Sulfoquinovose Catabolism in E. coli Strains: Compositional and Functional Divergence of yih Gene Cassettes
Abstract
1. Introduction
2. Results
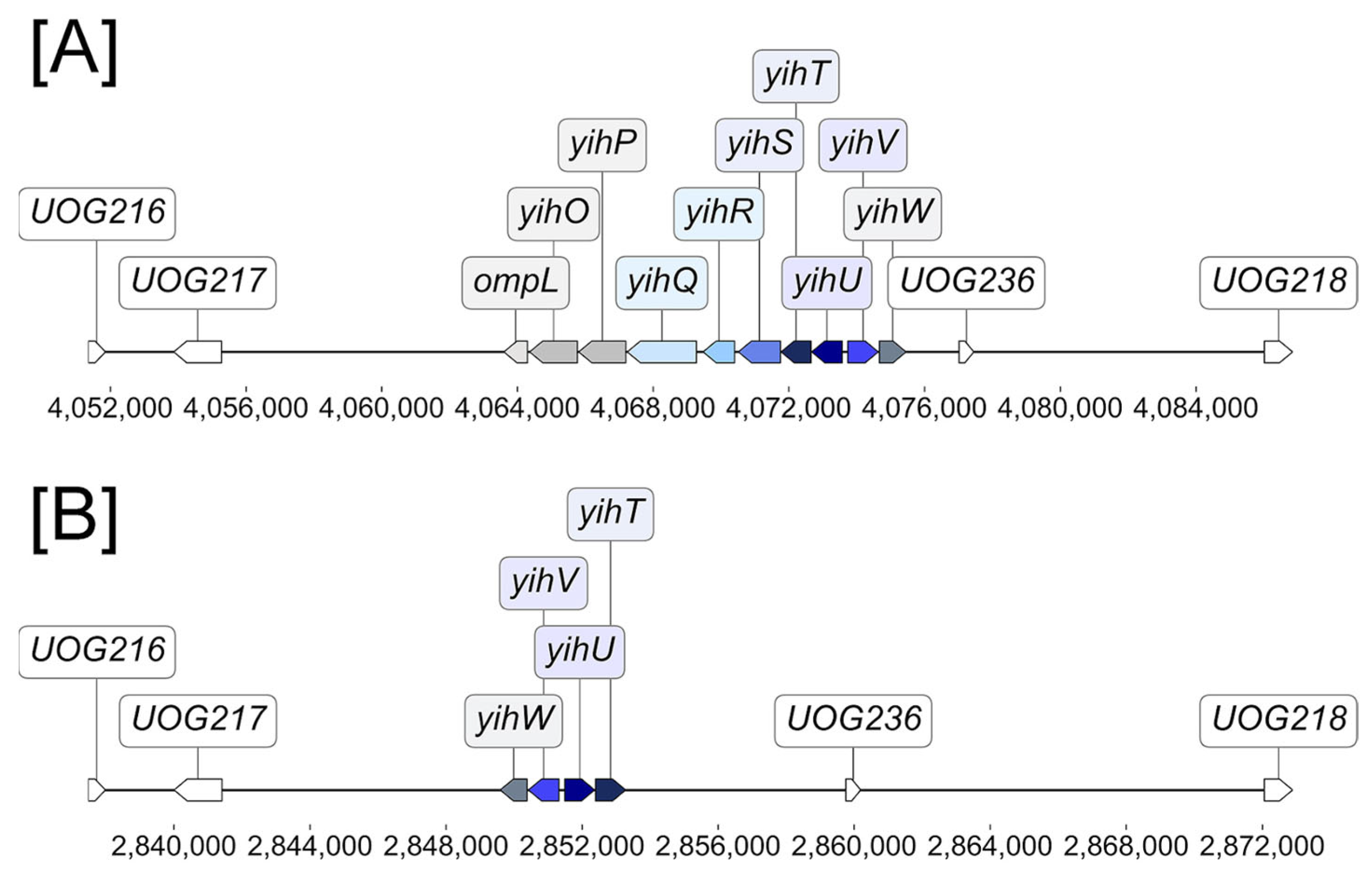
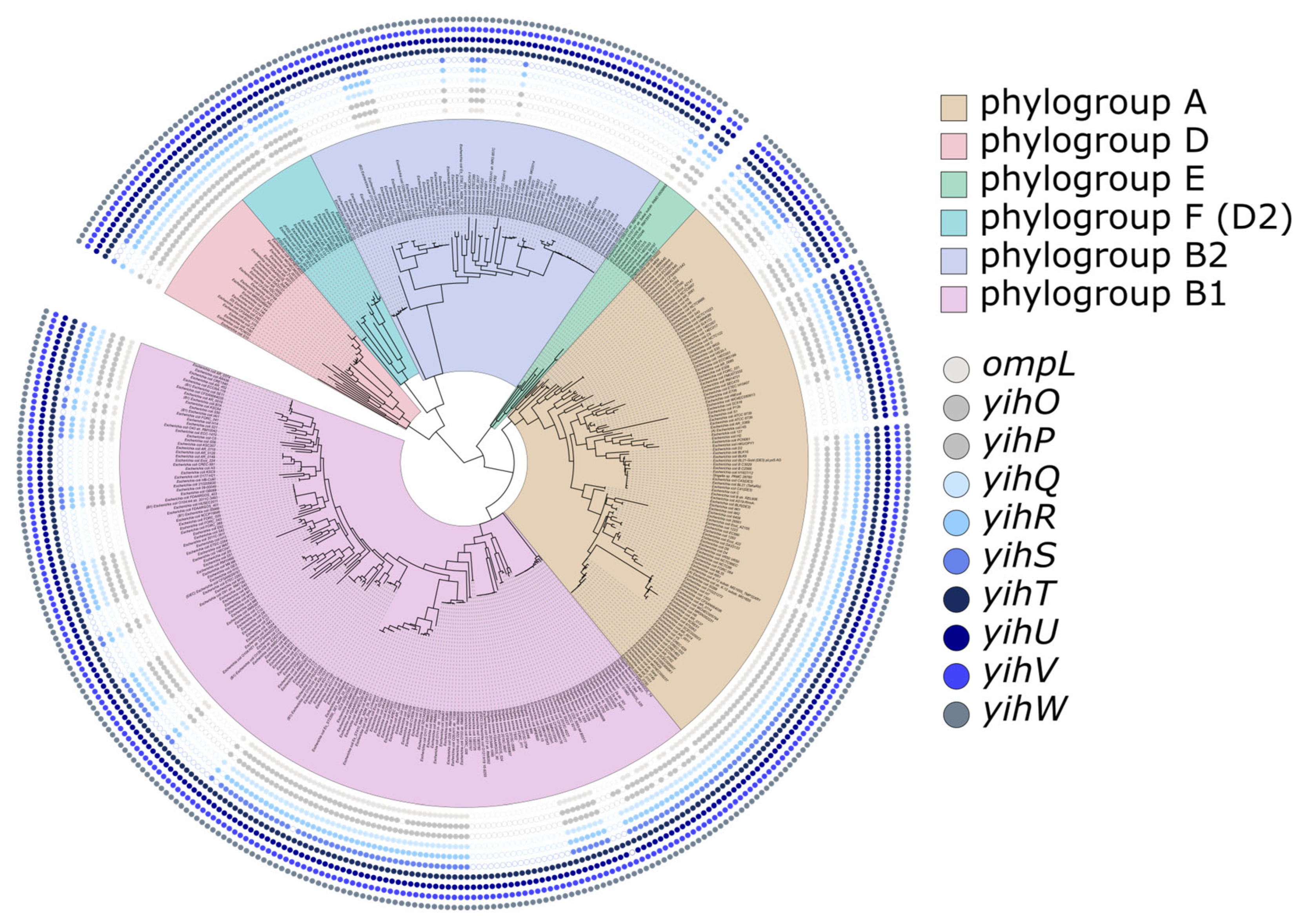
2.1. Long and Short yih Cassettes in E. coli Strains
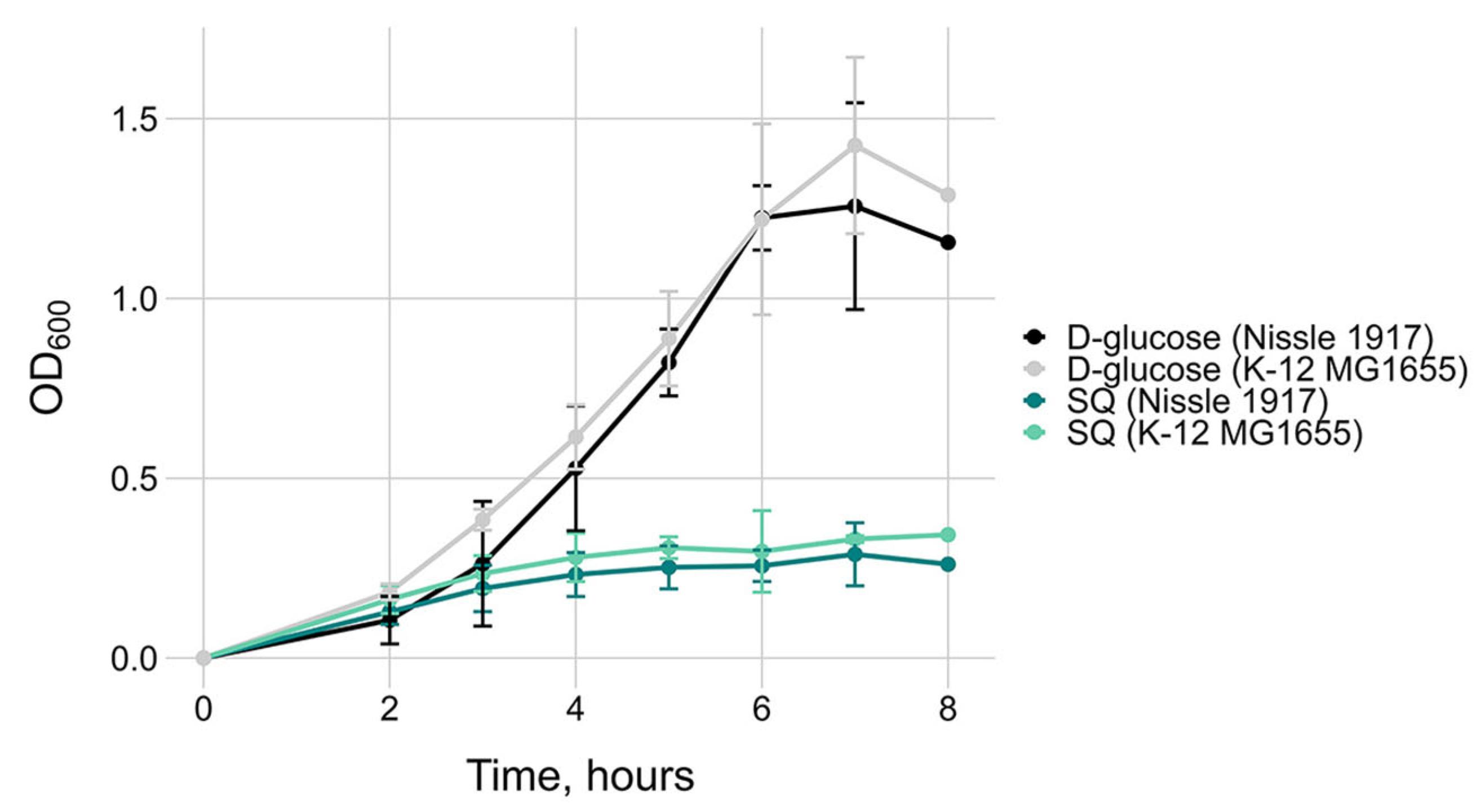
2.2. Transcriptional Changes in E. coli Strains with Short and Long yih Cassettes During Growth on Sulfoquinovose
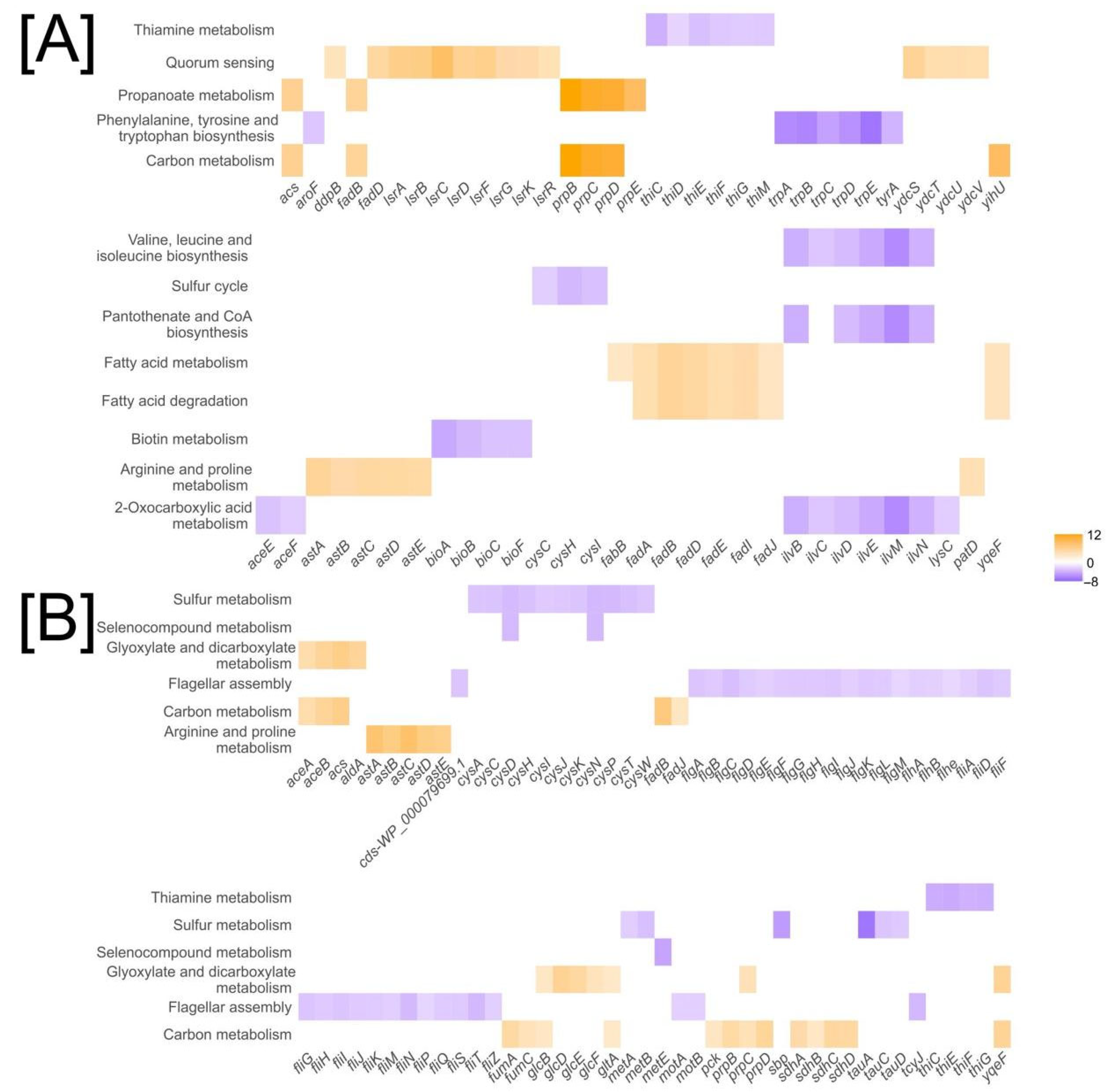
2.3. Shared and Divergent Gene Expression Patterns in E. coli K-12 and Nissle 1917 in Response to SQ
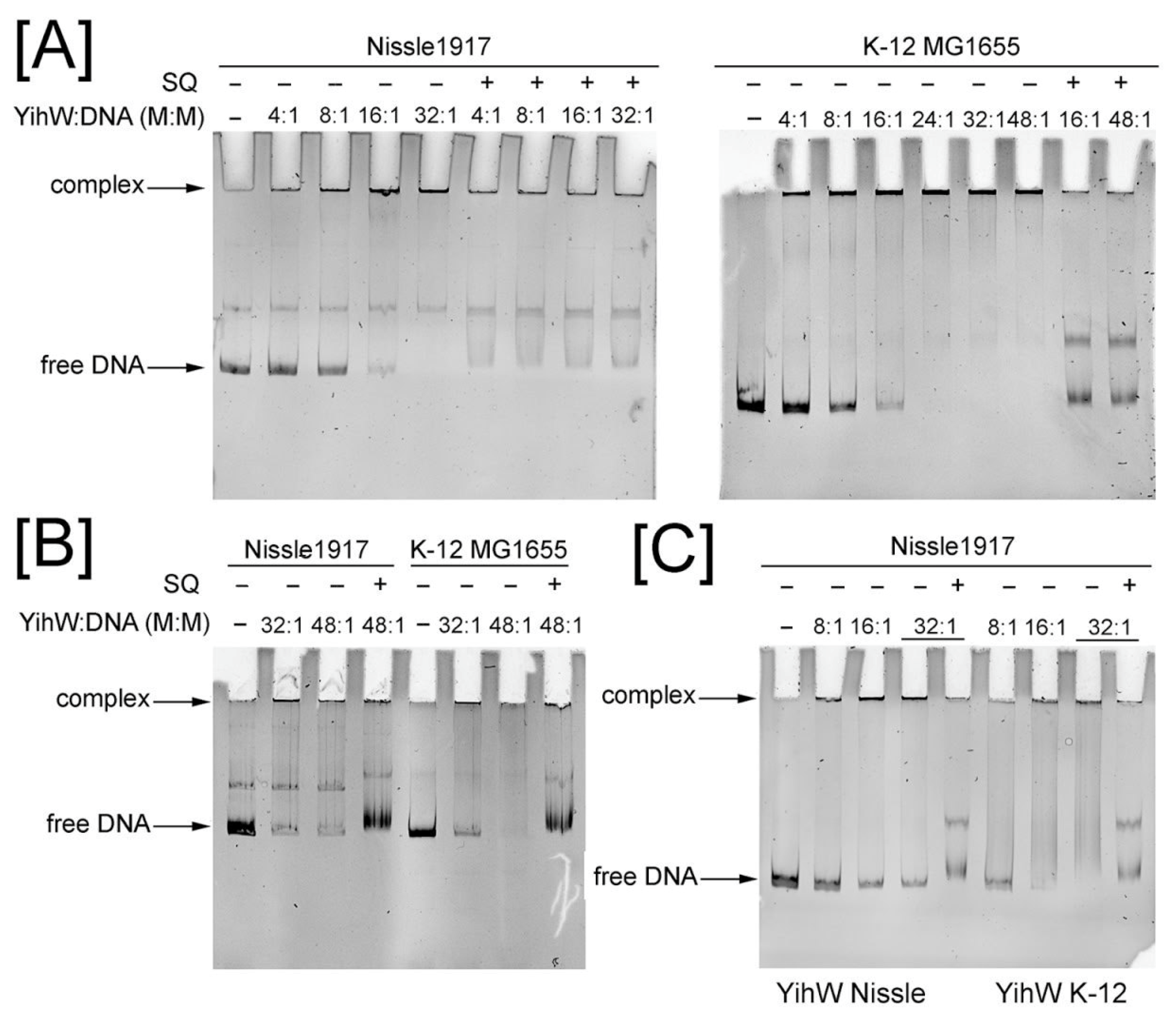
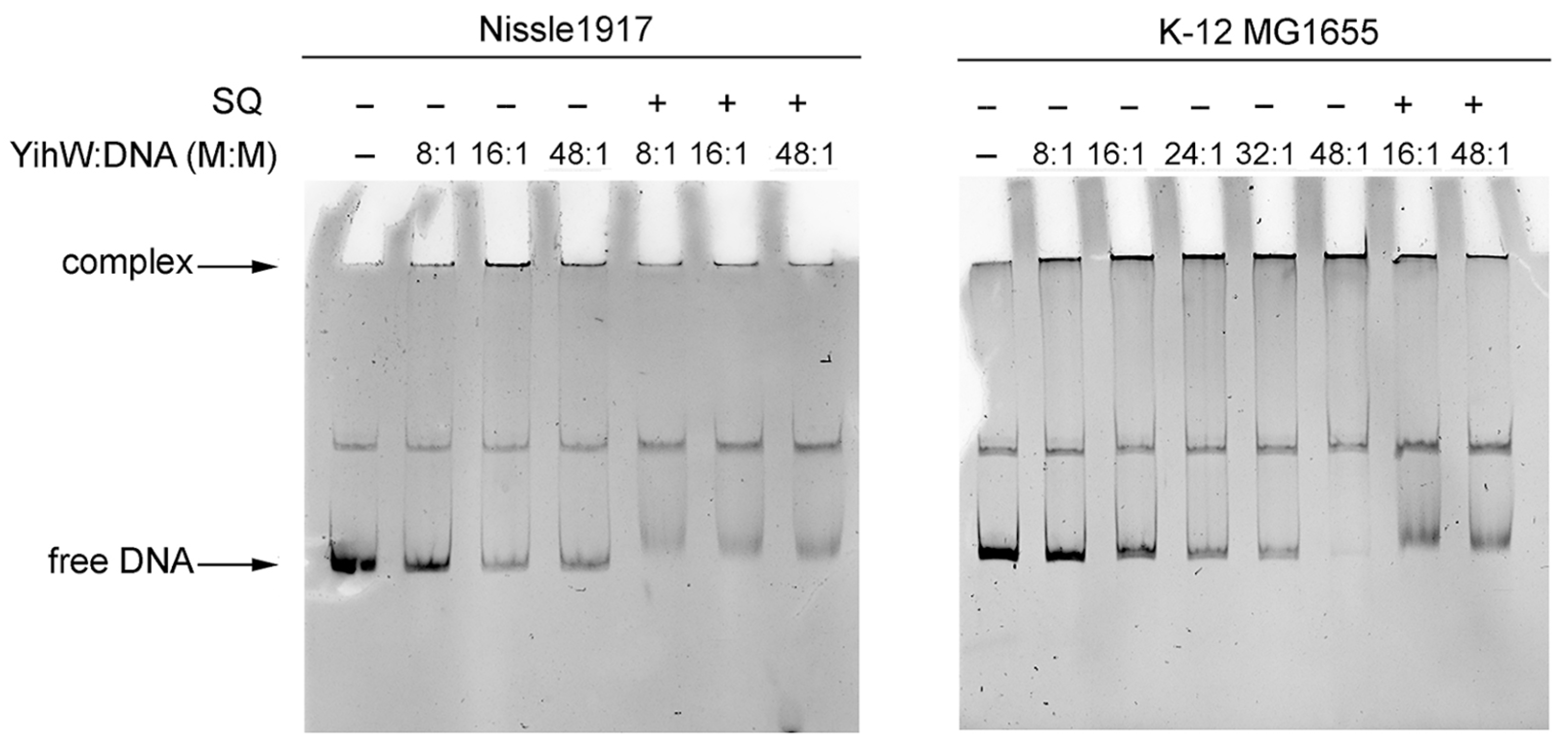
2.4. Binding of YihW (CsqR) to Intergenic Regions of Long and Short yih Cassettes
3. Discussion
4. Materials and Methods
4.1. Distribution and Composition of yih Cassettes in E. coli Strains
4.2. Strains and Growth Conditions
4.3. RNA Extraction and qRT-PCR
4.4. RNA Library Preparation and Sequencing
4.5. Analysis of RNA-Sequencing Data
4.6. YihW Expression and Purification
4.7. Electrophoretic Mobility Shift Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benson, A.A.; Daniel, H.; Wiser, R. A Sulfolipid in Plants. Proc. Natl. Acad. Sci. USA 1959, 45, 1582–1587. [Google Scholar] [CrossRef]
- Harwood, J.L.; Nicholls, R.G. The Plant Sulpholipid—A Major Component of the Sulphur Cycle. Biochem. Soc. Trans. 1979, 7, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Tong, Y.; Zhang, Y. New Mechanisms for Bacterial Degradation of Sulfoquinovose. Biosci. Rep. 2022, 42, BSR20220314. [Google Scholar] [CrossRef]
- Ye, Z.; Wei, Y.; Jiang, L.; Zhang, Y. Oxygenolytic Sulfoquinovose Degradation by an Iron-Dependent Alkanesulfonate Dioxygenase. iScience 2023, 26, 107803. [Google Scholar] [CrossRef]
- Felux, A.-K.; Spiteller, D.; Klebensberger, J.; Schleheck, D. Entner-Doudoroff Pathway for Sulfoquinovose Degradation in Pseudomonas Putida SQ1. Proc. Natl. Acad. Sci. USA 2015, 112, E4298–E4305. [Google Scholar] [CrossRef]
- Denger, K.; Weiss, M.; Felux, A.-K.; Schneider, A.; Mayer, C.; Spiteller, D.; Huhn, T.; Cook, A.M.; Schleheck, D. Sulphoglycolysis in Escherichia coli K-12 Closes a Gap in the Biogeochemical Sulphur Cycle. Nature 2014, 507, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Denger, K.; Huhn, T.; Hollemeyer, K.; Schleheck, D.; Cook, A.M. Sulfoquinovose Degraded by Pure Cultures of Bacteria with Release of C3-Organosulfonates: Complete Degradation in Two-Member Communities. FEMS Microbiol. Lett. 2012, 328, 39–45. [Google Scholar] [CrossRef]
- Mui, J.W.-Y.; De Souza, D.P.; Saunders, E.C.; McConville, M.J.; Williams, S.J. Remodeling of Carbon Metabolism during Sulfoglycolysis in Escherichia coli. Appl. Environ. Microbiol. 2023, 89, e0201622. [Google Scholar] [CrossRef]
- Kaznadzey, A.; Shelyakin, P.; Belousova, E.; Eremina, A.; Shvyreva, U.; Bykova, D.; Emelianenko, V.; Korosteleva, A.; Tutukina, M.; Gelfand, M.S. The Genes of the Sulphoquinovose Catabolism in Escherichia coli Are Also Associated with a Previously Unknown Pathway of Lactose Degradation. Sci. Rep. 2018, 8, 3177. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Yamamoto, K.; Nakano, M.; Watanabe, H.; Schleheck, D.; Ishihama, A. Regulatory Role of CsqR (YihW) in Transcription of the Genes for Catabolism of the Anionic Sugar Sulfoquinovose (SQ) in Escherichia coli K-12. Microbiology 2019, 165, 78–89. [Google Scholar] [CrossRef]
- Pérez-Rueda, E.; Collado-Vides, J. The Repertoire of DNA-Binding Transcriptional Regulators in Escherichia coli K-12. Nucleic Acids Res. 2000, 28, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Grozdanov, L.; Raasch, C.; Schulze, J.; Sonnenborn, U.; Gottschalk, G.; Hacker, J.; Dobrindt, U. Analysis of the Genome Structure of the Nonpathogenic Probiotic Escherichia coli Strain Nissle 1917. J. Bacteriol. 2004, 186, 5432–5441. [Google Scholar] [CrossRef]
- Seferbekova, Z.; Zabelkin, A.; Yakovleva, Y.; Afasizhev, R.; Dranenko, N.O.; Alexeev, N.; Gelfand, M.S.; Bochkareva, O.O. High Rates of Genome Rearrangements and Pathogenicity of Shigella spp. Front. Microbiol. 2021, 12, 628622. [Google Scholar] [CrossRef]
- Hua, Q.; Yang, C.; Oshima, T.; Mori, H.; Shimizu, K. Analysis of Gene Expression in Escherichia coli in Response to Changes of Growth-Limiting Nutrient in Chemostat Cultures. Appl. Environ. Microbiol. 2004, 70, 2354–2366. [Google Scholar] [CrossRef]
- Martínez-Gómez, K.; Flores, N.; Castañeda, H.M.; Martínez-Batallar, G.; Hernández-Chávez, G.; Ramírez, O.T.; Gosset, G.; Encarnación, S.; Bolivar, F. New Insights into Escherichia coli Metabolism: Carbon Scavenging, Acetate Metabolism and Carbon Recycling Responses during Growth on Glycerol. Microb. Cell Fact. 2012, 11, 46. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Roth, J.R. Selfish Operons: Horizontal Transfer May Drive the Evolution of Gene Clusters. Genetics 1996, 143, 1843–1860. [Google Scholar] [CrossRef]
- Kertesz, M.A. Riding the Sulfur Cycle—Metabolism of Sulfonates and Sulfate Esters in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2000, 24, 135–175. [Google Scholar] [CrossRef] [PubMed]
- Pavoncello, V.; Barras, F.; Bouveret, E. Degradation of Exogenous Fatty Acids in Escherichia coli. Biomolecules 2022, 12, 1019. [Google Scholar] [CrossRef]
- Brauer, M.J.; Yuan, J.; Bennett, B.D.; Lu, W.; Kimball, E.; Botstein, D.; Rabinowitz, J.D. Conservation of the Metabolomic Response to Starvation across Two Divergent Microbes. Proc. Natl. Acad. Sci. USA 2006, 103, 19302–19307. [Google Scholar] [CrossRef]
- Bouillet, S.; Bauer, T.S.; Gottesman, S. RpoS and the Bacterial General Stress Response. Microbiol. Mol. Biol. Rev. 2024, 88, e00151-22. [Google Scholar] [CrossRef]
- Schink, S.J.; Biselli, E.; Ammar, C.; Gerland, U. Death Rate of E. coli during Starvation Is Set by Maintenance Cost and Biomass Recycling. Cell Syst. 2019, 9, 64–73.e3. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Foster, K.R. Competition Sensing: The Social Side of Bacterial Stress Responses. Nat. Rev. Microbiol. 2013, 11, 285–293. [Google Scholar] [CrossRef]
- Kuhar, I.; Zgur-Bertok, D. Transcription Regulation of the Colicin K Cka Gene Reveals Induction of Colicin Synthesis by Differential Responses to Environmental Signals. J. Bacteriol. 1999, 181, 7373–7380. [Google Scholar] [CrossRef]
- Matin, A. The Molecular Basis of Carbon-starvation-induced General Resistance in Escherichia coli. Mol. Microbiol. 1991, 5, 3–10. [Google Scholar] [CrossRef]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and Transcriptomic Stress Response of Escherichia coli. Mol. Syst. Biol. 2010, 6, 364. [Google Scholar] [CrossRef]
- Li, J.; Attila, C.; Wang, L.; Wood, T.K.; Valdes, J.J.; Bentley, W.E. Quorum Sensing in Escherichia coli Is Signaled by AI-2/LsrR: Effects on Small RNA and Biofilm Architecture. J. Bacteriol. 2007, 189, 6011–6020. [Google Scholar] [CrossRef]
- Semsey, S.; Krishna, S.; Erdőssy, J.; Horváth, P.; Orosz, L.; Sneppen, K.; Adhya, S. Dominant Negative Autoregulation Limits Steady-State Repression Levels in Gene Networks. J. Bacteriol. 2009, 191, 4487–4491. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Horizontal Gene Transfer and the Evolution of Transcriptional Regulation in Escherichia coli. Genome Biol. 2008, 9, R4. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Dubchak, I.; Arkin, A.; Alm, E.; Gelfand, M.S. Reconstruction of Regulatory and Metabolic Pathways in Metal-Reducing Delta-Proteobacteria. Genome Biol. 2004, 5, R90. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Vitreschak, A.G.; Mironov, A.A.; Gelfand, M.S. Comparative Genomics of the Methionine Metabolism in Gram-Positive Bacteria: A Variety of Regulatory Systems. Nucleic Acids Res. 2004, 32, 3340–3353. [Google Scholar] [CrossRef] [PubMed]
- Leyn, S.A.; Suvorova, I.A.; Kholina, T.D.; Sherstneva, S.S.; Novichkov, P.S.; Gelfand, M.S.; Rodionov, D.A. Comparative Genomics of Transcriptional Regulation of Methionine Metabolism in Proteobacteria. PLoS ONE 2014, 9, e113714. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database Resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res 2025, 53, D20–D29. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Söding, J. MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Zulkower, V.; Rosser, S. DNA Features Viewer: A Sequence Annotation Formatting and Plotting Library for Python. Bioinformatics 2020, 36, 4350–4352. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Tutukina, M.N.; Potapova, A.V.; Cole, J.A.; Ozoline, O.N. Control of Hexuronate Metabolism in Escherichia coli by the Two Interdependent Regulators, ExuR and UxuR: Derepression by Heterodimer Formation. Microbiology 2016, 162, 1220–1231. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2014.
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Yan, G.-R.; He, Q.-Y. DOSE: An R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-Mapper: A Web Server for Precise Operon Identification in Bacterial and Archaeal Genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef] [PubMed]
- Rybina, A.A.; Glushak, R.A.; Bessonova, T.A.; Dakhnovets, A.I.; Rudenko, A.Y.; Ozhiganov, R.M.; Kaznadzey, A.D.; Tutukina, M.N.; Gelfand, M.S. Phylogeny and Structural Modeling of the Transcription Factor CsqR (YihW) from Escherichia coli. Sci. Rep. 2024, 14, 7852. [Google Scholar] [CrossRef]
- Igarashi, K.; Ishihama, A. Bipartite Functional Map of the E. coli RNA Polymerase Alpha Subunit: Involvement of the C-Terminal Region in Transcription Activation by cAMP-CRP. Cell 1991, 65, 1015–1022. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaznadzey, A.D.; Rybina, A.A.; Bessonova, T.A.; Korshunov, D.S.; Tutukina, M.N.; Gelfand, M.S. Sulfoquinovose Catabolism in E. coli Strains: Compositional and Functional Divergence of yih Gene Cassettes. Int. J. Mol. Sci. 2025, 26, 10351. https://doi.org/10.3390/ijms262110351
Kaznadzey AD, Rybina AA, Bessonova TA, Korshunov DS, Tutukina MN, Gelfand MS. Sulfoquinovose Catabolism in E. coli Strains: Compositional and Functional Divergence of yih Gene Cassettes. International Journal of Molecular Sciences. 2025; 26(21):10351. https://doi.org/10.3390/ijms262110351
Chicago/Turabian StyleKaznadzey, Anna D., Anna A. Rybina, Tatiana A. Bessonova, Dmitriy S. Korshunov, Maria N. Tutukina, and Mikhail S. Gelfand. 2025. "Sulfoquinovose Catabolism in E. coli Strains: Compositional and Functional Divergence of yih Gene Cassettes" International Journal of Molecular Sciences 26, no. 21: 10351. https://doi.org/10.3390/ijms262110351
APA StyleKaznadzey, A. D., Rybina, A. A., Bessonova, T. A., Korshunov, D. S., Tutukina, M. N., & Gelfand, M. S. (2025). Sulfoquinovose Catabolism in E. coli Strains: Compositional and Functional Divergence of yih Gene Cassettes. International Journal of Molecular Sciences, 26(21), 10351. https://doi.org/10.3390/ijms262110351




