Genome-Wide Identification of DlGRAS Family and Functional Analysis of DlGRAS10/22 Reveal Their Potential Roles in Embryogenesis and Hormones Responses in Dimocarpus longan
Abstract
1. Introduction
2. Results
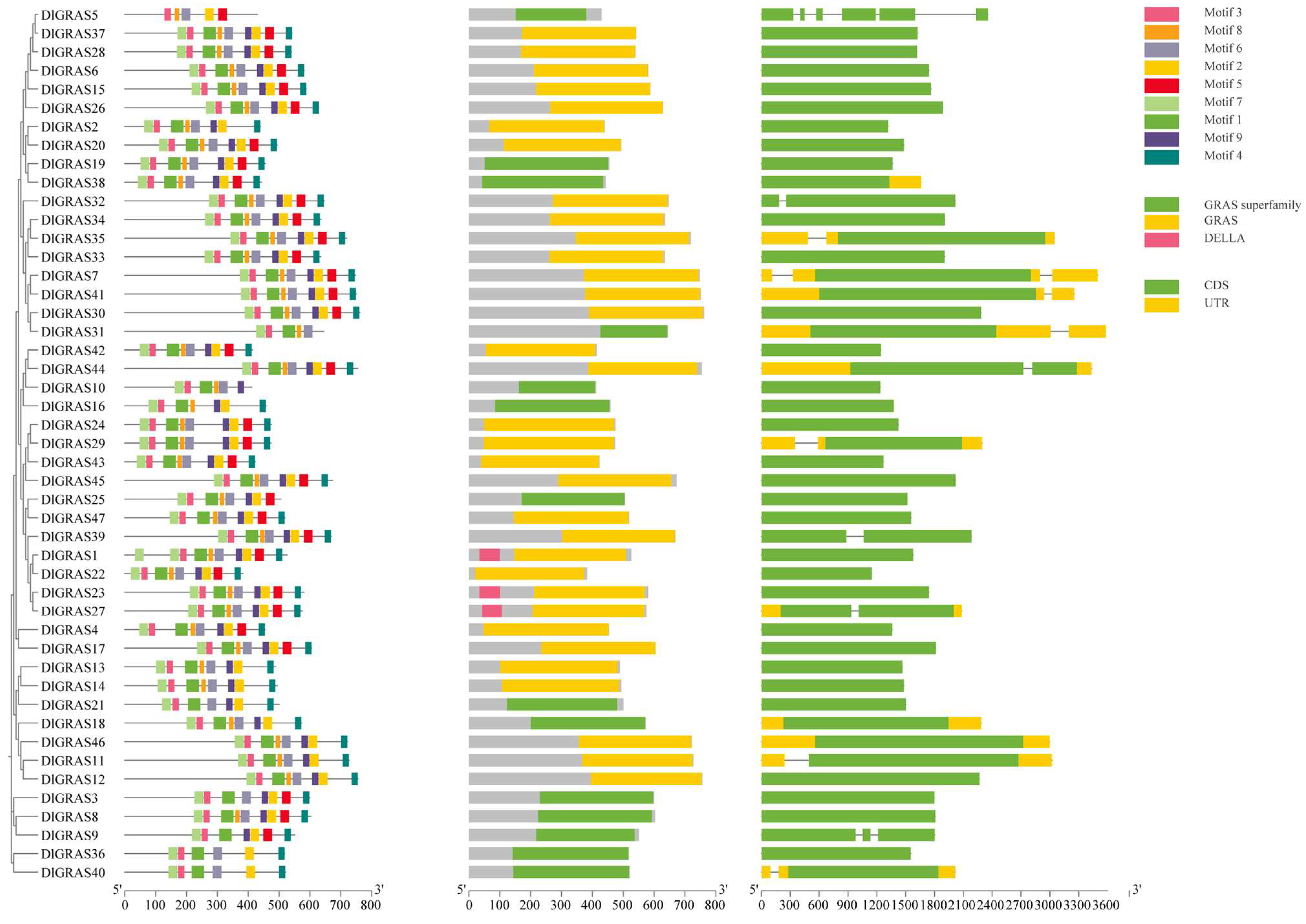
2.1. Diverse Physical and Chemical Properties of DlGRAS Family Members
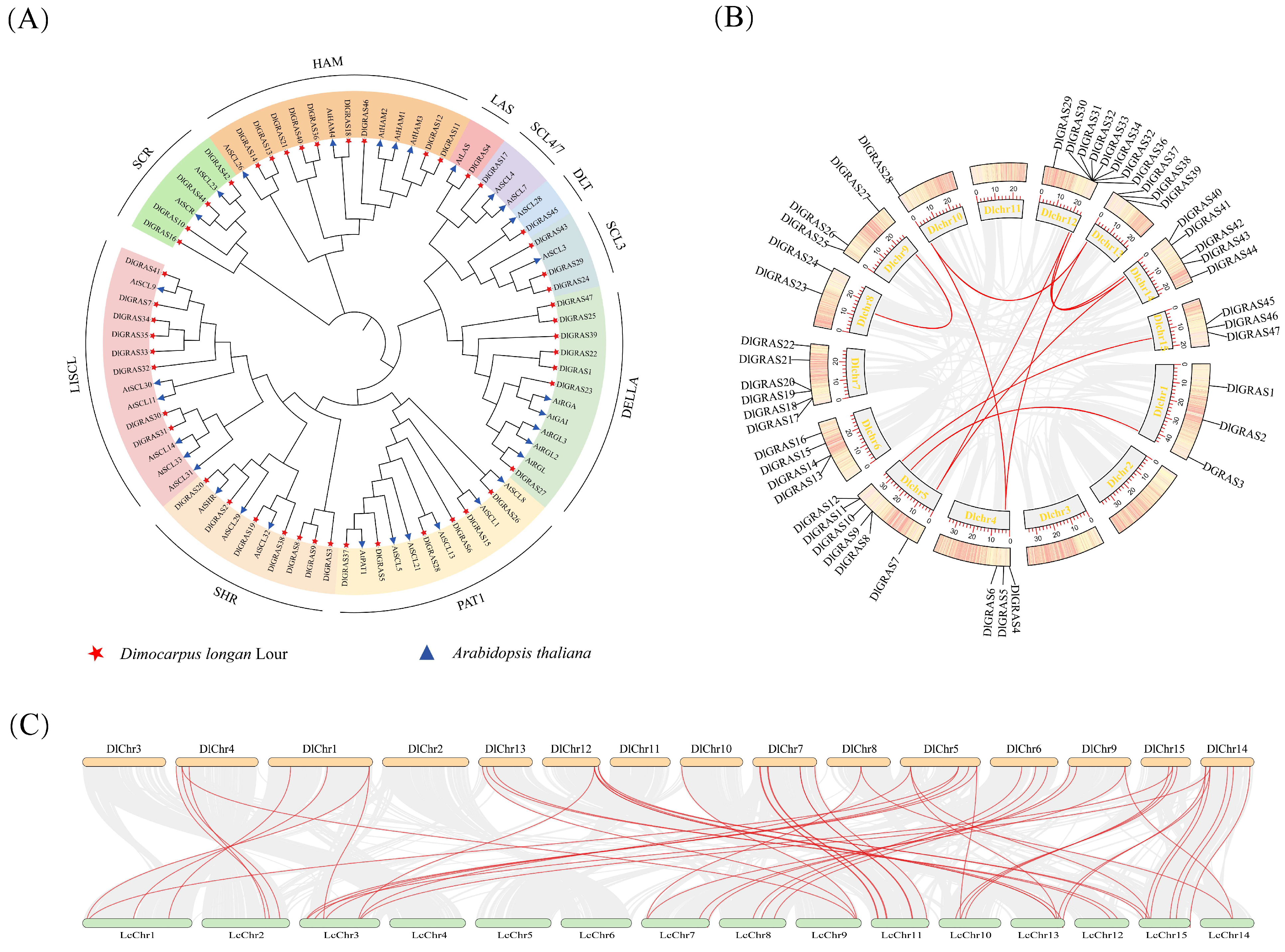
2.2. High Homology of DlGRAS Exists in Litchi and Longan
2.3. DlGRAS Exhibits Remarkable Conservation During Evolutionary Processes
2.4. Multiple Hormones and Transcription Factors Regulate DlGRAS Expression
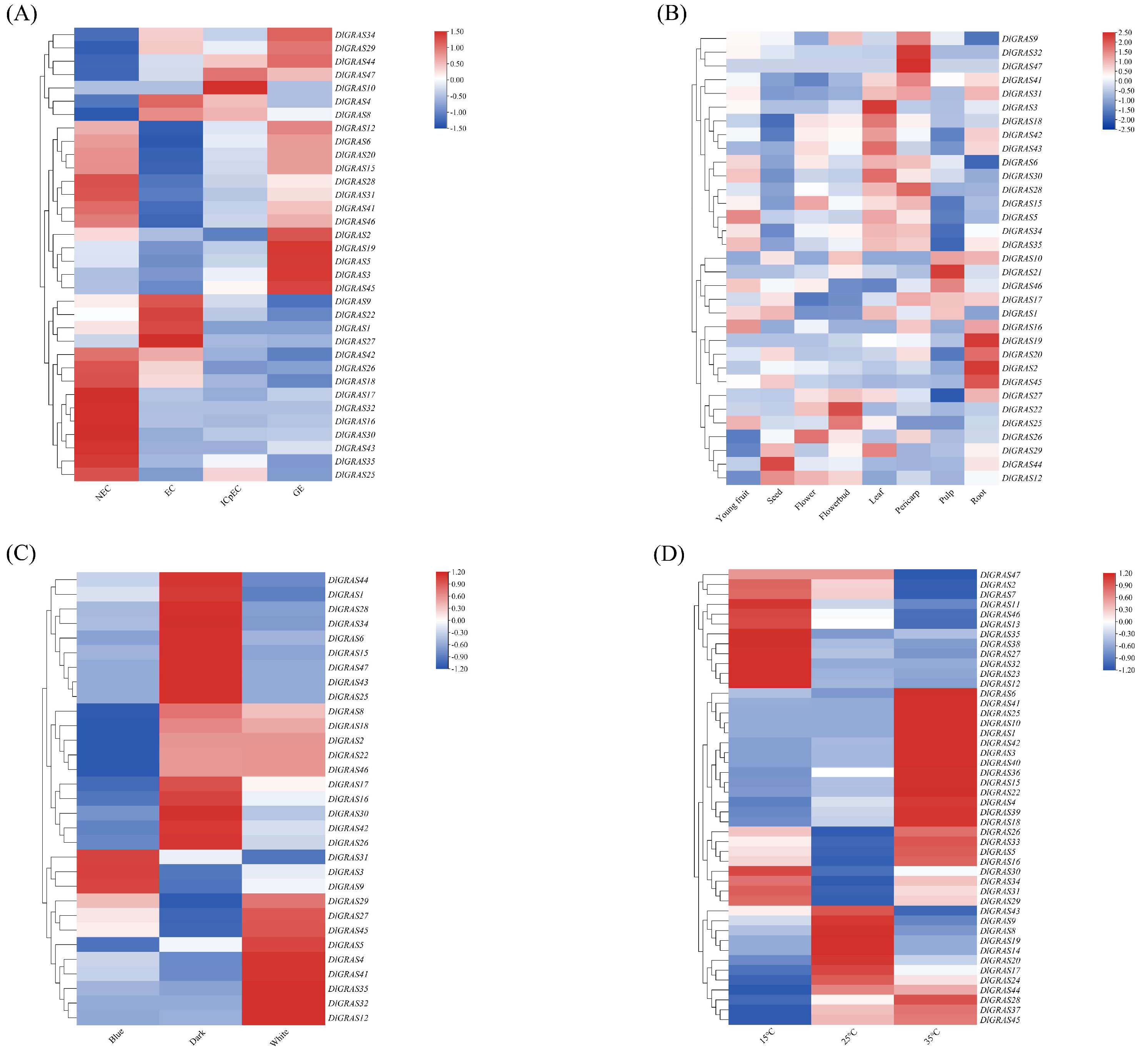
2.5. DlGRAS Transcription Factor Coordinates SE, Organogenesis and Stress Adaptation in Longan
2.6. Differential Regulation of Longan Embryo Development by DlGRAS Genes
2.7. DlGRAS Integrates GA3 and ABA Signaling to Regulate Longan SE
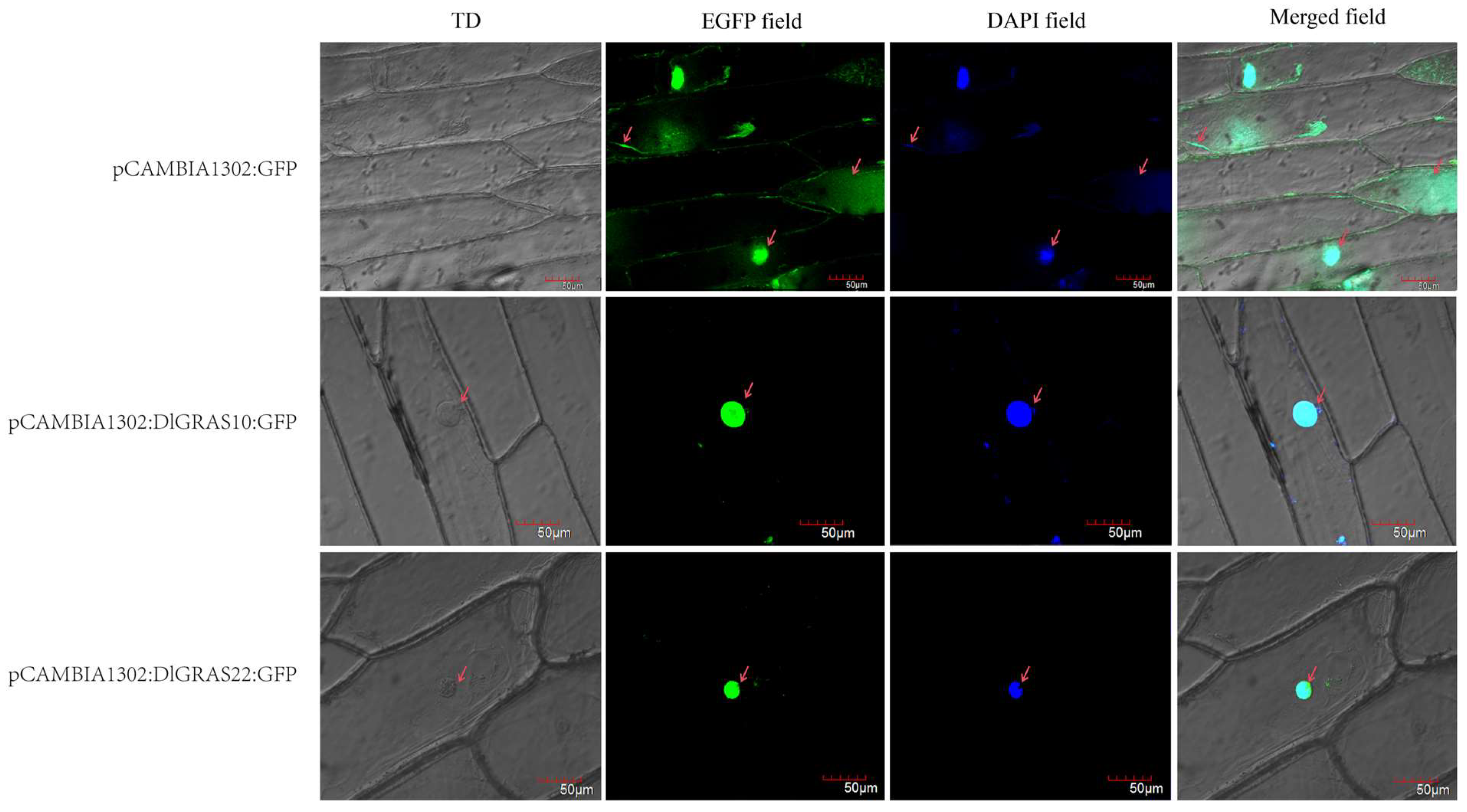
2.8. DlGRAS10 and DlGRAS22 Play an Important Role in the Nucleus
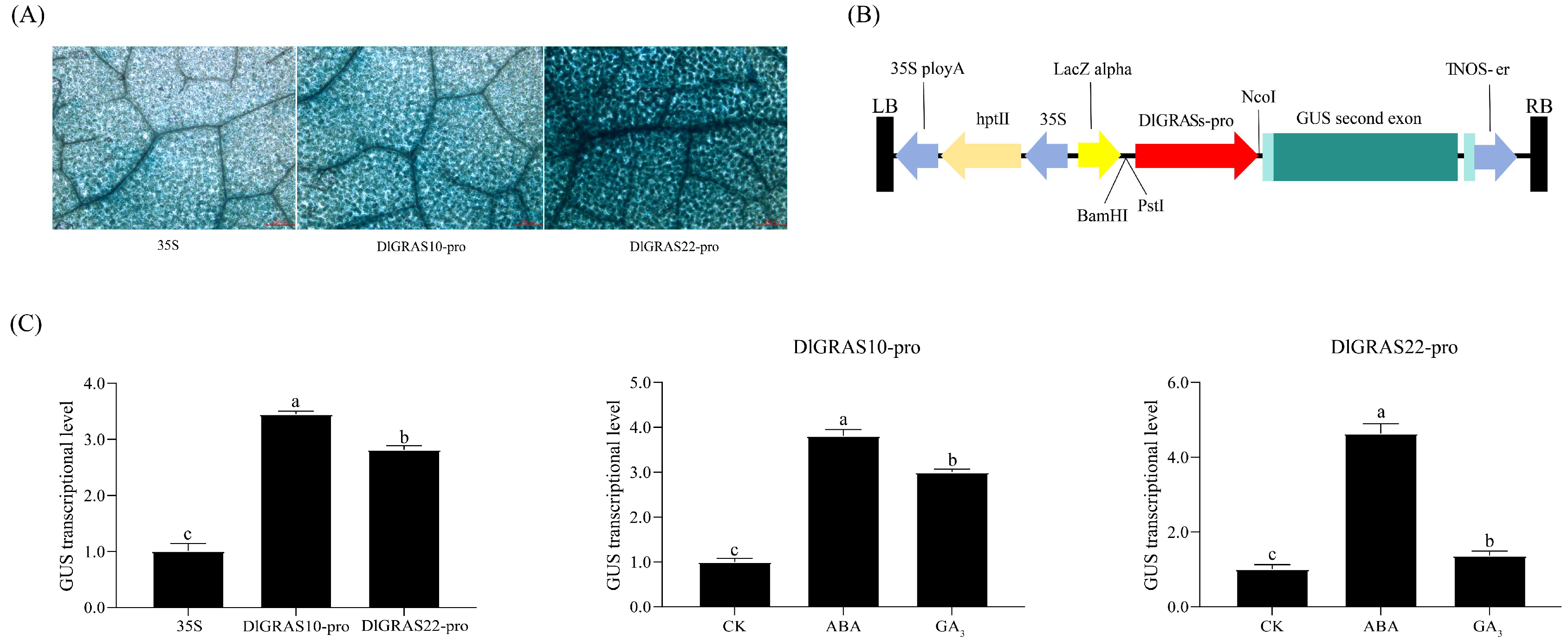
2.9. Exogenous GA3 and ABA Modulate DlGRAS10 and DlGRAS22 Promoter Activities During Longan SE
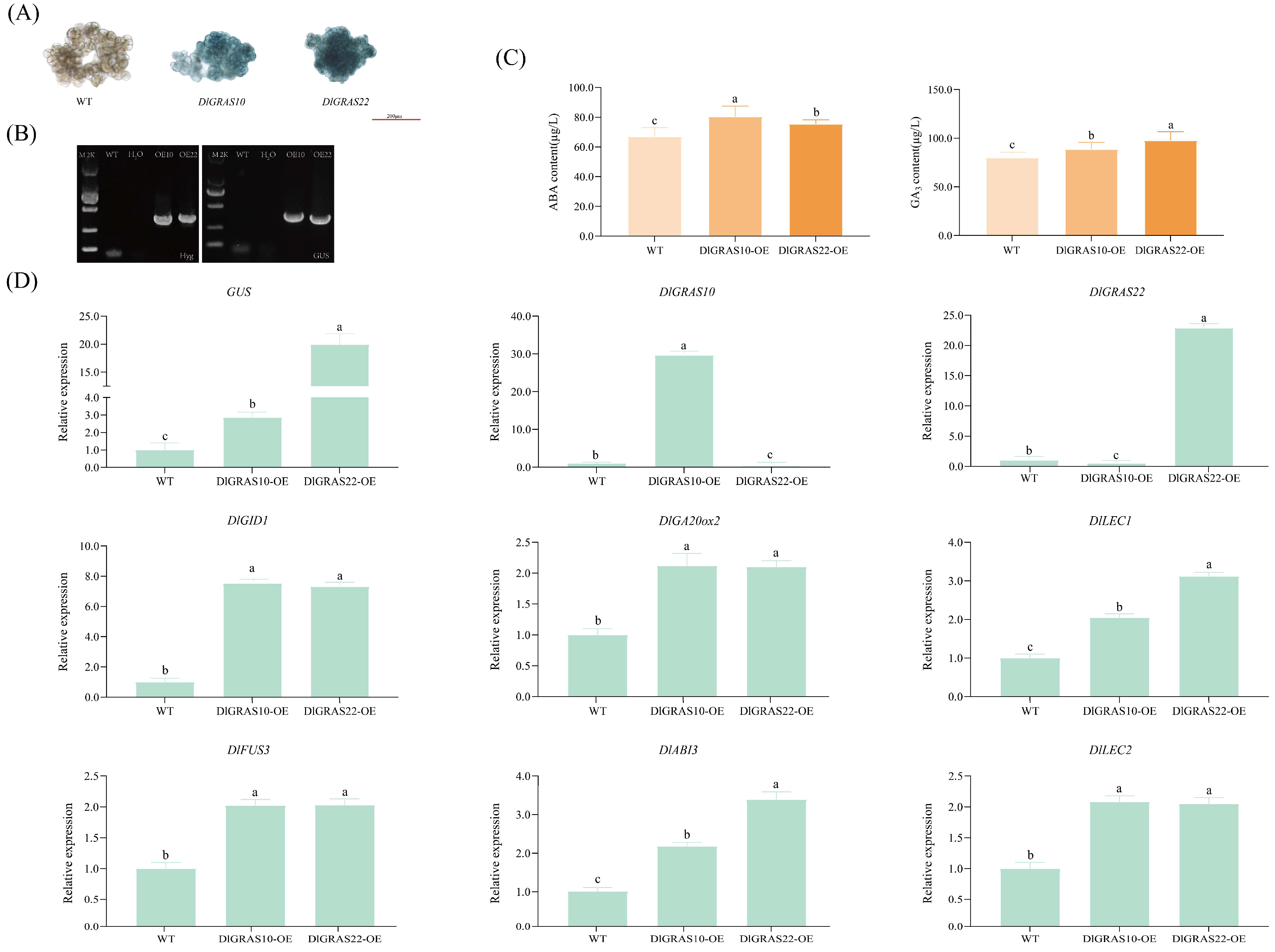
2.10. DlGRAS10/22 Transient Expression Activates Embryogenic Transcriptional Networks and Modulates Endogenous ABA/GA3 Homeostasis
3. Discussion
3.1. DlGRAS Family May Exhibit Evolutionary Conservation
3.2. DlGRAS Genes May Be Involved in Longan SE Through Hormone Responses
3.3. DlGRAS10/22 Affected Longan SE by Regulating SE-Related Genes and Hormone Synthesis

4. Materials and Methods
4.1. Materials
4.2. Identification of DlGRAS in Longan
4.3. Phylogenetic Relationship and Collinearity Analysis of DlGRAS in Longan
4.4. Analysis of Conserved Motifs, Domains and Gene Structure of DlGRAS
4.5. Transcription Factor Binding Site and Cis-Element Analysis of DlGRAS
4.6. Expression Analysis of DlGRAS During Early SE and Different Tissues/Light Qualities/Abiotic Stresses
4.7. RNA Extraction and qRT-PCR Analysis of DlGRAS
4.8. Subcellular Localization of DlGRAS10/12
4.9. Functional Analysis of DlGRAS10 and DlGRAS22 Promoters
4.10. DlGRAS10/22 Transiently Transformed to Longan EC
4.11. RNA, DNA Extraction and Molecular Identification of Transiently Transformed Longan EC
4.12. Expression Analysis of Transiently Transformed Longan EC
4.13. Determination of Endogenous Hormones in Transiently Transformed Longan EC
4.14. The Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Dl | Dimocarpus longan |
| aa | amino acid |
| NEC | non-embryogenic callus |
| EC | embryogenic callus |
| ICpEC | incomplete compact pro-embryogenic cultures |
| GE | globular embryos |
| SE | somatic embryogenesis |
| DNA | deoxyribonucleic acid |
| RNA | ribonucleic acid |
| cDNA | complementary DNA |
| qPCR | quantitative real-time PCR |
| CDS | coding sequence |
| UTRs | untranslated regions |
| bp | base pairs |
| g | gram |
| d | day |
| h | hour |
| min | minute |
| FPKM | Fragments Per Kilo-base of exon per Million fragments mapped |
| ABA | abscisic acid |
| GA | gibberellin |
| DAPI | 4’,6-diamidino-2-phenylindol |
| GFP | Green Fluorescent Protein |
| RNA-seq | RNA sequencing |
References
- Yang, L.; Chen, D.; Peng, Z.; Zhao, H.; Gao, Z. Cloning of Transcription Factor DlSCL6 from Dendrocalamus latiflorus and Its Ectopic Expression in Arabidopsis thaliana. Sci. Silvae Sin. 2014, 50, 52–57. [Google Scholar]
- Yuan, Y. Functional Study of VaPATI, a GRAS Gene from Vitis amerunsis, in Response to Abiotic Stresses. Doctor’s Thesis, University of Chinese Academy of Sciences (Wuhan Botanical Garden, Chinese Academy of Sciences), Wuhan, China, 2016. [Google Scholar]
- Morohashi, K.; Minami, M.; Takase, H.; Hotta, Y.; Hiratsuka, K. Isolation and Characterization of a Novel GRAS Gene That Regulates Meiosis-associated Gene Expression. J. Biol. Chem. 2003, 278, 20865–20873. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, Z.; Yu, R.; Wu, Z. Progress of the Structural and Functional Analysis of GRAS Gene in Plants. Mol. Plant Breed. 2019, 17, 6323–6331. [Google Scholar] [CrossRef]
- Aoyanagi, T.; Ikeya, S.; Kobayashi, A.; Kozaki, A. Gene Regulation via the Combination of Transcription Factors in the INDETERMINATE DOMAIN and GRAS Families. Genes 2020, 11, 613. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef]
- Lee, M.-H.; Kim, B.; Song, S.-K.; Heo, J.-O.; Yu, N.-I.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wan, X.; Zhang, X.; Zhang, J.; Zheng, C.; Yang, Q.; Yang, L.; Li, X.; Feng, L.; Zou, L.; et al. GRAS gene family in rye (Secale cereale L.): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. BMC Plant Biol. 2024, 24, 46. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-Wide Analysis of the GRAS Gene Family in Rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Rep. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Lu, H.; Xu, J.; Li, G.; Zhong, T.; Chen, D.; Lv, J. Genome-wide identification and expression analysis of GRAS gene family in Eucalyptus grandis. BMC Plant Biol. 2024, 24, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, Q.; Zhang, M.-M.; He, X.; Zhao, X.; Wang, L.; Lan, S.; Liu, Z.-J. Genome-Wide Identification and Expression Analysis of the GRAS Gene Family and Their Responses to Heat Stress in Cymbidium goeringii. Int. J. Mol. Sci. 2024, 25, 6363. [Google Scholar] [CrossRef]
- Khan, I.; Lubna; Asaf, S.; Jan, R.; Bilal, S.; Khan, A.L.; Kim, K.-M.; Al-Harrasi, A. Dynamic interplay of WRKY, GRAS, and ERF transcription factor families in tomato-endophytic fungal symbiosis: Insights from transcriptome and genome-wide analysis. Front. Plant Sci. 2023, 14, 1181227. [Google Scholar] [CrossRef]
- Tian, Z. Functional Study on Salt Tolerance of PAT1 Gene from Betula platyphylla. Master’s Thesis, Northeast Forestry University, Harbin, China, 2021. [Google Scholar]
- Delinur, A.; Feng, X.; Wang, Y. Study of the promoter properties of the HcSCL1313 gene for the transcription factor of Halostachys caspica (Bieb.) CA Mey. Xinjiang Agric. Sci. 2022, 59, 990–1000. [Google Scholar] [CrossRef]
- Wang, T.-T.; Yu, T.-F.; Fu, J.-D.; Su, H.-G.; Chen, J.; Zhou, Y.-B.; Chen, M.; Guo, J.; Ma, Y.-Z.; Wei, W.-L.; et al. Genome-Wide Analysis of the GRAS Gene Family and Functional Identification of GmGRAS37 in Drought and Salt Tolerance. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wang, P.; Yang, X.; Sun, S.; Wang, J.; Wang, J.; Li, X.; Li, D.; Wang, Y. The SCARECROW-LIKE transcription factor from Populus davidiana × P. bolleana simultaneously improved drought tolerance and plant growth through acetylation-dependent mechanisms. Plant Biotechnol. J. 2025, 23, 3650–3666. [Google Scholar] [CrossRef]
- Hou, J.; Xiao, H.; Yao, P.; Ma, X.; Shi, Q.; Yang, J.; Hou, H.; Li, L. Unveiling the mechanism of broad-spectrum blast resistance in rice: The collaborative role of transcription factor OsGRAS30 and histone deacetylase OsHDAC1. Plant Biotechnol. J. 2024, 22, 1740–1756. [Google Scholar] [CrossRef]
- Hernández-García, J.; Briones-Moreno, A.; Blázquez, M.A. Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 2021, 109, 46–54. [Google Scholar] [CrossRef]
- Fan, Y.; Yan, J.; Lai, D.; Yang, H.; Xue, G.; He, A.; Guo, T.; Chen, L.; Cheng, X.-B.; Xiang, D.-B.; et al. Genome-wide identification, expression analysis, and functional study of the GRAS transcription factor family and its response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2021, 22, 1–21. [Google Scholar] [CrossRef]
- Zhang, Q. Functional Study of Gibberellines Effects on the Initiation of Axillary Meristem in Arabidopsis. Master’s Thesis, ShanDong University, Jinan, China, 2018. [Google Scholar]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef]
- Hughes, T.E.; Sedelnikova, O.V.; Wu, H.; Becraft, P.W.; Langdale, J.A. Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development 2019, 146, dev177543. [Google Scholar] [CrossRef]
- Rich, M.K.; Courty, P.-E.; Roux, C.; Reinhardt, D. Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y. Functional Analysis of PtrDLT and PtrWOX4 Genes in the Development of Vascular Cambium in Populus trichocarpa. Master’s Thesis, Northeast Forestry University, Harbin, China, 2020. [Google Scholar]
- Ji, C.; Xu, L.; Li, Y.; Fu, Y.; Li, S.; Wang, Q.; Zeng, X.; Zhang, Z.; Zhang, Z.; Wang, W.; et al. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize. Mol. Plant 2022, 15, 468–487. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, F.; Wen, J.; Shi, X.; Wu, C.; Liu, M.; Wu, Z. Identification and Heat Resistance Analysis of SlGRAS4 Gene in Tomato. Acta Bot. Boreali-Occident. Sin. 2021, 41, 539–548. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, Z.; Li, F.; Yu, X.; Naeem, M.; Zhang, Y.; Chen, G. Manipulation of plant architecture and flowering time by down-regulation of the GRAS transcription factor SlGRAS26 in Solanum lycopersicum. Plant Sci. 2018, 271, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, L.; Huang, M.; He, X.; Yang, Y.; Liu, X.; Li, Y.; Hou, X. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 2018, 4, 289–298. [Google Scholar] [CrossRef]
- Lai, Z.; Pan, L.; Chen, Z. Establishment and maintenance of longan embryogenic cell lines. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 1997, 2, 33–40. [Google Scholar]
- Lai, Z.; Chen, Z. Somatic embryogenesis of high frequency from longan embryogenic calli. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 1997, 3, 271–276. [Google Scholar]
- Lin, Y.; Lai, Z. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]
- Chen, Y. Transcriptome and Proteomics Analysis During Early Somatic Embryogenesis and Expression, Functional Analysis of Flowering Time Related Genes in Dimocarpus longan Lour. Doctor’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2018. [Google Scholar]
- Sun, J.; Wu, C.; Guan, E.; Xiong, D.; Zhou, T.; Chen, Y.; Liu, X.; Zhao, Q.; Wang, D.; Li, Z.; et al. Genome-Wide Identification and Expression Analysis of GRAS Gene Family in Nicotiana tabacum L. Chin. Tob. Sci. 2024, 45, 7–18. [Google Scholar] [CrossRef]
- Chen, T.; Li, H.; Wang, P.; Jiang, Y.; Xiao, J.; Yan, L.; Wang, Y.; Fan, Y. Identification of GRAS Gene Family in Paper Mulberry and Analysis of Its Expression in Stem Development. Guangdong Agric. Sci. 2024, 51, 107–119. [Google Scholar] [CrossRef]
- Ortiz-Ramírez, C.; Guillotin, B.; Xu, X.; Rahni, R.; Zhang, S.; Yan, Z.; Araujo, P.C.D.; Demesa-Arevalo, E.; Lee, L.; Van Eck, J.; et al. Ground tissue circuitry regulates organ complexity in maize and Setaria. Science 2021, 374, 1247–1252. [Google Scholar] [CrossRef]
- Engstrom, E.M. HAM proteins promote organ indeterminacy. Plant Signal. Behav. 2012, 7, 227–234. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2007, 8, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Sverdlov, A.V.; Babenko, V.N.; Koonin, E.V. Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinform. 2005, 6, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lu, L.-L.; Ma, S.-Y.; Zhao, Y.-P.; Wu, N.; Li, W.-J.; Ma, L.; Kong, X.-H.; Xie, Z.-M.; Hou, Y.-X. Analysis of PAT1 subfamily members in the GRAS family of upland cotton and functional characterization of GhSCL13-2A in Verticillium dahliae resistance. Plant Cell Rep. 2023, 42, 487–504. [Google Scholar] [CrossRef]
- Wu, R.; Liu, W.; Liu, K.; Liang, G.; Wang, Y. Genome-Wide Identification and Expression of the GRAS Gene Family in Oat (Avena sativa L.). Agronomy 2023, 13, 1807. [Google Scholar] [CrossRef]
- Yu, L.; Hui, C.; Huang, R.; Wang, D.; Fei, C.; Guo, C.; Zhang, J. Genome-wide identification, evolution and transcriptome analysis of GRAS gene family in Chinese chestnut (Castanea mollissima). Front. Genet. 2023, 13, 1080759. [Google Scholar] [CrossRef]
- Kumari, P.; Gahlaut, V.; Kaur, E.; Singh, S.; Kumar, S.; Jaiswal, V. Genome-Wide Identification of GRAS Transcription Factors and Their Potential Roles in Growth and Development of Rose (Rosa chinensis). J. Plant Growth Regul. 2022, 42, 1505–1521. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Liu, D.; Guo, F.; Yang, Y.; Dong, T.; Zhang, Y.; Ma, C.; Tang, Z.; Li, F.; et al. Genome-wide survey and expression analysis of GRAS transcription factor family in sweetpotato provides insights into their potential roles in stress response. BMC Plant Biol. 2022, 22, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.D.; Fuster-Almunia, C.; Ocaña-Cuesta, J.; Alonso, J.M.; Pérez-Amador, M.A. RGL2 controls flower development, ovule number and fertility in Arabidopsis. Plant Sci. 2019, 281, 82–92. [Google Scholar] [CrossRef]
- Zhou, P.; Li, Q.; Xiong, M.; Fan, X.; Zhao, D.; Zhang, C. Advances in DELLA protein-mediated phytohormonal crosstalk in regulation of plant growth and development. Plant Physiol. J. 2020, 56, 661–671. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Wang, Y.; Lü, J.; Chen, D.; Zhang, J.; Qi, K.; Cheng, R.; Zhang, H.; Zhang, S. Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear. Genome 2018, 61, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. TRANSCRIPTIONAL REGULATORY NETWORKS IN CELLULAR RESPONSES AND TOLERANCE TO DEHYDRATION AND COLD STRESSES. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Mattoo, A.K. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 2018, 231, 318–328. [Google Scholar] [CrossRef]
- Kim, J.C.; Lee, S.H.; Cheong, Y.H.; Yoo, C.; Lee, S.I.; Chun, H.J.; Yun, D.; Hong, J.C.; Lee, S.Y.; Lim, C.O.; et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001, 25, 247–259. [Google Scholar] [CrossRef]
- Wang, J.-D.; Liu, Q.-Q.; Li, Q.-F. DELLA family proteins function beyond the GA pathway. Trends Plant Sci. 2025, 30, 352–355. [Google Scholar] [CrossRef]
- Fukazawa, J.; Mori, M.; Watanabe, S.; Miyamoto, C.; Ito, T.; Takahashi, Y. DELLA-GAF1 Complex Is a Main Component in Gibberellin Feedback Regulation of GA20 Oxidase. Plant Physiol. 2017, 175, 1395–1406. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Barthole, G.; Tremblais, G.; To, A.; Miquel, M.; Lepiniec, L.; Baud, S. Transcriptional Activation of Two Delta-9 Palmitoyl-ACP Desaturase Genes by MYB115 and MYB118 Is Critical for Biosynthesis of Omega-7 Monounsaturated Fatty Acids in the Endosperm of Arabidopsis Seeds. Plant Cell 2016, 28, 2666–2682. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a Key Gene Responsible for Bioactive Gibberellin Biosynthesis, Is Regulated during Embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef]
- Tang, M.; Gao, X.; Meng, W.; Lin, J.; Zhao, G.; Lai, Z.; Lin, Y.; Chen, Y. Transcription factors NF-YB involved in embryogenesis and hormones responses in Dimocarpus longan Lour. Front. Plant Sci. 2023, 14, 1255436. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, D.; Ma, X.; Xue, X.; Liu, M.; Xiao, X.; Lai, C.; Xu, X.; Chen, X.; Chen, Y.; et al. Genome-wide high-throughput chromosome conformation capture analysis reveals hierarchical chromatin interactions during early somatic embryogenesis. Plant Physiol. 2023, 193, 555–577. [Google Scholar] [CrossRef]
- Huala, E.; Dickerman, A.W.; Garcia-Hernandez, M.; Weems, D.; Reiser, L.; LaFond, F.; Hanley, D.; Kiphart, D.; Zhuang, M.; Huang, W.; et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef]
- Hu, G.; Feng, J.; Xiang, X.; Wang, J.; Salojärvi, J.; Liu, C.; Wu, Z.; Zhang, J.; Liang, X.; Jiang, Z.; et al. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 2022, 54, 73–83. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Nystrom, S.L.; McKay, D.J. Memes: A motif analysis environment in R using tools from the MEME Suite. PLOS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Lin, Y.; Min, J.; Lai, R.; Wu, Z.; Chen, Y.; Yu, L.; Cheng, C.; Jin, Y.; Tian, Q.; Liu, Q.; et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. GigaScience 2017, 6, 1–14. [Google Scholar] [CrossRef]
- Li, H.; Lyu, Y.; Chen, X.; Wang, C.; Yao, D.; Ni, S.; Lin, Y.; Chen, Y.; Zhang, Z.; Lai, Z. Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing. Int. J. Mol. Sci. 2019, 20, 441. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, Z.; Chen, X.; Zhang, S.; Lai, Z.; Lin, Y. Subcellular localization analysis and responses to exogenous ABA and SA in longan callus of DCL4. Chin. J. Trop. Crops 2021, 42, 1514–1519. [Google Scholar] [CrossRef]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, G.; Tang, M.; Wu, W.; Gao, W.; Xie, J.; Wang, J.; Lai, Z.; Lin, Y.; Chen, Y. Genome-Wide Identification of DlGRAS Family and Functional Analysis of DlGRAS10/22 Reveal Their Potential Roles in Embryogenesis and Hormones Responses in Dimocarpus longan. Int. J. Mol. Sci. 2025, 26, 10323. https://doi.org/10.3390/ijms262110323
Zhao G, Tang M, Wu W, Gao W, Xie J, Wang J, Lai Z, Lin Y, Chen Y. Genome-Wide Identification of DlGRAS Family and Functional Analysis of DlGRAS10/22 Reveal Their Potential Roles in Embryogenesis and Hormones Responses in Dimocarpus longan. International Journal of Molecular Sciences. 2025; 26(21):10323. https://doi.org/10.3390/ijms262110323
Chicago/Turabian StyleZhao, Guanghui, Mengjie Tang, Wanlong Wu, Wei Gao, Jinbing Xie, Jialing Wang, Zhongxiong Lai, Yuling Lin, and Yukun Chen. 2025. "Genome-Wide Identification of DlGRAS Family and Functional Analysis of DlGRAS10/22 Reveal Their Potential Roles in Embryogenesis and Hormones Responses in Dimocarpus longan" International Journal of Molecular Sciences 26, no. 21: 10323. https://doi.org/10.3390/ijms262110323
APA StyleZhao, G., Tang, M., Wu, W., Gao, W., Xie, J., Wang, J., Lai, Z., Lin, Y., & Chen, Y. (2025). Genome-Wide Identification of DlGRAS Family and Functional Analysis of DlGRAS10/22 Reveal Their Potential Roles in Embryogenesis and Hormones Responses in Dimocarpus longan. International Journal of Molecular Sciences, 26(21), 10323. https://doi.org/10.3390/ijms262110323







