Detection of Genes Associated with Polymyxin and Antimicrobial Peptide Resistance in Isolates of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility to the Peptide Antibiotics
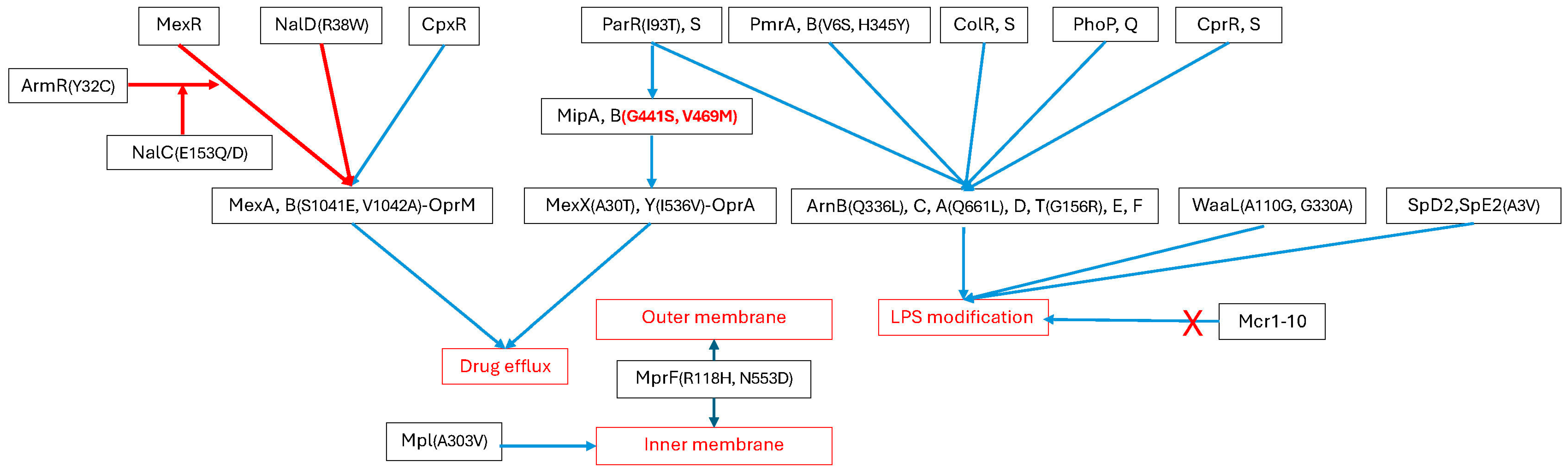
2.2. Possession of Genes and SNPs Associated with Polymxin Resistance
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antimicrobial Susceptibility Testing
4.3. Detection of Polymyxin Resistance Genes and SNPs
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMP | Antimicrobial peptide |
| MIC | Minimum inhibitory concentration |
| CARD | Comprehensive Antibiotic Resistance Database |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| SNP | Single nucleotide polymorphism |
| MDR | Multidrug resistant |
| XDR | Extensively drug resistant |
| LPS | Lipopolysaccharide |
| L-Ara4N | 4-amino-4-deoxy-L-arabinose |
References
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014. [Google Scholar]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef]
- Sathe, N.; Beech, P.; Croft, L.; Suphioglu, C.; Kapat, A.; Athan, E. Pseudomonas aeruginosa: Infections and novel approaches to treatment “Knowing the enemy” the threat of Pseudomonas aeruginosa and exploring novel approaches to treatment. Infect. Med. 2023, 2, 178–194. [Google Scholar] [CrossRef]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Yildiz, E.H.; Airiani, S.; Hammersmith, K.M.; Rapuano, C.J.; Laibson, P.R.; Virdi, A.S.; Hongyok, T.; Cohen, E.J. Trends in contact lens-related corneal ulcers at a tertiary referral center. Cornea 2012, 31, 1097–1102. [Google Scholar] [CrossRef]
- Ng, A.L.; To, K.K.; Choi, C.C.; Yuen, L.H.; Yim, S.M.; Chan, K.S.; Lai, J.S.; Wong, I.Y. Predisposing factors, microbial characteristics, and clinical outcome of microbial keratitis in a tertiary centre in Hong Kong: A 10-year experience. J. Ophthalmol. 2015, 2015, 769436. [Google Scholar] [CrossRef]
- Stapleton, F.; Keay, L.; Edwards, K.; Naduvilath, T.; Dart, J.K.; Brian, G.; Holden, B.A. The incidence of contact lens-related microbial keratitis in Australia. Ophthalmology 2008, 115, 1655–1662. [Google Scholar] [CrossRef]
- Morin, C.D.; Deziel, E.; Gauthier, J.; Levesque, R.C.; Lau, G.W. An organ system-based synopsis of Pseudomonas aeruginosa virulence. Virulence 2021, 12, 1469–1507. [Google Scholar] [CrossRef]
- Tummler, B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Research 2019, 8, 1371. [Google Scholar] [CrossRef]
- Kunz Coyne, A.J.; El Ghali, A.; Holger, D.; Rebold, N.; Rybak, M.J. Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 2022, 11, 661–682. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorli, L.; Luque, S.; Gomez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Montero, M.M.; Lopez Montesinos, I.; Knobel, H.; Molas, E.; Sorli, L.; Siverio-Pares, A.; Prim, N.; Segura, C.; Duran-Jorda, X.; Grau, S.; et al. Risk factors for mortality among patients with Pseudomonas aeruginosa bloodstream infections: What Is the influence of XDR phenotype on outcomes? J. Clin. Med. 2020, 9, 514. [Google Scholar] [CrossRef]
- Jurado-Martin, I.; Sainz-Mejias, M.; McClean, S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Liang, Y.; Li, J.; Xu, Y.; He, Y.; Jiang, B.; Wu, C.; Shan, B.; Shi, H.; Song, G. Genomic variations in polymyxin-resistant Pseudomonas aeruginosa clinical isolates and their effects on polymyxin resistance. Braz. J. Microbiol. 2023, 54, 655–664. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Enninful, G.N.; Kuppusamy, R.; Tiburu, E.K.; Kumar, N.; Willcox, M.D.P. Non-canonical amino acid bioincorporation into antimicrobial peptides and its challenges. J. Pept. Sci. 2024, 30, e3560. [Google Scholar] [CrossRef] [PubMed]
- Ridyard, K.E.; Overhage, J. The potential of human peptide LL-37 as an antimicrobial and anti-biofilm agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocincova, D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2011, 2, 118. [Google Scholar] [CrossRef]
- Lam, M.Y.; McGroarty, E.J.; Kropinski, A.M.; MacDonald, L.A.; Pedersen, S.S.; Hoiby, N.; Lam, J.S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J. Clin. Microbiol. 1989, 27, 962–967. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Moskowitz, S.M.; Brannon, M.K.; Dasgupta, N.; Pier, M.; Sgambati, N.; Miller, A.K.; Selgrade, S.E.; Miller, S.I.; Denton, M.; Conway, S.P.; et al. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2012, 56, 1019–1030. [Google Scholar] [CrossRef]
- Nummila, K.; Kilpeläinen, I.; Zähringer, U.; Vaara, M.; Helander, I.M. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coii are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 1995, 16, 271–278. [Google Scholar] [CrossRef]
- Fernández, L.; Jenssen, H.; Bains, M.; Wiegand, I.; Gooderham, W.J.; Hancock, R.E. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob. Agents Chemother. 2012, 56, 6212–6222. [Google Scholar] [CrossRef] [PubMed]
- Langendonk, R.F.; Neill, D.R.; Fothergill, J.L. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa: Implications for current resistance-breaking therapies. Front. Cell. Infect. Microbiol. 2021, 11, 665759. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: An overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef]
- Gahlot, D.K.; Patkowski, J.B.; Fernandez de Santaella, J.; Allsopp, L.P.; Pan, Z.; Filloux, A.; Larrouy-Maumus, G.; Francis, M.S.; Costa, T.R.D. Cpx-signalling in Yersinia pseudotuberculosis modulates Lipid-A remodelling and resistance to last-resort antimicrobials. npj Antimicrob. Resist. 2024, 2, 39. [Google Scholar] [CrossRef]
- Janet-Maitre, M.; Job, V.; Bour, M.; Robert-Genthon, M.; Brugiere, S.; Triponney, P.; Cobessi, D.; Coute, Y.; Jeannot, K.; Attree, I. Pseudomonas aeruginosa MipA-MipB envelope proteins act as new sensors of polymyxins. mBio 2024, 15, e0221123. [Google Scholar] [CrossRef]
- Huang, L.C.; Reins, R.Y.; Gallo, R.L.; McDermott, A.M. Cathelicidin-deficient (Cnlp -/-) mice show increased susceptibility to Pseudomonas aeruginosa keratitis. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4498–4508. [Google Scholar] [CrossRef]
- Wu, M.; McClellan, S.A.; Barrett, R.P.; Hazlett, L.D. Beta-defensin-2 promotes resistance against infection with P. aeruginosa. J. Immunol. 2009, 182, 1609–1616. [Google Scholar] [CrossRef]
- Wu, M.; McClellan, S.A.; Barrett, R.P.; Zhang, Y.; Hazlett, L.D. Beta-defensins 2 and 3 together promote resistance to Pseudomonas aeruginosa keratitis. J. Immunol. 2009, 183, 8054–8060. [Google Scholar] [CrossRef]
- Tummanapalli, S.S.; Willcox, M.D. Antimicrobial resistance of ocular microbes and the role of antimicrobial peptides. Clin. Exp. Optom. 2021, 104, 295–307. [Google Scholar] [CrossRef]
- Tajer, L.; Paillart, J.C.; Dib, H.; Sabatier, J.M.; Fajloun, Z.; Abi Khattar, Z. Molecular mechanisms of bacterial resistance to antimicrobial peptides in the modern era: An updated review. Microorganisms 2024, 12, 1259. [Google Scholar] [CrossRef]
- Kalaiselvan, P.; Konda, N.; Pampi, N.; Vaddavalli, P.K.; Sharma, S.; Stapleton, F.; Kumar, N.; Willcox, M.D.P.; Dutta, D. Effect of antimicrobial contact lenses on corneal infiltrative events: A randomized clinical trial. Transl. Vis. Sci. Technol. 2021, 10, 32. [Google Scholar] [CrossRef]
- Sharma, S.; Devkota, M.D.; Pokhrel, B.M.; Banjara, M.R. Detection of blaNDM-1,mcr-1 and MexB in multidrug resistant Pseudomonas aeruginosa isolated from clinical specimens in a tertiary care hospital of Nepal. BMC Microbiol. 2023, 23, 153. [Google Scholar] [CrossRef]
- Nitz, F.; de Melo, B.O.; da Silva, L.C.N.; de Souza Monteiro, A.; Marques, S.G.; Monteiro-Neto, V.; de Jesus Gomes Turri, R.; Junior, A.D.S.; Conceicao, P.C.R.; Magalhaes, H.J.C.; et al. Molecular detection of drug-resistance genes of blaOXA-23-blaOXA-51 and mcr-1 in clinical isolates of Pseudomonas aeruginosa. Microorganisms 2021, 9, 786. [Google Scholar] [CrossRef]
- Snesrud, E.; Maybank, R.; Kwak, Y.I.; Jones, A.R.; Hinkle, M.K.; McGann, P. Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e00679-18. [Google Scholar] [CrossRef]
- Ghassani, A.; Triponney, P.; Bour, M.; Plesiat, P.; Jeannot, K.; MucoMicrobes study Group. Mutations in genes lpxL1, bamA, and pmrB impair the susceptibility of cystic fibrosis strains of Pseudomonas aeruginosa to murepavadin. Antimicrob. Agents Chemother. 2024, 68, e0129823. [Google Scholar] [CrossRef]
- De Sousa, T.; Wang, H.Y.; Lin, T.W.; Canica, M.; Ramos, M.J.N.; Santos, D.; Silva, C.; Saraiva, S.; Beyrouthy, R.; Bonnet, R.; et al. Mutational analysis of colistin-resistant Pseudomonas aeruginosa isolates: From genomic background to antibiotic resistance. Pathogens 2025, 14, 387. [Google Scholar] [CrossRef]
- Lin, J.; Xu, C.; Fang, R.; Cao, J.; Zhang, X.; Zhao, Y.; Dong, G.; Sun, Y.; Zhou, T. Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob. Agents Chemother. 2019, 63, e00556-19. [Google Scholar] [CrossRef]
- Miller, A.K.; Brannon, M.K.; Stevens, L.; Johansen, H.K.; Selgrade, S.E.; Miller, S.I.; Hoiby, N.; Moskowitz, S.M. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2011, 55, 5761–5769. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, S.; Zhao, N.; Nong, C.; He, Y.; Bao, R. Pseudomonas aeruginosa two-component system CprRS regulates HigBA expression and bacterial cytotoxicity in response to LL-37 stress. PLoS Pathog. 2024, 20, e1011946. [Google Scholar] [CrossRef]
- Madden, D.E.; Baird, T.; Bell, S.C.; McCarthy, K.L.; Price, E.P.; Sarovich, D.S. Keeping up with the pathogens: Improved antimicrobial resistance detection and prediction from Pseudomonas aeruginosa genomes. Genome Med. 2024, 16, 78. [Google Scholar] [CrossRef]
- Poole, K.; Lau, C.H.; Gilmour, C.; Hao, Y.; Lam, J.S. Polymyxin susceptibility in Pseudomonas aeruginosa linked to the MexXY-OprM multidrug efflux system. Antimicrob. Agents Chemother. 2015, 59, 7276–7289. [Google Scholar] [CrossRef]
- Daigle, D.M.; Cao, L.; Fraud, S.; Wilke, M.S.; Pacey, A.; Klinoski, R.; Strynadka, N.C.; Dean, C.R.; Poole, K. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5441–5451. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Tian, Z.X.; Yi, X.X.; Cho, A.; O’Gara, F.; Wang, Y.P. CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-Type isolates of Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1005932. [Google Scholar] [CrossRef]
- Aguilar-Rodea, P.; Zuniga, G.; Cerritos, R.; Rodriguez-Espino, B.A.; Gomez-Ramirez, U.; Nolasco-Romero, C.G.; Lopez-Marceliano, B.; Rodea, G.E.; Mendoza-Elizalde, S.; Reyes-Lopez, A.; et al. Nucleotide substitutions in the mexR, nalC and nalD regulator genes of the MexAB-OprM efflux pump are maintained in Pseudomonas aeruginosa genetic lineages. PLoS ONE 2022, 17, e0266742. [Google Scholar] [CrossRef]
- Takrami, S.R.; Ranji, N.; Hakimi, F. New mutations in ciprofloxacin resistant strains of Pseudomonas aeruginosa isolated from Guilan Province, Northern Iran. Mol. Genet. Microbiol. Virol. 2018, 32, 218–223. [Google Scholar] [CrossRef]
- Abeyrathne, P.D.; Daniels, C.; Poon, K.K.; Matewish, M.J.; Lam, J.S. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 2005, 187, 3002–3012. [Google Scholar] [CrossRef]
- Johnson, L.; Mulcahy, H.; Kanevets, U.; Shi, Y.; Lewenza, S. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 2012, 194, 813–826. [Google Scholar] [CrossRef]
- Yang, B.; Liu, C.; Pan, X.; Fu, W.; Fan, Z.; Jin, Y.; Bai, F.; Cheng, Z.; Wu, W. Identification of novel PhoP-PhoQ regulated genes that contribute to polymyxin B tolerance in Pseudomonas aeruginosa. Microorganisms 2021, 9, 344. [Google Scholar] [CrossRef]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.J.; Proctor, R.A.; Sahl, H.G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus Infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; Tinelli, M.; Grundmann, H.; Akkerboom, V.; Monaco, M.; Del Grosso, M.; Errico, G.; Pantosti, A.; Friedrich, A.W. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprF and vraS genes: A new insight Into daptomycin resistance. Front. Microbiol. 2018, 9, 2705. [Google Scholar] [CrossRef]
- Yang, S.J.; Mishra, N.N.; Kang, K.M.; Lee, G.Y.; Park, J.H.; Bayer, A.S. Impact of multiple single-nucleotide polymorphisms within mprF on daptomycin resistance in Staphylococcus aureus. Microb. Drug Resist. 2018, 24, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Mishra, N.N.; Rubio, A.; Bayer, A.S. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 2013, 57, 5658–5664. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.S.; Mishra, N.N.; Chen, L.; Kreiswirth, B.N.; Rubio, A.; Yang, S.J. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2015, 59, 4930–4937. [Google Scholar] [CrossRef]
- Jiang, X.; Han, M.; Tran, K.; Patil, N.A.; Ma, W.; Roberts, K.D.; Xiao, M.; Sommer, B.; Schreiber, F.; Wang, L.; et al. An intelligent strategy with all-atom molecular dynamics simulations for the design of lipopeptides against multidrug-resistant Pseudomonas aeruginosa. J. Med. Chem. 2022, 65, 10001–10013. [Google Scholar] [CrossRef]
- Khan, M.; Stapleton, F.; Summers, S.; Rice, S.A.; Willcox, M.D.P. Antibiotic resistance characteristics of Pseudomonas aeruginosa isolated from keratitis in Australia and India. Antibiotics 2020, 9, 600. [Google Scholar] [CrossRef]
- Khan, M. Phenotypic and Genotypic Resistance Characteristics of Pseudomonas aeruginosa Isolates. Ph.D. Thesis, University of New South Wale, Sydney, Australia, 2020. [Google Scholar]
- DiGiandomenico, A.; Keller, A.E.; Gao, C.; Rainey, G.J.; Warrener, P.; Camara, M.M.; Bonnell, J.; Fleming, R.; Bezabeh, B.; Dimasi, N.; et al. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci. Transl. Med. 2014, 6, 262ra155. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Activity of antimicrobial peptides and ciprofloxacin against Pseudomonas aeruginosa biofilms. Molecules 2020, 25, 3843. [Google Scholar] [CrossRef]
- Khan, M.; Summers, S.; Rice, S.A.; Stapleton, F.; Willcox, M.D.P.; Subedi, D. Acquired fluoroquinolone resistance genes in corneal isolates of Pseudomonas aeruginosa. Infect. Genet. Evol. 2020, 85, 104574. [Google Scholar] [CrossRef]
- Khan, M.; Ma, K.; Wan, I.; Willcox, M.D. Ciprofloxacin resistance and tolerance of Pseudomonas aeruginosa ocular isolates. Contact Lens Anterior Eye 2023, 46, 101819. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Stapleton, F.; Willcox, M.D.P. Susceptibility of contact lens-related Pseudomonas aeruginosa keratitis isolates to multipurpose disinfecting solutions, disinfectants, and antibiotics. Transl. Vis. Sci. Technol. 2020, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Subedi, D.; Vijay, A.K.; Kohli, G.S.; Rice, S.A.; Willcox, M. Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep. 2018, 8, 15668. [Google Scholar] [CrossRef]
- Klockgether, J.; Munder, A.; Neugebauer, J.; Davenport, C.F.; Stanke, F.; Larbig, K.D.; Heeb, S.; Schock, U.; Pohl, T.M.; Wiehlmann, L.; et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 2010, 192, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, I.; Said, D.G.; Nubile, M.; Mastropasqua, L.; Dua, H.S. Cathelicidin-derived synthetic peptide improves therapeutic potential of vancomycin against Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 2190. [Google Scholar] [CrossRef]
- Munoz-Escudero, D.; Breazeale, S.D.; Lee, M.; Guan, Z.; Raetz, C.R.H.; Sousa, M.C. Structure and function of ArnD. A deformylase essential for lipid A modification with 4-amino-4-deoxy-l-arabinose and polymyxin resistance. Biochemistry 2023, 62, 2970–2981. [Google Scholar] [CrossRef]
- Puja, H.; Bolard, A.; Nogues, A.; Plesiat, P.; Jeannot, K. The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob. Agents Chemother. 2020, 64, e02033-19. [Google Scholar] [CrossRef]
- Furtado, G.H.; d’Azevedo, P.A.; Santos, A.F.; Gales, A.C.; Pignatari, A.C.; Medeiros, E.A. Intravenous polymyxin B for the treatment of nosocomial pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2007, 30, 315–319. [Google Scholar] [CrossRef]
| Strains | Minimum Inhibitory Concentrations (µg/mL) | |||
|---|---|---|---|---|
| Polymyxin B | Colistin | Mel4 | LL37 | |
| PA01—resistant | 128 | 128 | 128 | 64 |
| PA01—sensitive | 0.5 | 0.5 | 0.5 | 0.5 |
| PA189 | 0.5 | 4 | 64 | 32 |
| PA122 | 1 | 1 | 32 | 64 |
| PA9 | 1 | 1 | 32 | 16 |
| PA179 | 1 | 4 | 32 | 8 |
| PA214 | 1 | 4 | 32 | 32 |
| PA33 | 2 | 2 | 16 | 32 |
| PA182 | 2 | 2 | 16 | 16 |
| PA223 | 2 | 2 | 32 | 32 |
| PA224 | 2 | 2 | 128 | 32 |
| PA212 | 2 | 8 | 256 | 32 |
| PA213 | 2 | 8 | 32 | 16 |
| PA206 | 2 | 16 | 16 | 16 |
| PA196 | 4 | 2 | 128 | 64 |
| PA126 | 4 | 4 | 16 | 16 |
| PA31 | 4 | 128 | 128 | 256 |
| PA193 | 8 | 4 | 32 | 16 |
| PA209 | 16 | 8 | 32 | 256 |
| PA124 | 16 | 16 | 32 | 128 |
| PA229 | 64 | 128 | 32 | 32 |
| PA225 | 64 | 128 | 64 | 128 |
| PA226 | 64 | 128 | 64 | 128 |
| 6206 | 64 | 128 | 128 | 512 |
| PA222 | 128 | 64 | 32 | 256 |
| PA198 | 128 | 64 | 128 | 128 |
| PA227 | 128 | 64 | 256 | 256 |
| PA221 | 128 | 128 | 128 | 64 |
| ATCC 19660 | 128 | 256 | 64 | 256 |
| PA55 | 128 | 256 | 128 | 64 |
| PA56 | 256 | 4 | 32 | 16 |
| PA65 | 256 | 8 | 64 | 32 |
| PA217 | 256 | 64 | 16 | 256 |
| PA216 | 256 | 128 | 64 | 256 |
| PA220 | 256 | 128 | 256 | 32 |
| PA54 | 256 | 256 | 32 | 16 |
| PA219 | 256 | 256 | 128 | 256 |
| PA83 | 256 | 256 | 256 | 32 |
| PA123 | 512 | 512 | 16 | 64 |
| Statistical Parameters | Polymyxin B vs. Colistin | Polymyxin B vs. Mel4 | Colistin vs. Mel4 | Polymyxin B vs. LL37 | Colistin vs. LL-37 | Mel4 vs. LL-37 |
|---|---|---|---|---|---|---|
| 0.747 | 0.296 | 0.389 | 0.441 | 0.506 | 0.419 | |
| 2 | 0.558 | 0.088 | 0.151 | 0.195 | 0.256 | 0.176 |
| p value | <0.001 | 0.063 | 0.013 | 0.005 | 0.001 | 0.007 |
| Strains | pmrA | pmrB | phoP | phoQ | cprR | cprS | parR | parS | colR | colS | arnA | arnB | arnC | arnD | arnE | arnF | arnT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA189 | L71R | + | + | + | + | E386D, L411M | + | H398R | + | + | C312S, S313G | V302A, E376D | + | + | + | L114F | V20A, I509V |
| PA33 | + | S2P, A4T, V15I, G68S, | + | + | + | T16S | M59I, L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | V302A, E376D | + | + | + | + | H151Y, A267S, L337Q, T443A, I509V |
| PA182 | L71R | + | + | + | + | E386D, L411M | + | H398R | + | + | C312S, S313G | V302A | + | + | + | V14M | A267S, R445H, I509V |
| PA223 | + | + | + | + | + | + | + | H398R | + | + | C312S, S313G | V302A, E376D | + | + | + | + | C7W, H151Y, M274I, T443A, I509V |
| PA224 | L71R | S2P, A4T | + | + | + | + | + | H398R | + | + | + | V302A | + | E25D, F58L, G208S | + | V14M, A125T | S257R, R502Q, I509V, R521H |
| PA206 | D61E, L71R | A4T, P369A, A427T | + | + | E183D | E111D, A175V, N211H, T329S, E386D | T135A, L153R | A115E, V304I, E343D, H398R, Y407H | + | H353R | T42I, P57A, I138V, S313G, I388V, T636A | V302A, E376D | E35G, I309V, T316A | E25D, F58L | A109V | V14M | C7W, G14A, V89A, A116T, L163F, T166S, A214V, A404G, T443A, R502Q, I509V, S517G |
| PA126 | V34L, L71R | + | + | + | + | + | + | H398R | + | + | C312S, S313G, I388V, L591M | K286E, V302A | + | + | + | + | V266I, T443A, I509V |
| PA31 | + | S2P, A4T, V15I, G68S, | + | + | + | T16S | M59I, L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | V302A, E376D | + | F58L | + | + | H151Y, A267V, L337Q, T443A, I509V |
| PA193 | D61E, L71R | A4T, P369A, A427T | + | + | E183D | E111D, A175V, N211H, T329S, E386D | T135A, L153R | A115E, V304I, E343D, H398R, Y407H | + | + | T42I, P57A, I138V, S313G, I388V, T636A | V302A, E376D | + | F58L | + | + | D154E, V290L |
| PA225 | D61E, L71R | A4T, P369A, A427T | + | + | E183D | E111D, A175V, N211H, T329S, E386D | T135A, L153R | A115E, V304I, E343D, H398R, Y407H | + | + | T42I, P57A, I138V, S313G, I388V, T636A | A175V, V302A | + | G206C | + | V14M, A125T | R445H, R502Q, I509V |
| 6206 | + | S2P, A4T, V6A, V15I, G68S, | + | + | + | T16S, E386D | L153R, S170N | H398R | + | G285S | C312S, S313G, I388V | K286E, V302A, E376D | L254F | + | R28H | V14M | H151Y, T166I, A267V, I509V |
| PA198 | D61E, L71R | A4T, P369A, A427T | + | + | E183D | E111D, A175V, N211H, D220E, T329S, E386D | T135A, L153R | A115E, V304I, E343D, H398R, Y407H | + | + | T42I, P57A, I138V, S313G, I388V, T636A | V302A, E376D | + | + | + | + | H151Y, A267S, L337Q, T443A, I509V |
| PA227 | T31I | + | + | + | + | + | + | H398R | + | + | A170T, C312S, S313G, I388V | A175V, V302A | + | G206C | + | V14M, A125T | R445H, R502Q, I509V |
| PA221 | + | S2P, A4T, V6A, V15I, G68S, | + | + | + | T16S, E386D | L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | V302A | + | + | R28H | V14M | H151Y, T166I, A267V, I509V |
| ATCC 19660 | L71R | S2P, A4T | + | + | + | T16S, A88V, D153N, V159I, E386D | L153R, S170N | H398R | + | A316T | I138V, S313G, I388V, V564I | G72S, K286E, V302A, E376D | + | E25D, F58L, V123A | A109V | V14M | C7W, A214V, A225V, L337Q, I509V |
| PA55 | + | H345Y | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| PA217 | + | S2P, A4T, V6A, V15I, G68S, | + | + | + | T16S, D386E | L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | Q23L, K286E, V302A, E376D | + | F58L | R28H | + | C7W, H151Y, L337Q, T443A, I509V |
| PA216 | L71R | + | + | + | + | + | + | H398R | + | + | C312S, S313G | A259T, V302A, A316V, R340H | + | + | + | + | G156R, A267S |
| PA220 | L71R | S2P, A4T, V6A, V15I, G68S, | + | + | + | T16S, E386D | L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | V302A | + | + | R28H | V14M | H151Y, T166I, A265V, G338E, I509V |
| PA219 | + | S2P, A4T, V6A, V15I, G68S, | + | + | + | T16S, E386D | L153R, S170N | H398R | + | + | F80Y, C312S, S313G, I388V | V302A, E376D | + | F58L | + | + | H151Y, A267S, L337Q, T443A, I509V |
| PA123 | L71R | S2P, A4T | + | + | + | + | I93T | H398R | + | + | C312S, S313G, Q661L | V302A, Q336L | + | + | + | + | G156R, A267S, R502Q, I509V |
| Strains | mipA | mipB | armR | mexR | mexA | mexB | oprM | mexX | mexY | cpxR | nalC | nalD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA189 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, A32-del | N171S, H195Y, G152_R153insGG, A157_S158insAA, E406K, T413N | + | E126V, V132A | + | T90I | + | K329Q, W358R | A254G, Q282R, T543A | + | G71E, S209R | + |
| PA33 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | D225E, P328L, I350V, S368R, K387R, E406K, I427V, R441S, D494G, T496A, S507P | T13I, S21T, Y32C | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A, G589A, Q840E | + | G71E, D79E, S209R | + |
| PA182 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, H82Q, N171S, H195Y, G152_R153insGG, A157_S158insAA, E406K, I427V, D494G, T496A, S507P, G525S | + | E126V | + | T90I | + | K329Q, L331V, W358R | T543A | + | G71E, S209R | + |
| PA223 | K2R, T3S, A4P, G14S, T20V, A21S, Y26D, L28I, L30P, G31S, S33-del | E40K, N171S, H195Y, H246Q, G255S, A258_S259insAA, I350V, K387R, E406K, P410S, I427V, D494G, T496A, S507P, G525S | + | + | + | T90I | + | K329Q, L331V, W358R | T543A | + | G71E, S209R | + |
| PA224 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, S33-del | N171S, H195Y, G152_R153insGG, A157_S158insAA, P343R, E406K | + | E126V | + | T90I | + | K329Q, L331V, W358R | T543A, Q840E | + | G71E, S209R, P210L | + |
| PA206 | no significant homology | N171S, H195Y, G250S, G58_L59insT, V263T, A341T, K387R, R401P, A402T, E406K, D412E, I427V, F440I, M457A, D494G, T496V, S507P, G550E, T551A, V567I, Q595H, P602H, W603X | T5A, S21T | A103G, E126V | + | T90I, N248K | + | L12P, A30T, V322L, K329Q, L331V, G344D, W358R | T543A, Q840E, G1035D, N1036T, Q1039R, I1040T | + | G71E, Q182K, Q208A, S209D, P210-, A211-, Q212-, G213- | + |
| PA126 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, A32-del | N171S, H195Y, P343R, R401del, E456K | + | + | K289R | T90I | + | K329Q, L331V, W358R | T543A | + | G71E | + |
| PA31 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, H195Y, D125E, P328L, I350V, S368R, K387R, E406K, I427V, G441S, D494G, T496V, S507P | T13I, S21T, Y32C | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A, G589A | + | G71E, E153Q, S209R | + |
| PA193 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, S33-del | D494G, T496A, S507P | T5A, S21T | A103G, E126V | + | T90I, N248K | + | K329Q, L331V, W358R | Q282R, T543A, V980I | + | G71E, Q182K, Q208A, S209D, P210-, A211-, Q212-, G213- | + |
| PA225 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32V, S33Q, N34-del | S8P, S368R, K287R, E406K, I427V, Y430H, V469M, D494Q, T496A, S507P | T5A, S21T | A103G, E126V | + | T90I, N248K | + | K329Q, W358R | T543A | + | G71E, Q182K, Q208A, S209D, P210-, A211-, Q212-, G213- | + |
| 6206 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, A28V, H195Y, D225E, I350V, K387R, E406K, I427V, V469M, D494G, T496A, S507P | S21T, Y32C | E126V | + | T90I | + | A30T, K329Q, L331V, W358R | I536V, T543A | S80A, L92R | G71E, E153Q, S209R | + |
| PA198 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, H195Y, D125E, P328L, I350V, S368R, K387R, E406K, I427V, G441S, D494G, T496A, S507P | + | A103G, E126V | + | T90I, N248K | + | A30T, K329Q, L331V, W358R | I536V, T543A, G589A, Q840E, N1036T | + | G71E, Q182K, Q208A, S209D, P210-, A211-, Q212-, G213- | + |
| PA227 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32V, S33Q, N34-del | S8P, S368R, K287R, E406K, I427V, Y430H, V469M, D494Q, T496A, S507P | + | + | + | T90I | + | K329Q, W358R | T543A | + | G71E, E153D, A186T | + |
| PA221 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, H195Y, D125E, I350V, K387R, E406K, I427V, V469M, D494G, T496A, S507P | S21T, Y32C | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A | + | G71E, E153Q, S209R | R38W |
| ATCC 19660 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32V, S33Q, N34-del | S8P, A28V, F47L, H95Y, D225E, Q228M, G255S, R288H, I350V, K387R, E406K, R423H, I427V, V469M, D494G, T496A, S507P, G524S | S21T, G23E, Y32C | E126V | + | T90I, I186V, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A, Q840E | + | G71E, A145V, S209R | + |
| PA55 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, S33-del | + | + | + | + | T90I | + | + | + | + | + | + |
| PA217 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, S33-del | S8P, A28V, D225E, A258_S259insAA, I350V, R356H, S368R, K387R, E406K, V469M, D494G, T496A, S507P | + | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | T543A | + | G71E, E153Q, S209R | R38W |
| PA216 | no significant homology | N180S, H195Y, P343R, R401del, E406K | + | + | + | T90I | + | K329Q, L331V, W358R | T543A | + | G71E, A186T | + |
| PA220 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, A28V, H195Y, D225E, I350V, K387R, E406K, I427V, V469M, D494G, T496A, S507P | S21T, Y32C | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A | + | G71E, E153Q, S209R | R38W |
| PA219 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, A32-del | S8P, H195Y, D225E, P328L, I350V, S368R, K387R, E406K, I427V, G441S, D494G, T496A, S507P | T13I, S21T, Y32C | E126V | + | T90I, S1041E, V1042A | + | A30T, K329Q, L331V, W358R | I536V, T543A, N1036T | + | G71E, E153Q, S209R | R38W |
| PA123 | K2R, T3S, A4P, G14S, T20V, Y26D, L28I, L30P, G31S, A32-del | N171S, H195Y, H246Q, A258_S259insAA | + | + | + | T90I | + | K329Q, L331V, W358R | T543A, Q840E | + | G71E, S209R | + |
| Strains | oprH | papP | mpl | slyB | ppgS | ppgH | speD2 | speE2 | waaL | PA5005 | rsmA | mprF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA189 | + | M18L, S56G, M62L | M297V | + | D553N | A40D, R110H, A165S, K209Q, Q230K, P388S | + | + | S8T, T93A, R147Q | H399N | + | S81G, A748V |
| PA33 | + | M18L, M62L | + | + | G81S, H187R, D553N, Q792K | A40D, A50V, K209Q, P388S | + | T271A | S8T, T93A, R147Q, L179F, G330A, I398T | + | + | R187H, N554D, N670S, A731T, A748V, D772E, K793Q |
| PA182 | + | M18L, S56G, M62L | + | + | G81S, H187R, L199S, D553N, G587S, A731T, E772D | A40D, A165S, K209Q, P388S | + | + | S8T, R147Q | + | + | G587S, A731T, A748V, K793Q |
| PA223 | + | M18L, S56G, M62L | + | + | G81S, H187R, L199S, D553N, G587S | A40D, G95S, A165S, K209Q, Q230K | + | + | S8T, Y58F, L61I, R64Q, G71R, F75L, F78I, S83A, S90L, T98A, L102F, A109V, F112L, A115V, A116G, E121Q, L124E, K127R, T128N, A129I, I139F, S140A, A143V, L145V, L146V, R147H, Y149H, W150L, D151Q, A152T, N153H, P154_L156insAW, L155M, T158S, A178V, L179V, A182V, P189H, I190L, I197L, L201I, G203C, G204C, I207L, A208S, V216I, G217A, A221C, M223G, V226L, L227V, D230N, R231Q, A234T, A237V, L238I, A239G, L242A, A243L, A245M, L246I, L247V, G248A, L251F, Y252N, V255L, I256V, A261V, A269S, D270E, A271S, S276G, V292K, S294A, I316V, V322S, G330V, S332A, K337R, S338D, A340M, A354S, L363M, P364S, M377L, I385V, Q387K | H399N | + | G587S, A748V, D772E, K793Q |
| PA224 | + | M18L, S56G, M62L | M297V | + | G81S, H187R, L199S, D553N, G587S, A731T, E772D | A40D, A165S, K209Q, Q230K, P388S | + | + | S8T, T93A, R147Q | H399N | + | G587S, A731T, A748V, K793Q |
| PA206 | + | M18L, S56G, M62L, S74G | M297V | + | D70E, V103I, H187R, V192I, A545V, G587S, A731E | A40D, A109T, S190R, S203T, K209Q, Q230K | + | T271A | S8T, Y58F, L61I, R64Q, G71R, F75L, F78I, S83A, S90L, T98A, L102F, A109V, F112L, A115V, A116G, E121Q, L124E, K127R, T128N, A129I, I139F, S140A, A143V, L145V, L146V, R147H, Y149H, W150L, D151Q, A152T, N153H, P154_L156insAW, L155M, T158S, A178V, L179V, A182V, P189H, I190L, I197L, L201I, G203C, G204C, I207L, A208S, V216I, G217A, A221C, M223G, V226L, L227V, A228V, D230N, R231Q, A234T, A237V, L238I, A239G, L242A, A243L, A245M, L246I, L247A, G248A, L251F, Y252N, V255L, I256V, A261V, A269S, D270E, A271S, S276G, V292K, S294A, I316V, V322S, G330V, S332A, K337R, S338D, A340M, A354S, L363M, P364S, M377L, I385V, Q387K | Q396K, H399N, R403Q | + | D71E, S82G, V104I, V93I, S100L, V546A, N554D, G587S, A731E, A748V, D772E, K793Q |
| PA126 | + | M18L, S56G, M62L | + | + | G81S, D175N, D553N, G587S | A40D, G95S, A165S, K209Q | + | + | S8T, T93A, R147Q | H399N | + | D175N, R187H, S199L, G587S, A748V, D772E, K793Q |
| PA31 | + | M18L, M62L | D411A | + | G81S, L199S, N670S, A731T | A40D, A50V, K209Q, P388S | + | V217I, T271A | S8T, T93A, R147Q, L179F, G330A, I398T | H399N | + | R187H, N554D, N670S, A731T, A748V, D772E, K793Q |
| PA193 | + | S56G, M62L | M297V | + | H65R, H187R, L199S, G304S, D553N, E772D, Q792K | A40D, R110H, A165S, K209Q, Q230K, P388S | + | + | S8T, R147Q, L281F | H399N | + | H65R, S81G, G304S, A758V |
| PA225 | + | S56G, M62L | M297V | + | H187R, L199S, D553N, I740M, E772D | A40D, R110H, A165S, K209Q, Q230K, P388S | + | + | S8T, T93A, R147Q, L179F, S276G, G330A | H399N | + | S81G, I740M, A748V, K793Q |
| 6206 | + | M18L, M62L | + | + | P14L, G81S, L199S, D553N, G587S | A40D, R110H, A165S, K209Q, P231A, P388S | + | + | S8T, T93A, A129V, R147Q, L179F, I256F, S276G, G330A | D23E, H399D | + | P13L, R187H, G587S, A748V, D772E, K793Q |
| PA198 | + | M18L, M62L | + | + | G81S, L199S, N670S, A731T | A40D, A50V, K209Q, P388S | + | V217I, T271A | S8T, T93A, R147Q, L179F, G330A, I398T | H399N | + | R187H, N554D, N670S, A731T, A748V, D772E, K793Q |
| PA227 | + | M18L, S56G, M62L | M297V, A303V | + | H187R, L199S, D553N, I740M, E772D | A40D, R110H, A165S, K209Q, Q230K, P388S | + | + | S8T, R147Q, L281F, S276G, G330A | H399N | + | S81G, I740M, A748V, K793Q |
| PA221 | + | M18L, M62L | + | + | G81S, L199S, D553N, G587S | A40D, R110H, A165S, K209Q, P231A, P388S | + | + | S8T, T93A, A129V, R147Q, L179F, I256F, S276G, G330A | D23E, H399D | + | R187H, G587S, A748V, D772E, K793Q |
| ATCC 19660 | + | + | + | + | E616G, A731Y | K209Q | + | + | S8T, T93A, R147Q, L179F, G330A | H399N | + | S81G, R187H, S199L, N553D, E616G, A731T, A748V, D772E, K793Q |
| PA55 | + | M18L, S56G, M62L | + | + | G81S, H187R, L199S, D553N, G587S, E772D | A40D, A165S, K209Q, P388S | + | + | + | + | + | G587S, A748V, K793Q |
| PA217 | + | M18L, M62L | + | + | L199S, A270V | A40D, K209Q, P388S | + | C161S, A165T, V217T, S244A, T271A, E277D, S244A, T271A, E277D, P326A, E235G | S8T, R16H, Y58F, L61I, R64Q, G71R, F75L, F78I, S83A, S90L, T98A, L102F, A109V, F112L, A115V, A116G, E121Q, L124E, K127R, T128N, A129I, I139F, S140A, A143V, L145V, L146V, R147H, Y149H, W150L, D151Q, A152T, N153H, P154_L156insAW, L155M, T158S, A178V, L179V, A182V, P189H, I190L, I197L, L201I, G203C, G204C, I207L, A208S, V216I, G217A, A221C, M223G, V226L, L227V, D230N, R231Q, A234T, A237V, L238I, A239G, L242A, A243L, A245M, L246I, L247V, G248A, L251F, Y252N, V255L, I256V, A261V, A269S, D270E, A271S, S276G, V292K, S294A, I316V, V322S, G330I, S332A, K337R, S338D, A340M, A354S, L363M, P364S, M377L, I385V, Q387K | H399N, N400D | + | S81G, R187H, A270V, N553D, A748V, D772E, K793Q |
| PA216 | + | M18L, S56G, M62L | M297V, A303V | + | A12T, G81S, H187R, L199S, L243M | A40D, K209Q | + | + | S8T, Y58F, L61I, G71R, F75L, F78I, S83A, S90L, T98A, L102F, A109V, F112L, A116G, E121Q, L124E, K127R, T128A, A129L, V138T, I139L, S140A, A143V, R147H, Y148F, Y149H, W150I, D151Q, A152S, N153P, P154_L156insAW, L155M, T158S, A178V, A182V, P189H, I190A, I197L, L201I, G203C, G204C, I207L, V216I, T219A, A221C, L222M, M223A, V226L, A234T, A237V, L238I, A239G, L240I, A241V, L242V, A245L, L247V, G248V, L251F, L252V, Y253N, V255L, T257I, A261V, A269S, D270E, A271S, S276G, V292K, S294A, I316V, V322S, G330V, S332A, K337R, S338D, A340M, A354S, L363M, P364S, M377L, I385V, Q387K | H399N | + | A12T, L242M, N553D, A731T, A748V, D772E, K793Q |
| PA220 | + | M18L, M62L | + | + | G81S, L199S, D553N, G587S | A40D, R110H, A165S, K209Q, P231A, P388S | + | T271A | S8T, T93A, A129V, R147Q, L179F, I256F, S276G, G330A | D23E, H399D | + | R188H, G587S, D772E, K793Q |
| PA219 | + | M18L, M62L | + | + | G81S, L199S, N670S, A731T | A40D, A50V, K209Q, P388S | + | V217I, T271A | S8T, T93A, R147Q, L179F, G330A, I398T | H399N | + | R188H, N553D, N670S, A731T, A748V, D772E, K793Q |
| PA123 | + | M18L, S56G, M62L | M297V, V358I | + | G81S, H187R, L199S, D553N, G587S, E772D | A40D, A165S, K209Q, P388S | + | A3V | S8T, Y58F, L61I, R64Q, G71R, F75L, F78I, S83A, S90L, T98A, L102F, A109V, A110G, F112L, A115V, A116G, E121Q, L124E, K127R, T128N, A129I, I139F, S140A, A143V, L145V, L146V, R147H, Y149H, W150L, D151Q, A152T, N153H, P154_L156insAW, L155M, T158S, A178V, L179V, A182V, P189H, I190L, I197L, L201I, G203C, G204C, I207L, A208S, V216I, G217A, A221C, M223G, V226L, L227V, D230N, R231Q, A234T, A237V, L238I, A239G, L242A, A243L, A245M, L246I, L247V, G248A, L251F, Y252N, V255L, I256V, A261V, A269S, D270E, A271S, S276G, V292K, S294A, I316V, V322S, G330V, S332A, K337R, S338D, A340M, A354S, L363M, P364S, M377L, I385V, Q387K | H399N | + | G587S, A748V, K793Q |
| Strain Number | Infection Type, Site of Isolation, Country and Date of Isolation | Known Antibiotic Resistance Characteristics | Biosample Number (NCBI) |
|---|---|---|---|
| PA01— resistant | Mutant derived from PAO1-sensitive [57] | Chloramphenicol, Colistin [57] | Not available |
| PA01— sensitive | Wound, Unknown, Australia, 1954 | Chloramphenicol [57,66] | SAMN02603714 |
| PA189 | Keratitis, Eye, India, 2017 | CIP S, LEV S, GEN S, TOB R, PIP S, IMI R, CFT S [63] | SAMN13340385 |
| PA122 | Keratitis, Eye, Australia, 2006 | Not known | Not available |
| PA9 | Keratitis, Eye, Australia, 1994 | Not known | Not available |
| PA179 | Keratitis, Eye, Australia, 2006 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI S, CFT S [64] | Not available |
| PA214 | Keratitis, Eye, India, 2017 | Not known | Not available |
| PA33 | Keratitis, Eye, India, 1998 | CIP R, LEV R, GEN R, TOB R, PIP R, IMI R, CFF R [63] | SAMN08435058 |
| PA182 | Keratitis, Eye, Australia, 2006 | CIP S, LEV S, GEN S, TOB S, PIP S, IMI R, CTF S [63] | SAMN13340383 |
| PA223 | Keratitis, Eye, Australia, 2018 | CIP R, LEV S, GEN S, TOB S, PIP R, IMI S, CFT I [63] | SAMN16123412 |
| PA224 | Keratitis, Eye, Australia, 2018 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI R, CFT I [63] | SAMN16123413 |
| PA212 | Keratitis, Eye, India, 2017 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI R, CFT S [58] | Not available |
| PA213 | Keratitis, Eye, India, 2017 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI I, CFT I [58] | Not available |
| PA206 | Keratitis, Eye, India, 2017 | CIP I, LEV S, GEN S, TOB S, PIP S, IMI S, CFT S [59] | SAMN13340389 |
| PA196 | Keratitis, Eye, India, 2017 | CIP S, LEV S, GEN S, TOB S, PIP S, IMI R, CFT S [59] | Not available |
| PA126 | Keratitis, Eye, Australia, 2006 | CIP S, LEV S, GEN S, TOB S, PIP S, IMI R, CFT R [64] | SAMN13340377 |
| PA31 | Keratitis, Eye, India, 1998 | CIP R, LEV R, MOX R, GEN R, CFT I, CEFE I, IMI I, TIC I, AZT I [65] | SAMN08435056 |
| PA193 | Keratitis, Eye, India, 2017 | CIP S, LEV S, GEN S, TOB S, PIP S, IMI S, CFT S [58] | SAMN13340386 |
| PA209 | Keratitis, Eye, India, 2017 | CIP I, LEV I, GEN S, TOB S, PIP R, IMI S, CFT S [63] | Not available |
| PA124 | Keratitis, Eye, Australia, 2006 | CIP I, LEV S, GEN S, TOB S, PIP S, IMI R, CFT S [63] | Not available |
| PA229 | Keratitis, Eye, Australia, 2018 | CIP S, LEV S, GEN S, TOB S, PIP S, IMI R, CFT S [63] | Not available |
| PA225 | Keratitis, Eye, Australia, 2018 | CIP R, LEV R, GEN S, TOB S, PIP S, IMI R, CEF S [63] | SAMN16123414 |
| PA226 | Keratitis, Eye, Australia, 2018 | CIP I, LEV I, GEN S, TOB S, PIP R, IMI R, CFT S [63] | Not available |
| 6206 | Keratitis, Eye, USA, 1995 | CIP S, TOB S [60,61] | SAMN12437401 |
| PA222 | Keratitis, Eye, Australia, 2017 | CIP S, LEV I, GEN S, TOB S, PIP S, IMI R, CFT R [63] | Not available |
| PA198 | Keratitis, Eye, India, 2017 | CIP R, LEV R, MOX R [62] | SAMN13340387 |
| PA227 | Keratitis, Eye, Australia, 2018 | CIP R, LEV R, GEN S, TOB S, PIP S, IMI R, CFT I [63] | SAMN16123415 |
| PA221 | Keratitis, Eye, India, 2017 | CIP R, LEV R, GEN R, TOB R, PIP R, IMI R, CFT R [63] | SAMN13340395 |
| ATCC 19660 | Unknown, 1965 | CIP I, GEN S [61,67] | SAMN01918025 |
| PA55 | Cystic Fibrosis, Sputum, Australia, 2003 | CIP S, LEV S, MOX S, CFT R, CEFE R, IMI R, TIC R, AZT S [65] | SAMN08435068 |
| PA56 | Cystic Fibrosis, Sputum, Australia, 2003 | Not known | Not available |
| PA65 | Cystic Fibrosis, Sputum, Australia, 2003 | Not known | Not available |
| PA217 | Keratitis, Eye, India, 2017 | CIP R, LEV R, GEN S, TOB S, PIP R, IMI R, CFF R [63] | SAMN13340391 |
| PA216 | Keratitis, Eye, India, 2017 | CIP R, LEV R, GEN S, TOB S, PIP R, IMI R, CFT R [63] | SAMN13340390 |
| PA220 | Keratitis, Eye, India, 2017 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI R, CFT R [63] | SAMN13340394 |
| PA54 | Keratitis, Eye, Australia, 2003 | Not known | Not available |
| PA219 | Keratitis, Eye, India, 2017 | CIP R, LEV S, GEN S, TOB S, PIP S, IMI R, CFT I [63] | SAMN13340393 |
| PA83 | Cystic Fibrosis, Sputum, Australia, 2003 | Not known | Not available |
| PA123 | Keratitis, Eye, Australia, 2006 | CIP I, LEV I, GEN S, TOB S, PIP S, IMI I, CFT I [63] | SAMN13340376 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damtie, M.A.; Vijay, A.K.; Willcox, M.D.P. Detection of Genes Associated with Polymyxin and Antimicrobial Peptide Resistance in Isolates of Pseudomonas aeruginosa. Int. J. Mol. Sci. 2025, 26, 10499. https://doi.org/10.3390/ijms262110499
Damtie MA, Vijay AK, Willcox MDP. Detection of Genes Associated with Polymyxin and Antimicrobial Peptide Resistance in Isolates of Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2025; 26(21):10499. https://doi.org/10.3390/ijms262110499
Chicago/Turabian StyleDamtie, Meseret Alem, Ajay Kumar Vijay, and Mark Duncan Perry Willcox. 2025. "Detection of Genes Associated with Polymyxin and Antimicrobial Peptide Resistance in Isolates of Pseudomonas aeruginosa" International Journal of Molecular Sciences 26, no. 21: 10499. https://doi.org/10.3390/ijms262110499
APA StyleDamtie, M. A., Vijay, A. K., & Willcox, M. D. P. (2025). Detection of Genes Associated with Polymyxin and Antimicrobial Peptide Resistance in Isolates of Pseudomonas aeruginosa. International Journal of Molecular Sciences, 26(21), 10499. https://doi.org/10.3390/ijms262110499







