CD5 Expression in CTCL and Its Implications for Anti-CD5 CAR T-Cell Therapy
Abstract
1. Introduction
2. Results
2.1. Written Results
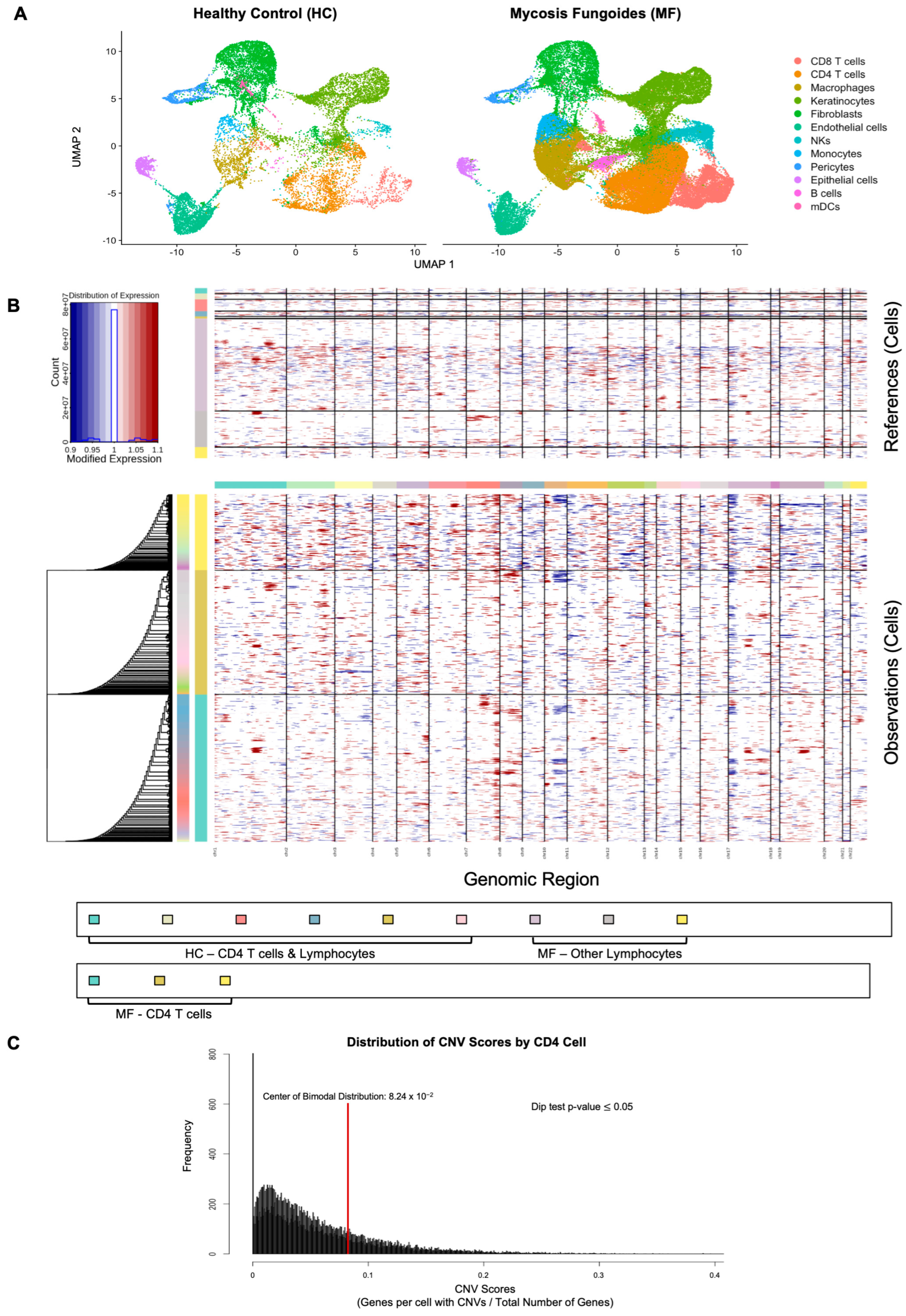
2.1.1. CD4 T Cells Are Prominent in MF Skin Lesions
2.1.2. Malignant CD4 Cell Identification by Copy Number Variants
2.1.3. CD5 Expression Across All Cell Clusters
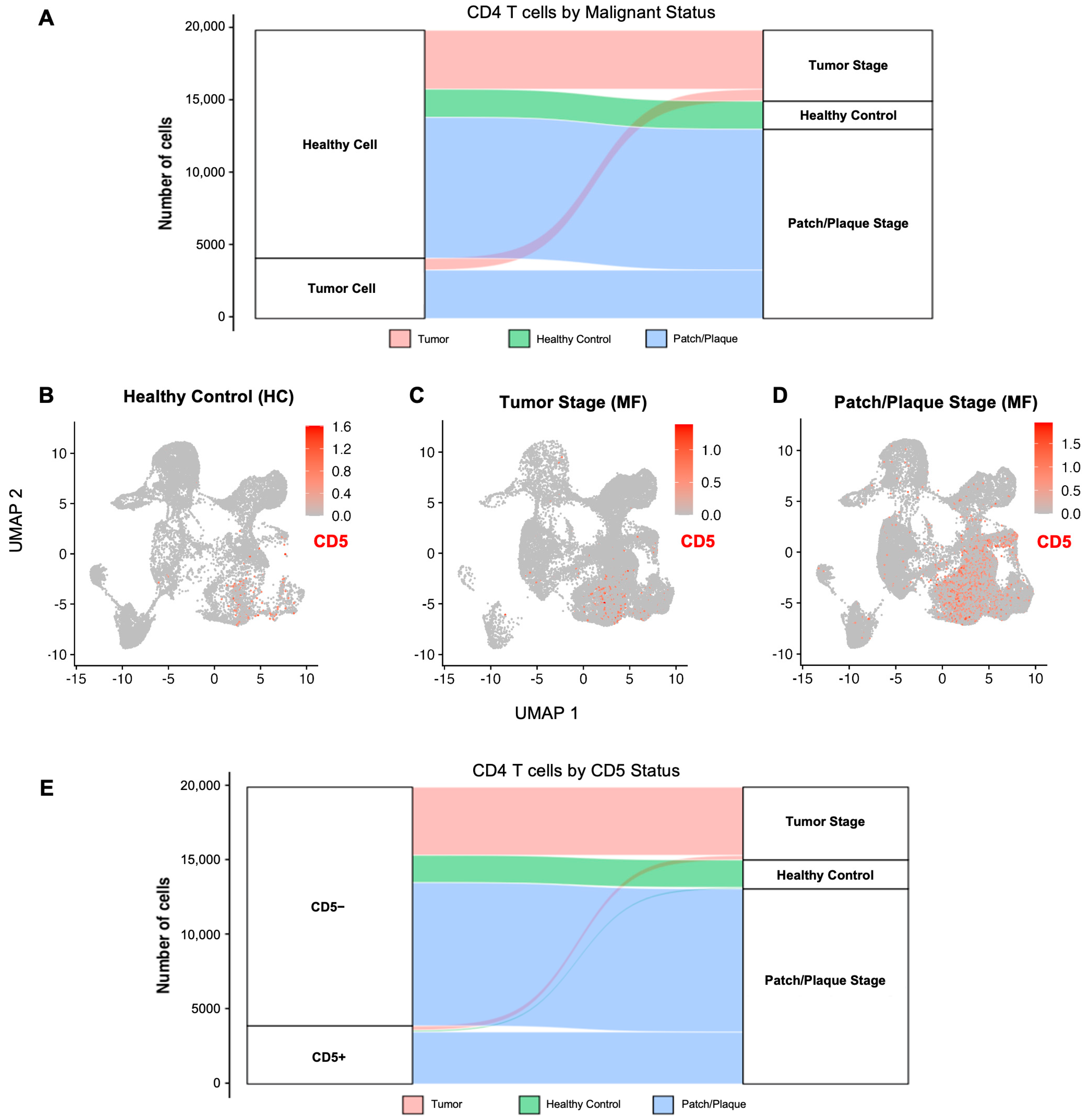
2.1.4. CD5 Expression in CD4 T Cells from MF Patients Versus Healthy Controls
2.1.5. CD5 Expression in Malignant CD4 T Cells from Patch/Plaque Versus Tumor-Stage MF Lesions
2.1.6. CD5 Expression in Other Lymphocytes in MF
3. Discussion
4. Materials and Methods
4.1. Sample Selection and Data Acquisition
4.2. Computational Environment
4.3. Preprocessing and Quality Control
4.4. Normalization and Feature Selection
4.5. Data Integration and Clustering
4.6. Cell Type Annotation
4.7. Estimating and Inferring Malignant CD4 T Cells
4.8. Differential Gene Expression Analysis
4.9. Visualization
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CTCL | Cutaneous T-cell lymphoma |
| MF | Mycosis Fungoides |
| CAR | Chimeric Antigen Receptor |
| scRNA-seq | Single-cell RNA sequencing |
| UMAP | Uniform Manifold Approximation and Projection |
| UMI | Unique Molecular Identifier |
| GEO | Gene Expression Omnibus |
| IHC | Immunohistochemistry |
| CD | Cluster of Differentiation |
| mDC | Myeloid Dendritic Cell |
| PCA | Principal Component Analysis |
| CCA | Canonical Correlation Analysis |
| HC | Healthy Control |
| RNA | Ribonucleic Acid |
| R | Statistical computing language R |
References
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019, 133, 1703–1714, Erratum in Blood 2019, 134, 1112. [Google Scholar] [CrossRef] [PubMed]
- Pulitzer, M. Cutaneous T-cell lymphoma. Clin. Lab. Med. 2017, 37, 527–546. [Google Scholar] [CrossRef] [PubMed]
- Jawed, S.I.; Myskowski, P.L.; Horwitz, S.; Moskowitz, A.; Querfeld, C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): Part I. Diagnosis: Clinical and histopathologic features and new molecular and biologic markers. J. Am. Acad. Dermatol. 2014, 70, 205.e1–205.e16, quiz 221–222. [Google Scholar] [CrossRef] [PubMed]
- Tabbekh, M.; Mokrani-Hammani, B.; Bismuth, G.; Mami-Chouaib, F. T-cell modulatory properties of CD5 and its role in antitumor immune responses. Oncoimmunology 2013, 2, e22841. [Google Scholar] [CrossRef] [PubMed]
- Soldevila, G.; Raman, C.; Lozano, F. The immunomodulatory role of CD5 on lymphocyte receptor in health and disease. Curr. Opin. Immunol. 2011, 23, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Elghawy, O.; Cao, M.; Xu, J.; Landsburg, D.J.; Svoboda, J.; Nasta, S.D.; Chong, E.A.; Schuster, S.J.; Thomas, C.J.; Carter, J.S.; et al. Prevalence and prognostication of CD5+ mature T-cell lymphomas. Cancers 2024, 16, 3430. [Google Scholar] [CrossRef] [PubMed]
- Beygi, S.; Duran, G.E.; Fernandez-Pol, S.; Rook, A.H.; Kim, Y.H.; Khodadoust, M.S. Resistance to mogamulizumab is associated with loss of CCR4 in cutaneous T-cell lymphoma. Blood 2022, 139, 3732–3736. [Google Scholar] [CrossRef] [PubMed]
- To, V.; Evtimov, V.J.; Jenkin, G.; Pupovac, A.; Trounson, A.O.; Boyd, R.L. CAR-T cell development for cutaneous T cell lymphoma: Current limitations and potential treatment strategies. Front. Immunol. 2022, 13, 968395. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.C.; Rouce, R.H.; Mamonkin, M. Antitumor efficacy and safety of unedited autologous CD5.CAR T cells in relapsed/refractory mature T-cell lymphomas. Blood 2024, 143, 1231–1241, Erratum in Blood 2024, 144, 1349. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.T.; Akilov, O.E. Exhausted Markers in Cutaneous T-Cell Lymphoma: The Face that Launched a Thousand Ships. J. Investig. Dermatol. 2022, 142 Pt A, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tan, Y.; Shan, L.; Seery, S.; Deng, B.; Ling, Z.; Xu, J.; Duan, J.; Wang, Z.; Wang, K.; et al. Allogeneic CD5-specific CAR-T therapy for relapsed/refractory T-ALL: A phase 1 trial. Nat. Med. 2025, 31, 126–136, Erratum in Nat. Med. 2025, 31, 352. [Google Scholar] [CrossRef] [PubMed]
- Gaydosik, A.M.; Tabib, T.; Geskin, L.J.; Bayan, C.A.; Conway, J.F.; Lafyatis, R.; Fuschiotti, P. Single-cell lymphocyte heterogeneity in advanced cutaneous T-cell lymphoma skin tumors. Clin. Cancer Res. 2019, 25, 4443–4454. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Alkon, N.; Rindler, K.; Rojahn, T.B.; Shaw, L.E.; Porkert, S.; Weninger, W.; Trautinger, F.; Stingl, G.; Tschandl, P.; et al. Single-cell RNA sequencing profiling in a patient with discordant primary cutaneous B-cell and T-cell lymphoma reveals micromilieu-driven immune skewing. Br. J. Dermatol. 2021, 185, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Rindler, K.; Bauer, W.M.; Jonak, C.; Wielscher, M.; Shaw, L.E.; Rojahn, T.B.; Thaler, F.M.; Porkert, S.; Simonitsch-Klupp, I.; Weninger, W.; et al. Single-cell RNA sequencing reveals tissue compartment-specific plasticity of Mycosis Fungoides tumor cells. Front. Immunol. 2021, 12, 666935. [Google Scholar] [CrossRef] [PubMed]
- Rindler, K.; Jonak, C.; Alkon, N.; Thaler, F.M.; Kurz, H.; Shaw, L.E.; Stingl, G.; Weninger, W.; Halbritter, F.; Bauer, W.M.; et al. Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma. Mol. Cancer 2021, 20, 124. [Google Scholar] [CrossRef] [PubMed]
- Chennareddy, S.; Rindler, K.; Ruggiero, J.R.; Alkon, N.; Cohenour, E.R.; Tran, S.; Weninger, W.; Griss, J.; Jonak, C.; Brunner, P.M. Single-cell RNA sequencing comparison of CD4+, CD8+ and T-cell receptor γδ+ cutaneous T-cell lymphomas reveals subset-specific molecular phenotypes. Br. J. Dermatol. 2025, 192, 269–282, Erratum in Br. J. Dermatol. 2025, 192, e2. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Satija, R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 2022, 23, 27. [Google Scholar] [CrossRef] [PubMed]
| Cases of CTCL Used for scRNA-Seq | ||||
|---|---|---|---|---|
| Case No. | GEO ID | Lesion | Clinical Stage | Pathological Stage |
| 1 | GSM5280111 | Patch/Plaque | IB | T2bN0M0B0 |
| 2 | GSM5047045 | Patch/Plaque | IVB | T3N3M1B1 |
| 3 | GSM5261812 | Patch/Plaque | IIB | T2bN0M0B0 |
| 4 | GSM5534588 | Patch/Plaque | IA | T1bN0M0B0 |
| 5 | GSM5534589 | Patch/Plaque | IA | T1aN0M0B0 |
| 6 | GSM8255139 | Healthy Control | - | - |
| 7 | GSM8255140 | Healthy Control | - | - |
| 8 | GSM8255141 | Healthy Control | - | - |
| 9 | GSM8255142 | Healthy Control | - | - |
| 10 | GSM8255143 | Tumor | IVA | TNM Unknown, B2 |
| 11 | GSM8255144 | Tumor | IVA | TNM Unknown, B2 |
| 12 | GSM8255145 | Tumor | IIB | TNM Unknown, B0 |
| 13 | GSM3679033 | Tumor | IVA | T4NxB2M0 |
| 14 | GSM3679034 | Tumor | IIB | T3N0B0M0 |
| 15 | GSM3679035 | Tumor | IIB | T3NxB0M0 |
| 16 | GSM3679036 | Tumor | IIB | T3N0B0M0 |
| 17 | GSM3679037 | Tumor | IVA | T4NxM0B0 |
| Canonical Markers for scRNA-Seq | ||
|---|---|---|
| Cell Type | Marker | No. of Cells |
| B cells | MS4A1 | 977 |
| Endothelial cells | PECAM1 | 5447 |
| Epithelial cels | KRT18 | 1596 |
| Fibroblasts | COL1A1 | 10,930 |
| Keratinocytes | KRT1, KRT5, KRT14 | 17,851 |
| NKs | GNLY | 3224 |
| Macrophages | CD16, LYZ | 7887 |
| Monocytes | CD14, LYZ | 2421 |
| Pericytes | RGS5 | 1805 |
| CD4 T cells | CD3D, IL7R, CCR7 | 19,909 |
| CD8 T cells | CD8A | 8289 |
| mDCs | CD83 | 788 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wardeh, L.; Williams, M.; Prestwood, C.; Wolner, Z.; Nikbakht, N. CD5 Expression in CTCL and Its Implications for Anti-CD5 CAR T-Cell Therapy. Int. J. Mol. Sci. 2025, 26, 10411. https://doi.org/10.3390/ijms262110411
Wardeh L, Williams M, Prestwood C, Wolner Z, Nikbakht N. CD5 Expression in CTCL and Its Implications for Anti-CD5 CAR T-Cell Therapy. International Journal of Molecular Sciences. 2025; 26(21):10411. https://doi.org/10.3390/ijms262110411
Chicago/Turabian StyleWardeh, Leena, Madeline Williams, Courtney Prestwood, Zachary Wolner, and Neda Nikbakht. 2025. "CD5 Expression in CTCL and Its Implications for Anti-CD5 CAR T-Cell Therapy" International Journal of Molecular Sciences 26, no. 21: 10411. https://doi.org/10.3390/ijms262110411
APA StyleWardeh, L., Williams, M., Prestwood, C., Wolner, Z., & Nikbakht, N. (2025). CD5 Expression in CTCL and Its Implications for Anti-CD5 CAR T-Cell Therapy. International Journal of Molecular Sciences, 26(21), 10411. https://doi.org/10.3390/ijms262110411





