Moonlighting Proteins: Some Hypotheses on the Structural Origin of Their Multifunctionality
Abstract
1. Introduction
1.1. Structural and Regulatory Mechanisms of Moonlighting
1.2. Evolutionary Origins: Key Unanswered Questions
2. Results and Discussion
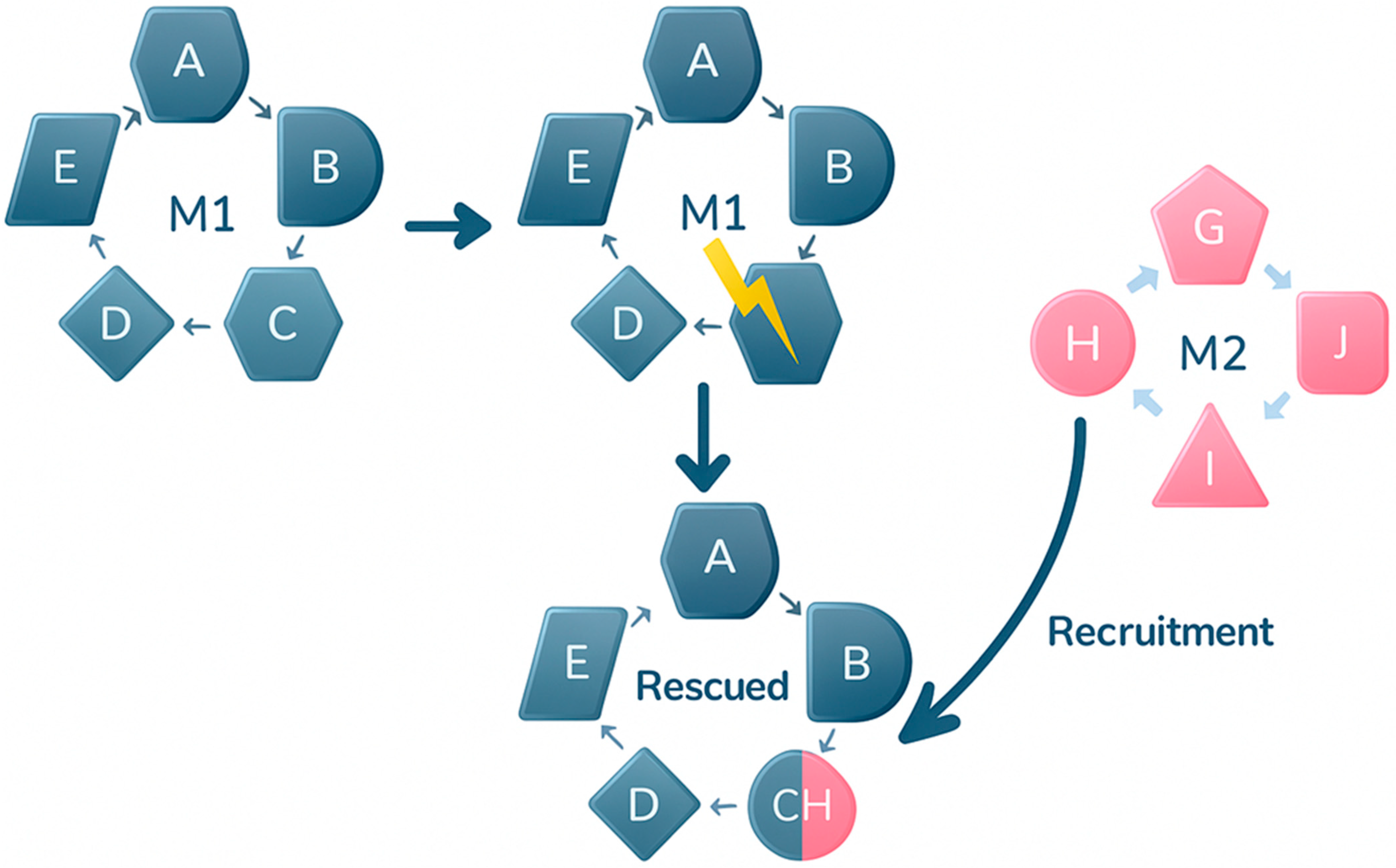
2.1. Mechanistic Hypotheses for Moonlighting Acquisition
2.2. Strategies for Acquiring Moonlighting Functions: Exploiting Intrinsic Properties
2.3. Redox-Sensitive Functional Switching: Exploiting Catalytic Residues
2.4. Oligomerization as a Conformational Switch
2.5. Fold-Switching Proteins: Subtle Conformational Changes Beyond Oligomerization
2.6. Why Is Not Moonlighting More Widespread?
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NOGD | Non-Orthologous Gene Displacement |
| NHIE | Non-Homologous Isofunctional Enzymes |
| FSP | Fold-Switching Proteins |
| IDR | Intrinsically disordered regions |
| PTM | Postraductional modifications |
| Prx | Peroxiredoxins |
| ROS | Reactive oxygen species |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
References
- Huberts, D.H.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta 2010, 1803, 520–525. [Google Scholar] [CrossRef]
- Copley, S.D. Moonlighting is mainstream: Paradigm adjustment required. Bioessays 2012, 34, 578–588. [Google Scholar] [CrossRef]
- Jeffery, C.J. An introduction to protein moonlighting. Biochem. Soc. Trans. 2014, 42, 1679–1683. [Google Scholar] [CrossRef]
- Singh, N.; Bhalla, N. Moonlighting Proteins. Annu. Rev. Genet. 2020, 54, 265–285. [Google Scholar] [CrossRef]
- Hernandez, S.; Ferragut, G.; Amela, I.; Perez-Pons, J.; Pinol, J.; Mozo-Villarias, A.; Cedano, J.; Querol, E. MultitaskProtDB: A database of multitasking proteins. Nucleic Acids Res. 2014, 42, D517–D520. [Google Scholar] [CrossRef] [PubMed]
- Franco-Serrano, L.; Hernandez, S.; Calvo, A.; Severi, M.A.; Ferragut, G.; Perez-Pons, J.; Pinol, J.; Pich, O.; Mozo-Villarias, A.; Amela, I.; et al. MultitaskProtDB-II: An update of a database of multitasking/moonlighting proteins. Nucleic Acids Res. 2018, 46, D645–D648. [Google Scholar] [CrossRef]
- Franco-Serrano, L.; Huerta, M.; Hernandez, S.; Cedano, J.; Perez-Pons, J.; Pinol, J.; Mozo-Villarias, A.; Amela, I.; Querol, E. Multifunctional Proteins: Involvement in Human Diseases and Targets of Current Drugs. Protein J. 2018, 37, 444–453. [Google Scholar] [CrossRef]
- Franco-Serrano, L.; Sanchez-Redondo, D.; Najar-Garcia, A.; Hernandez, S.; Amela, I.; Perez-Pons, J.A.; Pinol, J.; Mozo-Villarias, A.; Cedano, J.; Querol, E. Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors. Microorganisms 2021, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Huerta, M.; Franco-Serrano, L.; Amela, I.; Perez-Pons, J.A.; Pinol, J.; Mozo-Villarias, A.; Querol, E.; Cedano, J. Role of Moonlighting Proteins in Disease: Analyzing the Contribution of Canonical and Moonlighting Functions in Disease Progression. Cells 2023, 12, 235. [Google Scholar] [CrossRef]
- Henderson, B.; Martin, A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011, 79, 3476–3491. [Google Scholar] [CrossRef] [PubMed]
- Franco-Serrano, L.; Cedano, J.; Perez-Pons, J.A.; Mozo-Villarias, A.; Pinol, J.; Amela, I.; Querol, E. A hypothesis explaining why so many pathogen virulence proteins are moonlighting proteins. Pathog. Dis. 2018, 76, fty046. [Google Scholar] [CrossRef]
- Yoshida, N.; Oeda, K.; Watanabe, E.; Mikami, T.; Fukita, Y.; Nishimura, K.; Komai, K.; Matsuda, K. Protein function. Chaperonin turned insect toxin. Nature 2001, 411, 44. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. The role of posttranslational modification in moonlighting glyceraldehyde-3-phosphate dehydrogenase structure and function. Amino Acids 2021, 53, 507–515. [Google Scholar] [CrossRef]
- Tompa, P.; Szasz, C.; Buday, L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005, 30, 484–489. [Google Scholar] [CrossRef]
- Macossay-Castillo, M.; Marvelli, G.; Guharoy, M.; Jain, A.; Kihara, D.; Tompa, P.; Wodak, S.J. The Balancing Act of Intrinsically Disordered Proteins: Enabling Functional Diversity while Minimizing Promiscuity. J. Mol. Biol. 2019, 431, 1650–1670. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Walker, D.R.; Koonin, E.V. Analogous enzymes: Independent inventions in enzyme evolution. Genome Res. 1998, 8, 779–790. [Google Scholar] [CrossRef]
- Omelchenko, M.V.; Galperin, M.Y.; Wolf, Y.I.; Koonin, E.V. Non-homologous isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 2010, 5, 31. [Google Scholar] [CrossRef]
- Porter, L.L.; Looger, L.L. Extant fold-switching proteins are widespread. Proc. Natl. Acad. Sci. USA 2018, 115, 5968–5973. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.K.; Porter, L.L. Functional and Regulatory Roles of Fold-Switching Proteins. Structure 2021, 29, 6–14. [Google Scholar] [CrossRef]
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997, 91, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Rhee, S.G. Redox signaling: Hydrogen peroxide as intracellular messenger. Exp. Mol. Med. 1999, 31, 53–59. [Google Scholar] [CrossRef]
- Angelucci, F.; Saccoccia, F.; Ardini, M.; Boumis, G.; Brunori, M.; Di Leandro, L.; Ippoliti, R.; Miele, A.E.; Natoli, G.; Scotti, S.; et al. Switching between the alternative structures and functions of a 2-Cys peroxiredoxin, by site-directed mutagenesis. J. Mol. Biol. 2013, 425, 4556–4568. [Google Scholar] [CrossRef]
- Sirover, M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: Biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 2011, 1810, 741–751. [Google Scholar] [CrossRef]
- Perica, T.; Marsh, J.A.; Sousa, F.L.; Natan, E.; Colwell, L.J.; Ahnert, S.E.; Teichmann, S.A. The emergence of protein complexes: Quaternary structure, dynamics and allostery. Colworth Medal Lecture. Biochem. Soc. Trans. 2012, 40, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 105–153. [Google Scholar] [CrossRef]
- Jeffery, C.J. Molecular mechanisms for multitasking: Recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 2004, 14, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hsiao, H.H.; Bubunenko, M.; Weber, G.; Court, D.L.; Gottesman, M.E.; Urlaub, H.; Wahl, M.C. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell 2008, 32, 791–802. [Google Scholar] [CrossRef]
- Jones, S.; Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Keskin, O.; Gursoy, A.; Ma, B.; Nussinov, R. Principles of protein-protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 2008, 108, 1225–1244. [Google Scholar] [CrossRef]
- Copley, S.D. An evolutionary perspective on protein moonlighting. Biochem. Soc. Trans. 2014, 42, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A.; Teichmann, S.A. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 2015, 84, 551–575. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Comeau, D.C.; Connor, R.; DiCuccio, M.; Farrell, C.M.; Feldgarden, M.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2024, 52, D33–D43. [Google Scholar] [CrossRef]

| FSD PDB Code | Canonical Function and Species | Moonlighting Biological Function | Function Mapping | UniProt Code |
|---|---|---|---|---|
| 5aoeB/5Iy6B | Pneumolysin | Lipid binding & cytolysin | C-ter undecapeptide for membrane binding and cell lysis | Q04IN8 |
| S. pneumoniae | The domains rearrange upon membrane insertion, in particular, alpha-helices in D3 refold to form the transmembrane beta-strands | |||
| The relative position of domain D4 changes, allowing it to interact with host membranes | ||||
| 426–437 | ||||
| 3gmhL/2vfxL | Spindle checkpoint protein Mad2 dimer Human | DNA & RNA binding protein Transcriptional regulation | At 195–205 interaction with CDC20 | Q13257 |
| At 16–191 HORMA domain | ||||
| 2frhA/1fzpD | SarA Transcription regulator | 23 S-diacylglycerol cysteine | P31306 | |
| Staphyocccus aureus | 23–663 Oligopeptide-binding protein SarA | |||
| 4rr2D/319qB | Primase | Bypass oxidative lesions in DNA | Active site at 44 and 109 | P49642 |
| Human | Binding site at 109 | |||
| 3ifaA/5et5A | Fructose-1,6-biphosphatase | Cytoskeleton and Acrosomal membrane | Active sites at 43, 188, 230 | P04075 |
| Human | Binding sites at 272–274, 301, 304, 364 | |||
| 4fu4C/4g0Az | Collagenase 3 | Gene expression regulation | 100–121 Collagenase-like 1 | P08253 |
| Human | 397–465 Collagenase-like 2 | |||
| 222–236 Collagen binding | ||||
| Fibronectin types at 228–276, 286–334, 344–392 | ||||
| Hemopexin at 472–516, 517–563, 565–613, 614–660 | ||||
| TIMP2 binding at 414–460 | ||||
| 4dxtA/4dxrA | Sun2 (linker of nucleoskeleton and cytoskeleton) | Signal transduction. Gametogenesis gene-expression | 507–717 interaction with SYNE1 and SYNE2 | Q9UH99 |
| Human | ||||
| 2hdmA/2n54B | Lymphotactin/XCL1 | Chemotaxis & inflammatory response | 22–114 Lymphotactin | P47992 |
| (activates the G protein-coupled receptor XCR1) | 4–89 Small inducible cytokine A | |||
| Human | 22–114 cathogene G3DSA family | |||
| 32–88 TED domain | ||||
| 1uxmK/2namA | OxidoreductaseSOD1 | RNA-binding protein | 2–152 cooper/zing SODC | P00441 |
| Human | 2–151 TED domain | |||
| 3ejhA/3m7pA | Fibronectin | Serin protease. Macrophage activation | Glomerulopathy. Spndylometaphyseal dysplasia | P02751 |
| Human | 907–1172 DNA binding | |||
| 52–272 fibrin & heparin binding | ||||
| 1ceeB/2k42A | CDC42 (Clathrin-mediated endocytosis) | Mitotic spindle orientation | 53–88 & 276–311 EF Hands | Q6PJ79 |
| Human | ||||
| 2n0aD/2kkwA | Alpha synuclein (tubulin polymerization) | Dopamine regulation | 1–116 interaction LIMK1 | O94811 |
| Human | 52–207 pfam PF05517 domain | |||
| 49–142 TED domain | ||||
| 2ougC/2IcIA | RfaH | Translational regulation | 3–96 Pfam PF02357 domain | P0AFW0 |
| Escherichia coli |
| Protein Canonical Function and Species | Moonlighting Biological Function(s) | UniProt/PDB Codes |
|---|---|---|
| Superoxide dismutase SOD1 | RNA-binding protein | P00441/1AZV |
| Human | ||
| Cyclooxygenase1 | Heme-dependent peroxidase | P23219/6Y3C |
| Human | ||
| Prostaglandin G/H Synthase 1 | Cycloxygenase | P23219/6Y3C |
| Human | ||
| Peroxiredoxin-6 | Phospholipase aiPLA2 | P30041/5b6m_A |
| Human | ||
| Serine/threonine protein phosphatase | Dephosphorylating substrates. Chromatin structure. RNA-binding protein | P36873/1IT6 |
| Peroxidase | Phospholipase aiPLA2 | P30041/5b6m_A |
| Human | ||
| Fructose 1,6-biphosphate aldolase | Cytoskeleton and Acrosomal membrane | P04075/1zai_A |
| Human | ||
| S3 Ribosomal protein | DNA repair | P23396/1WH9 |
| Human | NF-KappB-mediated transcription | |
| Galactosidase | Cell adhesión and migration | Q6NVH9/1kjl_A |
| Human | ||
| Glucose-6-phosphate isomerase | Neuroleukin | G6PI_HUMAN/1JLH |
| Human | ||
| Gluthatione S-transferase | Sperm head proteins involved in zona pellucida binding | Q6FGJ9/Q6FGJDB |
| Human | ||
| Peptidyl-prolyl isomerase | Cytokine | P62937/1ak4_A |
| Human |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cedano, J.; Huerta, M.; Mozo-Villarias, A.; Querol, E. Moonlighting Proteins: Some Hypotheses on the Structural Origin of Their Multifunctionality. Int. J. Mol. Sci. 2025, 26, 10375. https://doi.org/10.3390/ijms262110375
Cedano J, Huerta M, Mozo-Villarias A, Querol E. Moonlighting Proteins: Some Hypotheses on the Structural Origin of Their Multifunctionality. International Journal of Molecular Sciences. 2025; 26(21):10375. https://doi.org/10.3390/ijms262110375
Chicago/Turabian StyleCedano, Juan, Mario Huerta, Angel Mozo-Villarias, and Enrique Querol. 2025. "Moonlighting Proteins: Some Hypotheses on the Structural Origin of Their Multifunctionality" International Journal of Molecular Sciences 26, no. 21: 10375. https://doi.org/10.3390/ijms262110375
APA StyleCedano, J., Huerta, M., Mozo-Villarias, A., & Querol, E. (2025). Moonlighting Proteins: Some Hypotheses on the Structural Origin of Their Multifunctionality. International Journal of Molecular Sciences, 26(21), 10375. https://doi.org/10.3390/ijms262110375






