Shu, H.Y.; Lin, G.H. Biological roles of indole-3-acetic acid in Acinetobacter baumannii. Microbiol. Res. 2018, 216, 30–39. [Google Scholar] [CrossRef]
- Wei, C.F.; Tsai, Y.H.; Tsai, S.H.; Lin, C.S.; Chang, C.J.; Lu, C.C.; Huang, H.-C.; Lai, H.-C. Cross-talk between bacterial two-component systems drives stepwise regulation of flagellar biosynthesis in swarming development. Biochem. Biophys. Res. Commun. 2017, 489, 70–75. [Google Scholar] [CrossRef]
- Urano, H.; Umezawa, Y.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cooperative regulation of the common target genes between H2O2-sensing YedVW and Cu2+-sensing CusSR in Escherichia coli. Microbiology 2015, 161, 729–738. [Google Scholar] [CrossRef]
- Urano, H.; Yoshida, M.; Ogawa, A.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cross-regulation between two common ancestral response regulators, HprR and CusR, in Escherichia coli. Microbiology 2017, 163, 243–252. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef]
- Petrova, O.E.; Sauer, K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009, 5, e1000668. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Lin, G.H.; Chen, H.P.; Huang, J.H.; Liu, T.T.; Lin, T.K.; Wang, S.J.; Tseng, C.-H.; Shu, H.-Y. Identification and characterization of an indigo-producing oxygenase involved in indole 3-acetic acid utilization by Acinetobacter baumannii. Antonie Van Leeuwenhoek 2012, 101, 881–890. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Barbieri, C.M.; Stock, A.M. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 2008, 376, 73–82. [Google Scholar] [CrossRef]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef]
- Kadeřábková, N.; Mahmood, A.J.S.; Mavridou, D.A.I. Antibiotic susceptibility testing using minimum inhibitory concentration (MIC) assays. NPJ Antimicrob. Resist. 2024, 2, 37. [Google Scholar] [CrossRef]
- Chen, S.J.; Shu, H.Y.; Lin, G.H. Regulation of tert-Butyl Hydroperoxide Resistance by Chromosomal OhrR in A. baumannii ATCC 19606. Microorganisms 2021, 9, 629. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).